The Effects of Preservatives on Antibiotic- and Preservative-Resistant Microbes and Nitrogen/Sulfur Cycle Associated Microbial Communities in Freshwater River Sediments
Abstract
1. Introduction
2. Results
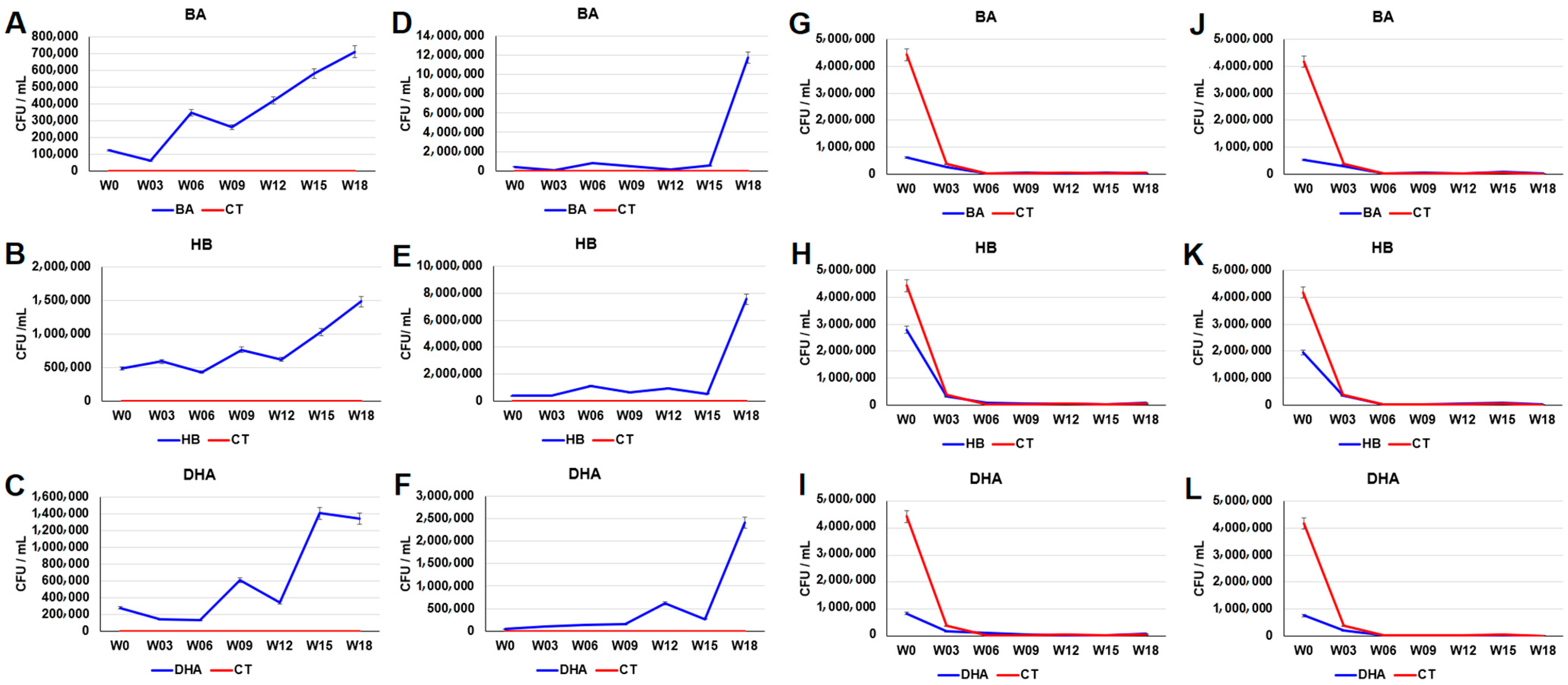
2.1. Increase in the Tetracycline-, Sulfamethoxazole- and Preservative-Resistant Microbes in Sediments
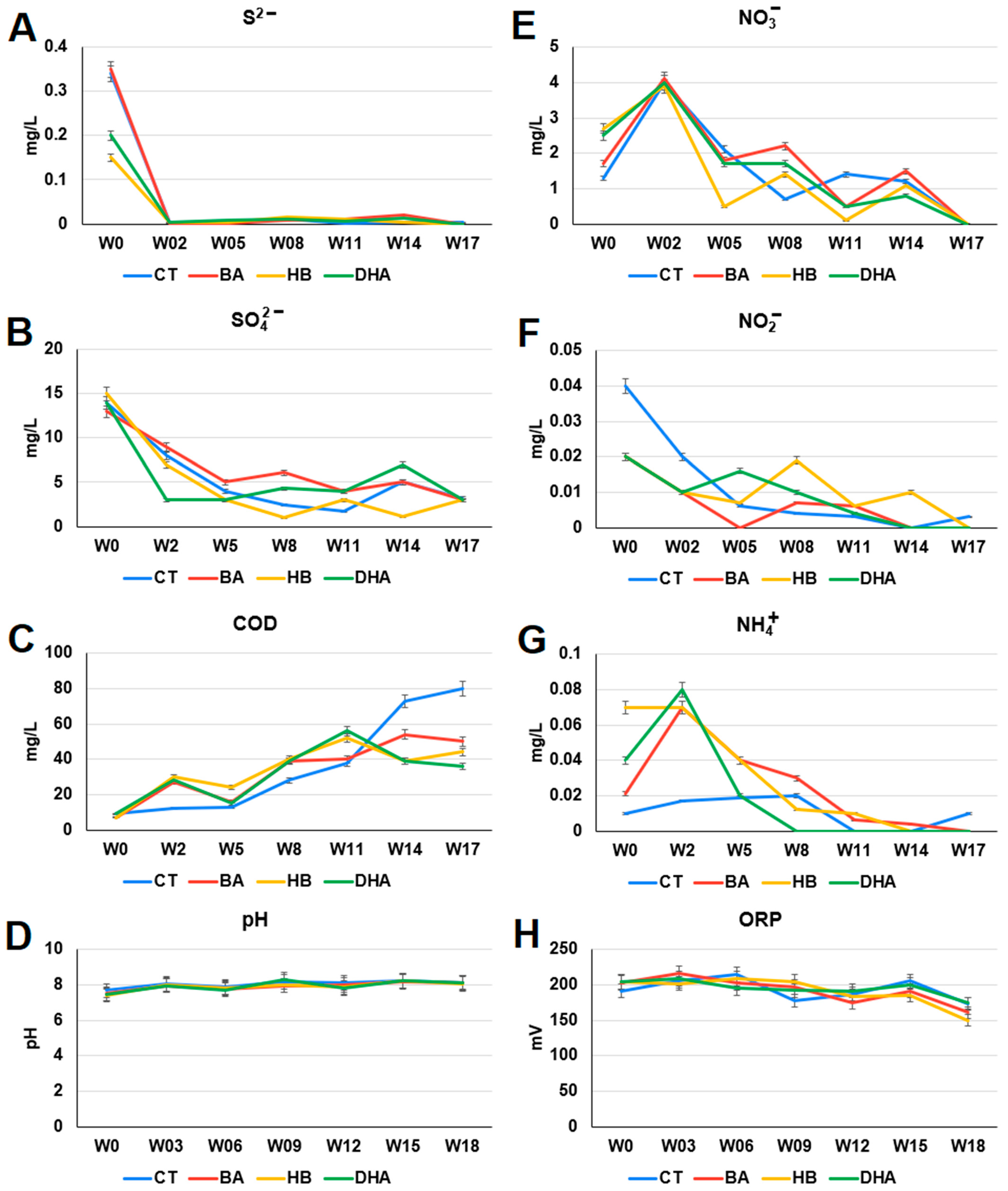
2.2. Analysis of Chemical Compositions and Oxidation-Reduction Potential (ORP) in Waters
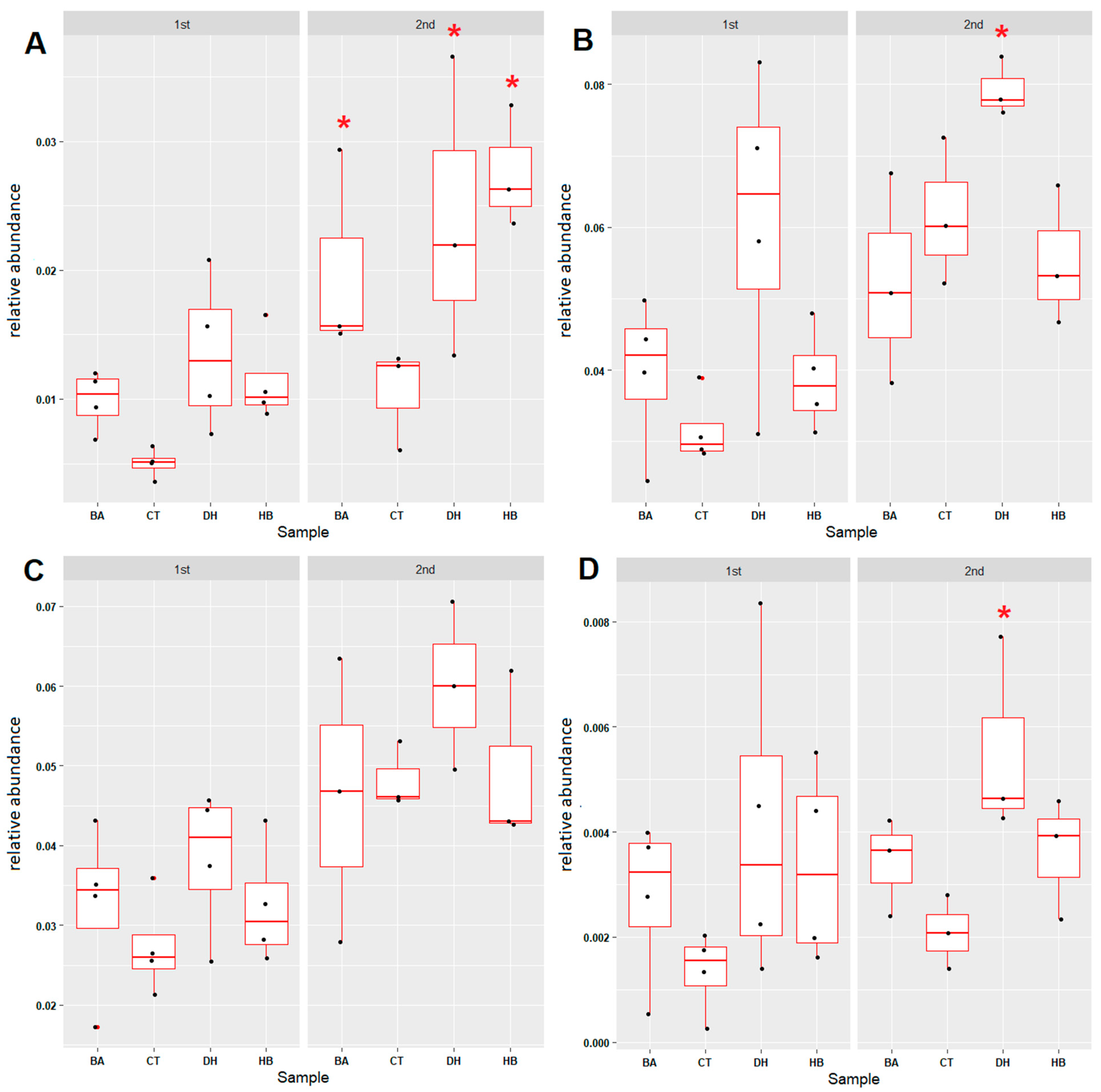
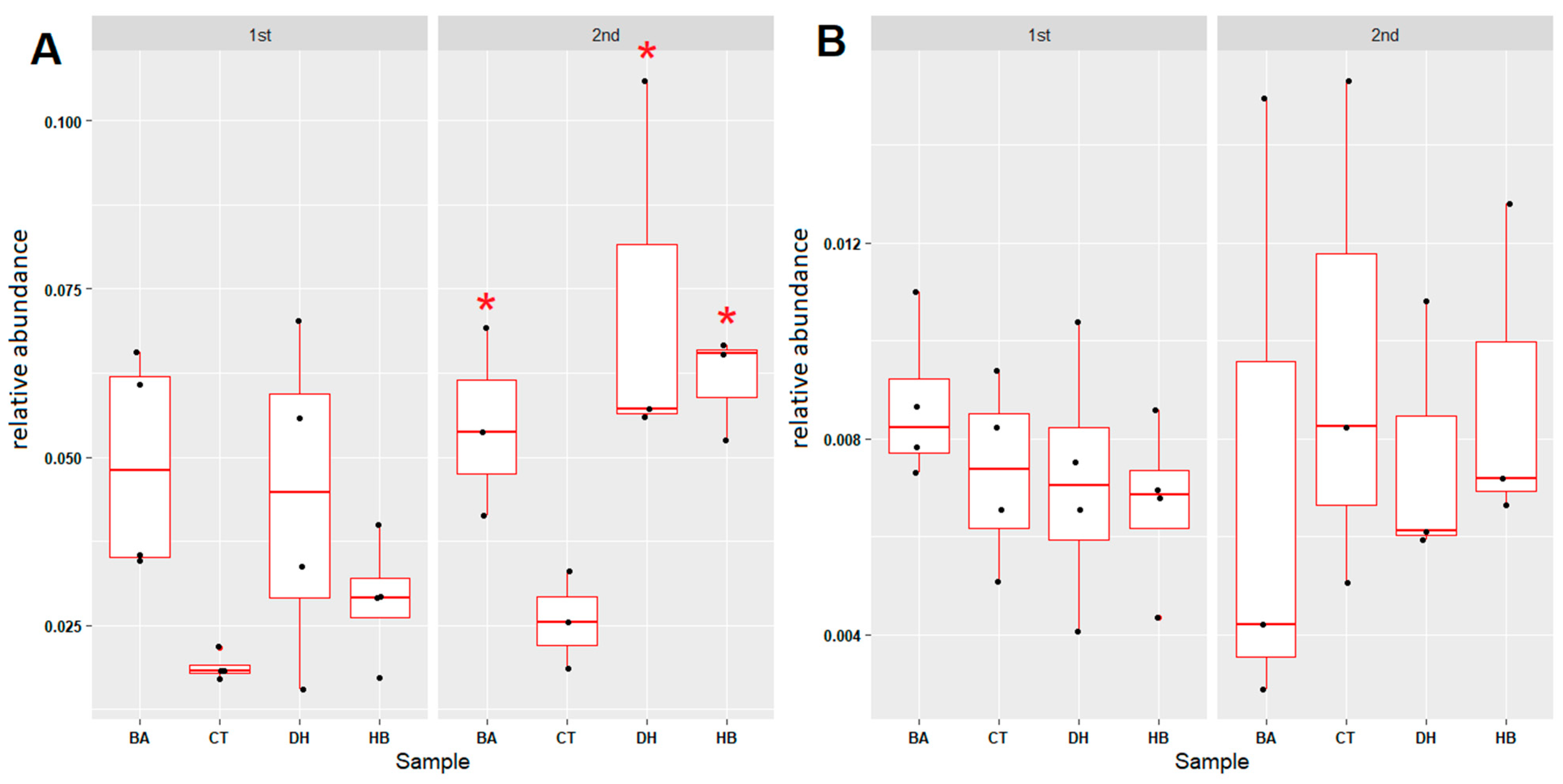
2.3. Analysis of Microbial Community Compositions
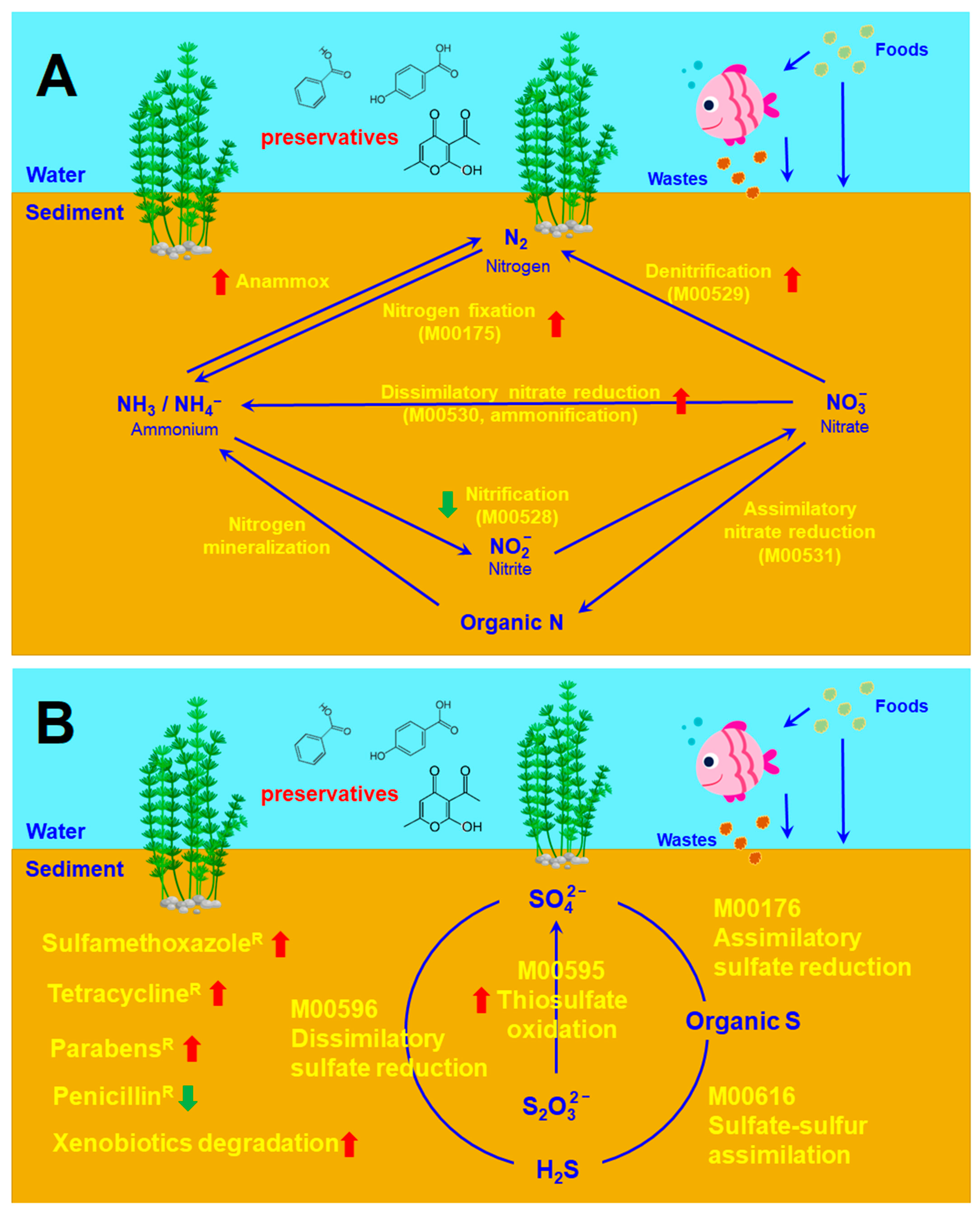
2.4. Microbial Community Associated with Nitrogen Cycle
2.5. Microbial Community Associated with Sulfur Cycle
2.6. Microbial Community Associated with Xenobiotic Degradation and Pathogenic Bacteria
3. Discussions
4. Materials and Methods
4.1. Chemicals
4.2. Experimental Design
4.3. Microbial Cultures and Plate Counts
4.4. Analysis of Chemical Compositions in Water
4.5. HPLC Analysis of Residual Preservatives in Water
4.6. DNA Extraction, and 16S rRNA Amplicon Sequencing
4.7. Microbiome Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Del Olmo, A.; Calzada, J.; Nuñez, M. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy. Crit. Rev. Food Sci. Nutr. 2017, 57, 3084–3103. [Google Scholar] [CrossRef]
- Johnson, W.; Bergfeld, W.F.; Belsito, D.V.; Hill, R.A.; Klaassen, C.D.; Liebler, D.C.; Marks, J.G.; Shank, R.C.; Slaga, T.J.; Snyder, P.W.; et al. Safety Assessment of Benzyl Alcohol, Benzoic Acid and its Salts, and Benzyl Benzoate. Int. J. Toxicol. 2017, 36, 5S–30S. [Google Scholar] [CrossRef]
