Abstract
Helicobacter pylori (H. pylori) antibiotic resistance is the leading cause for unsuccessful eradication therapy. After one or more failures, the chance of encountering secondary antibiotic resistance increases. The aim of this study was to characterize genotypic secondary resistance in a cohort of southern Italian H. pylori patients with at least one previous failure. Such patients collected stool samples using a dedicated kit (THD fecal testTM), and bacterial DNA was extracted and amplified using RT-PCR. Resistance to clarithromycin, amoxicillin, metronidazole, levofloxacin, and tetracycline was assessed using a high-resolution melting curve. We enrolled 50 patients. A total of 72% of patients failed one previous antibiotic course, 16% failed two, 10% failed three, and 2% failed four. The rate of secondary antibiotic resistance was 16% for clarithromycin, 18% for metronidazole, 14% for amoxicillin, 14% for levofloxacin, and 2% for tetracycline. Among the eight clarithromycin-resistant patients, five (62.5%) previously received a clarithromycin-based regimen. The same rate was 33.3% (3/9) for metronidazole. The only tetracycline-resistant patient had received Pylera. In conclusion, our data seem to show that, even though secondary resistance is not very high, resistance to clarithromycin could be very likely related to previous exposure to this antibiotic.
1. Introduction
Helicobacter pylori (H. pylori) is the leading cause of gastritis, peptic ulcers, and other extra-gastric diseases, including iron deficiency anemia, vitamin B12 deficiency, and idiopathic thrombocytopenic purpura [1,2,3,4]. Furthermore, it is a well-recognized class I carcinogen, involved in the pathogenesis of gastric cancer [5,6,7,8]. The therapy of H. pylori is based on the combination of several antibiotics. In first-line treatment, the most commonly used regimens are sequential, concomitant, triple, and bismuth-based quadruple (BQT) therapy [9,10,11]. However, antibiotic resistance is a rising phenomenon, and it may explain most cases of therapy failure. Resistance to clarithromycin, for example, strongly decreases the effectiveness of conventional triple therapy [12,13,14], and this is the reason why current guidelines no longer recommend it in geographical areas with high resistance prevalence, thus suggesting the use of other regimen such as BQT or concomitant [15]. Even resistance to levofloxacin, an antibiotic that is used in second-line treatment, is dramatically increasing [16,17,18], and this may reduce the eradication probability of some rescue regimens.
Indeed, after one failure, second-line regimens are required [19]. Even though empirical rescue therapies are still advocated by guidelines, determination of antibiotic resistance may be a promising strategy to achieve success [20,21]. Indeed, after one failure, onset of novel resistance due to exposure to antibiotics (secondary resistance) is quite frequent and may hamper the eradication of the bacterium [22].
The aim of this study was to characterize genotypic secondary resistances in a cohort of southern Italian H. pylori patients with at least one previous failure.
2. Results
Fifty patients were enrolled. The male/female ratio was 17/33, and the mean age was 53.3 ± 13.6 years. In total, 72% failed one previous antibiotic course, 16% failed two, 10% failed three, and 2% failed four. The most previously failed regimens were the 3-in-1 pill BQT (PyleraTM), in 11 cases, followed by sequential therapy (n = 9), triple therapy with clarithromycin (n = 7), concomitant therapy (n = 5), and triple therapy with levofloxacin (n = 3). Some other patients had undergone other therapies not listed by the current guidelines (e.g., triple therapy with only amoxicillin and tetracycline or metronidazole, sequential therapy with levofloxacin), while the remaining ones did not recall previous therapies. Three out of fifty patients (6%) had a previous early therapy termination due to side effects.