- Casado, F.J.; Sánchez, A.H.; De Castro, A.; Rejano, L.; Beato, V.M.; Montaño, A. Fermented vegetables containing benzoic and ascorbic acids as additives: Benzene formation during storage and impact of additives on quality parameters. J. Agric. Food Chem. 2011, 59, 2403–2409. [Google Scholar] [CrossRef] [PubMed]
- Guynot, M.E.; Ramos, A.J.; Sanchis, V.; Marín, S. Study of benzoate, propionate, and sorbate salts as mould spoilage inhibitors on intermediate moisture bakery products of low pH (4.5–5.5). Int. J. Food Microbiol. 2005, 101, 161–168. [Google Scholar] [CrossRef]
- Heydaryinia, A.; Veissi, M.; Sadadi, A. A comparative study of the effects of the two preservatives, sodium benzoate and potassium sorbate on Aspergillus niger and Penicillium notatum. Jundishapur J. Microb. 2011, 4, 301–307. [Google Scholar]
- Liu, J.; Li, X.; Jia, Z.; Zhang, T.; Wang, X. Effect of benzoic acid on soil microbial communities associated with soilborne peanut diseases. Appl. Soil Ecol. 2017, 110, 34–42. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Liu, L.; Zhang, Y.; Yuan, J. Microbial synthesis of 4-hydroxybenzoic acid from renewable feedstocks. Food Chem. 2021, 3, 100059. [Google Scholar] [CrossRef]
- Wang, S.; Bilal, M.; Hu, H.; Wang, W.; Zhang, X. 4-Hydroxybenzoic acid-a versatile platform intermediate for value-added compounds. Appl. Microbiol. Biotechnol. 2018, 102, 3561–3571. [Google Scholar] [CrossRef] [PubMed]
- Krömer, J.O.; Nunez-Bernal, D.; Averesch, N.J.; Hampe, J.; Varela, J.; Varela, C. Production of aromatics in Saccharomyces cerevisiae—A feasibility study. J. Biotechnol. 2013, 163, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Bohra, B.S.; Pandey, N.; Tatrari, G.; Rana, S.; Sahoo, N.G. The effects of functionalized graphene oxide on the thermal and mechanical properties of liquid crystalline polymers. Soft Matter. 2022, 18, 3981–3992. [Google Scholar] [CrossRef]
- Sytar, O.; Brestic, M.; Rai, M.; Shao, H. Plant phenolic compounds for food, pharmaceutical and cosmetics production. J. Med. Plant Res. 2012, 20126, 2526–2539. [Google Scholar]
- Canavez, A.D.P.M.; de Oliveira Prado Corrêa, G.; Isaac, V.L.B.; Schuck, D.C.; Lorencini, M. Integrated approaches to testing and assessment as a tool for the hazard assessment and risk characterization of cosmetic preservatives. J. Appl. Toxicol. 2021, 41, 1687–1699. [Google Scholar] [CrossRef]
- Li, Z.P.; Zhou, Y.M.; Chen, M.; Xie, Z.H.; Lu, Q.S.; Wang, T. A study on effect of antimildew agent dehydrogenated sodium acetate. Cereal Feed Ind. 2008, 7, 32–34. [Google Scholar]
- Tang, X.; Ouyang, Q.; Jing, G.; Shao, X.; Tao, N. Antifungal mechanism of sodium dehydroacetate against Geotrichum citri-aurantii. World J. Microbiol. Biotechnol. 2018, 34, 29. [Google Scholar] [CrossRef]
- Duan, X.F.; Jing, G.X.; Fan, F.; Tao, N.G. Control of postharvest green and blue molds of citrus fruit by application of sodium dehydroacetate. Postharvest Biol. Technol. 2016, 113, 17–19. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, X.; Zhu, K.; Ding, S.; Shao, B. Sodium dehydroacetate induces cardiovascular toxicity associated with Ca2+ imbalance in zebrafish. Ecotoxicol. Environ. Saf. 2021, 208, 111613. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhao, X.; Zhu, K.; Ding, S.; Shao, B. Sodium dehydroacetate exposure decreases locomotor persistence and hypoxia tolerance in zebrafish. Environ. Res. 2021, 195, 110276. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Y.; Gao, L.; Liu, J.; Cai, Y. Occurrence, fate and risk assessment of parabens and their chlorinated derivatives in an advanced wastewater treatment plant. J. Hazard. Mater. 2015, 300, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Mortimer, M.; Cheng, H.; Sang, N.; Guo, L.H. Parabens as chemicals of emerging concern in the environment and humans: A review. Sci. Total Environ. 2021, 778, 146150. [Google Scholar] [CrossRef]
- Izawa, T.; Nakayama, K.; Uchida, N.; Nojima, K. Photoreactivities of the antiseptics dehydroacetic acid and sodium dehydroacetate used in cosmetics. Chem. Pharm. Bull. 2018, 66, 581–584. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, S.; Li, S.; Zhang, L.; Wang, G.; Zhang, L.; Wang, J.; Li, Z. The cycle of nitrogen in river systems: Sources, transformation, and flux. Environ. Sci. Process Impacts 2018, 20, 863–891. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Liu, T.; Xia, X.; Xia, N. Effect of particle size and composition of suspended sediment on denitrification in river water. Sci. Total Environ. 2016, 541, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Giblin, A.; Tobias, C.; Song, B.; Weston, N.; Banta, G.T.; Rivera-Monroy, V.H. The importance of dissimilatory nitrate reduction to ammonium (DNRA) in the nitrogen cycle of coastal ecosystems. Oceanography 2013, 26, 124–131. [Google Scholar] [CrossRef]