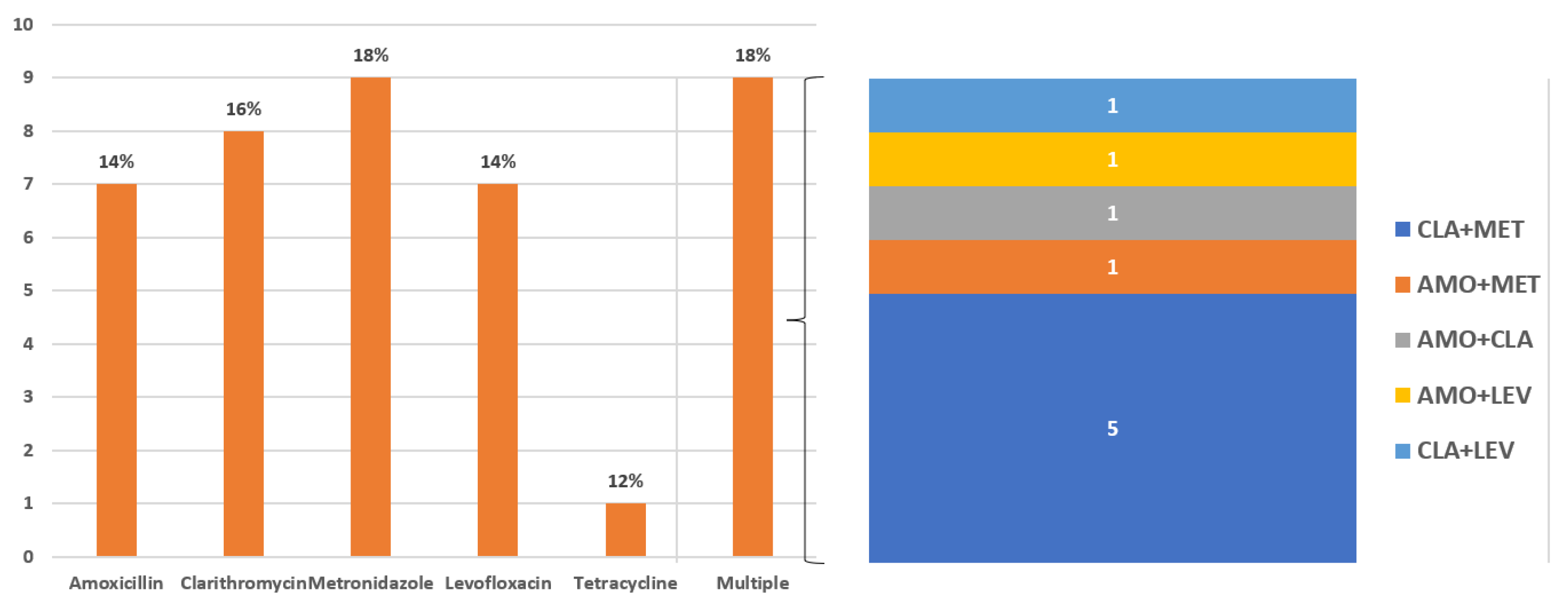
The rate of secondary antibiotic resistance was 16% for clarithromycin (n = 8), 18% for metronidazole (n = 9), 14% for amoxicillin (n = 7), 14% for levofloxacin (n = 7), and 2% for tetracycline (n = 1). Multi-resistance was detected in nine patients (18%) and double resistance to clarithromycin + metronidazole was the most common resistance found (n = 5). Results are depicted in Figure 1.
Figure 1.
Rates of secondary antibiotic resistance. Multi-resistance is detailed in the right panel of the figure.
Among patients with previous failure to an amoxicillin-containing regimen (Table 1), amoxicillin resistance was found in 4 out of 20 patients (20%). Among patients with previous failure adhering to a clarithromycin-containing regimen, resistance to such an antibiotic was detected in 6 out of 19 patients (31.6%). Among patients with previous failure in adhering to a metronidazole-containing regimen (n = 21), metronidazole resistance was found in three cases (14.3%). Failed levofloxacin-based therapy was administered in three patients, but resistance was found in none of them (0%). Finally, among patients with previous failure adhering to a tetracycline-containing therapy, resistance to this antibiotic was detected in 1 out of 12 patients (8.3%), as reported in Table 1.