- Roose-Amsaleg, C.; Laverman, A.M. Do antibiotics have environmental side-effects? Impact of synthetic antibiotics on biogeochemical processes. Environ. Sci. Pollut. Res. Int. 2016, 23, 4000–4012. [Google Scholar] [CrossRef] [PubMed]
- Conkle, J.L.; White, J.R. An initial screening of antibiotic effects on microbial respiration in wetland soils. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 2012, 47, 1381–1390. [Google Scholar] [CrossRef]
- Lotti, T.; Cordola, M.; Kleerebezem, R.; Caffaz, S.; Lubello, C.; van Loosdrecht, M.C. Inhibition effect of swine wastewater heavy metals and antibiotics on anammox activity. Water Sci. Technol. 2012, 66, 1519–1526. [Google Scholar] [CrossRef]
- Podnecky, N.L.; Fredheim, E.G.A.; Kloos, J.; Sørum, V.; Primicerio, R.; Roberts, A.P.; Rozen, D.E.; Samuelsen, Ø.; Johnsen, P.J. Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nat. Commun. 2018, 9, 3673. [Google Scholar] [CrossRef]
- Imamovic, L.; Sommer, M.O. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013, 5, 204ra132. [Google Scholar] [CrossRef]
- Imamovic, L.; Ellabaan, M.M.H.; Dantas Machado, A.M.; Citterio, L.; Wulff, T.; Molin, S.; Krogh Johansen, H.; Sommer, M.O.A. Drug-Driven Phenotypic Convergence Supports Rational Treatment Strategies of Chronic Infections. Cell 2018, 172, 121–134.e14. [Google Scholar] [CrossRef]
- Barbosa, C.; Römhild, R.; Rosenstiel, P.; Schulenburg, H. Evolutionary stability of collateral sensitivity to antibiotics in the model pathogen Pseudomonas aeruginosa. eLife 2019, 8, e51481. [Google Scholar] [CrossRef]
- Barbosa, C.; Trebosc, V.; Kemmer, C.; Rosenstiel, P.; Beardmore, R.; Schulenburg, H.; Jansen, G. Alternative Evolutionary Paths to Bacterial Antibiotic Resistance Cause Distinct Collateral Effects. Mol. Biol. Evol. 2017, 34, 2229–2244. [Google Scholar] [CrossRef] [PubMed]
- Maltas, J.; Wood, K.B. Pervasive and diverse collateral sensitivity profiles inform optimal strategies to limit antibiotic resistance. PLoS Biol. 2019, 17, e3000515. [Google Scholar] [CrossRef]
- Bhardwaj, P.; Hans, A.; Ruikar, K.; Guan, Z.; Palmer, K.L. Reduced Chlorhexidine and Daptomycin Susceptibility in Vancomycin-Resistant Enterococcus faecium after Serial Chlorhexidine Exposure. Antimicrob. Agents Chemother. 2017, 62, e01235-17. [Google Scholar] [CrossRef]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Mechanisms of Increased Resistance to Chlorhexidine and Cross-Resistance to Colistin following Exposure of Klebsiella pneumoniae Clinical Isolates to Chlorhexidine. Antimicrob. Agents Chemother. 2016, 61, e01162-16. [Google Scholar] [CrossRef]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B.; et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Lamrabet, O.; Martin, M.; Lenski, R.E.; Schneider, D. Changes in Intrinsic Antibiotic Susceptibility during a Long-Term Evolution Experiment with Escherichia coli. mBio 2019, 10, e00189-19. [Google Scholar] [CrossRef]
- Bello-López, J.M.; Cabrero-Martínez, O.A.; Ibáñez-Cervantes, G.; Hernández-Cortez, C.; Pelcastre-Rodríguez, L.I.; Gonzalez-Avila, L.U.; Castro-Escarpulli, G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms 2019, 7, 363. [Google Scholar] [CrossRef] [PubMed]
- Pantosti, A.; Sanchini, A.; Monaco, M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007, 2, 323–334. [Google Scholar] [CrossRef]
- Park, T.J.; Lee, J.H.; Lee, M.S.; Park, C.H.; Lee, C.H.; Moon, S.D.; Chung, J.; Cui, R.; An, Y.J.; Yeom, D.H.; et al. Development of water quality criteria of ammonia for protecting aquatic life in freshwater using species sensitivity distribution method. Sci. Total Environ. 2018, 634, 934–940. [Google Scholar] [CrossRef]
- Liu, S.; Yang, F.; Gong, Z.; Meng, F.; Chen, H.; Xue, Y.; Furukawa, K. Application of anaerobic ammonium-oxidizing consortium to achieve completely autotrophic ammonium and sulfate removal. Bioresour. Technol. 2008, 99, 6817–6825. [Google Scholar] [CrossRef]
- Rios-Del Toro, E.E.; Valenzuela, E.I.; López-Lozano, N.E.; Cortés-Martínez, M.G.; Sánchez-Rodríguez, M.A.; Calvario-Martínez, O.; Sánchez-Carrillo, S.; Cervantes, F.J. Anaerobic ammonium oxidation linked to sulfate and ferric iron reduction fuels nitrogen loss in marine sediments. Biodegradation 2018, 29, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Xie, G.J.; Xing, D.F.; Liu, B.F.; Ding, J.; Cao, G.L.; Ren, N.Q. Sulfate dependent ammonium oxidation: A microbial process linked nitrogen with sulfur cycle and potential application. Environ. Res. 2021, 192, 110282. [Google Scholar] [CrossRef] [PubMed]
- Tomei, M.C.; Mosca Angelucci, D.; Clagnan, E.; Brusetti, L. Anaerobic biodegradation of phenol in wastewater treatment: Achievements and limits. Appl. Microbiol. Biotechnol. 2021, 105, 2195–2224. [Google Scholar] [CrossRef]
- Li, M.; Ning, P.; Sun, Y.; Luo, J.; Yang, J. Characteristics and Application of Rhodopseudomonas palustris as a Microbial Cell Factory. Front. Bioeng. Biotechnol. 2022, 10, 897003. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, C.-S.; Cao, X.-D.; Lee, W.-C.; Yang, C.-W. The Effects of Preservatives on Antibiotic- and Preservative-Resistant Microbes and Nitrogen/Sulfur Cycle Associated Microbial Communities in Freshwater River Sediments. Antibiotics 2023, 12, 1082. https://doi.org/10.3390/antibiotics12071082
Liao C-S, Cao X-D, Lee W-C, Yang C-W. The Effects of Preservatives on Antibiotic- and Preservative-Resistant Microbes and Nitrogen/Sulfur Cycle Associated Microbial Communities in Freshwater River Sediments. Antibiotics. 2023; 12(7):1082. https://doi.org/10.3390/antibiotics12071082
Chicago/Turabian StyleLiao, Chien-Sen, Xuan-Di Cao, Wei-Chen Lee, and Chu-Wen Yang. 2023. "The Effects of Preservatives on Antibiotic- and Preservative-Resistant Microbes and Nitrogen/Sulfur Cycle Associated Microbial Communities in Freshwater River Sediments" Antibiotics 12, no. 7: 1082. https://doi.org/10.3390/antibiotics12071082
APA StyleLiao, C.-S., Cao, X.-D., Lee, W.-C., & Yang, C.-W. (2023). The Effects of Preservatives on Antibiotic- and Preservative-Resistant Microbes and Nitrogen/Sulfur Cycle Associated Microbial Communities in Freshwater River Sediments. Antibiotics, 12(7), 1082. https://doi.org/10.3390/antibiotics12071082