Table 1.
Proportions of secondary antibiotic resistance according to the type of previously failed therapy.
On the other side, among the seven amoxicillin-resistant patients, three (42.8%) had received amoxicillin-containing therapy. Among the nine amoxicillin-resistant patients, three (33.3%) had received a metronidazole-based regimen. Among the eight amoxicillin-resistant subjects, five (62.5%) had received a clarithromycin-containing regimen. None of the seven levofloxacin-resistant patients had received a therapy based on such an antibiotic. Finally, the only tetracycline-resistant patient had failed PyleraTM.
3. Discussion
Treatment of H. pylori infection is becoming a complex issue worldwide due to the spread of antimicrobial resistance [23]. Argueta et al. in 2021 found a resistance rate for clarithromycin and levofloxacin very close to 30% in the United States, thus confirming what was previously reported in southern European countries and observed in Italy in the last 4 years [24].
As is known, H. pylori infection can be diagnosed using non-invasive tests such as of the H. pylori antigen in stool samples, a UBT (Urea Breath Test), and serology, as well as invasive tests such as histology for conventional diagnosis, culture, and a PCR (polymerase chain reaction) both for a diagnosis and for the evaluation of antibiotic sensitivity. Invasive tests require endoscopy [25,26,27].
The first susceptibility test has conventionally been based on culturing and susceptibility testing in H. pylori isolates, although it is recommended by current guidelines only after repeated treatment failure [15]. Indeed, it is almost impossible to use this method for first-line treatment selection, as a relatively high rate of false-negative results, often resulting in low sensitivity, has thus far weighed on the use of the test on a large scale. This complexity is mainly due to the need to create and maintain a micro-aerophilic environment, in which the bacterium can grow. Other factors that have so far prevented the widespread spread of H. pylori culture are the following problems related to the methodology: the number of gastric biopsies, time-consuming endoscopic procedures, conditions, and interval of transportation of biopsy samples, characteristics of the laboratory, long and unpredictable times required to obtain the result of the investigation [25,28,29]. Of note, culturing does not detect the heteroresistant status of H. pylori, e.g., the simultaneous presence of sensitive and resistant strains [26,30,31].
As an alternative to bacterial culture and susceptibility testing, techniques based on real-time polymerase chain reaction (RT-PCR) testing (genotypic resistance detection) have been developed [32]. They are based on the principle of amplifying and detecting the point mutations responsible for antibiotic resistance in H. pylori DNA isolated from gastric biopsy samples. These culture-free approaches are accurate in revealing minimal traces of resistant genotypic strains as well as uncovering the heteroresistant state. Furthermore, the ability to evaluate resistant mutant genotypes using PCR not only on fresh specimens, but also on archived paraffin-embedded biopsy specimens, which has been shown to provide an equally reliable substrate for DNA analysis as fresh material, has further emphasized the importance and usefulness of these methods [33,34]. Based on what has been reported, molecular tests unquestionably offer advantages and guarantee feasibility compared to cultures even if they are not used in clinical practice. It is presumable that the need for an invasive endoscopic procedure was the most important limit to their diffusion. Therefore, a next step was represented by the attempt to overcome this drawback through an in-depth and appropriate analysis. A further advancement in this area was represented by the possibility of detecting point mutations that confer resistance to antibiotics in fecal samples of bacterial DNA. From 2003 to 2021, several studies were performed on fecal H. pylori DNA using a real-time polymerase chain reaction (RT-PCR) for the diagnosis of infection and/or antibiotic susceptibility, and all showed high rates of sensitivity and specificity when culturing or PCRs on gastric biopsy specimens were used as the gold standard [35].
Therefore, genetic mutations conferring resistance can be detected in stools, representing an excellent substrate for studying sensitivity to H. pylori antibiotics [36]. Indeed, there is growing evidence of the potential future availability of non-invasive investigations capable of detecting resistance to H. pylori antibiotics, such as clarithromycin and quinolones, which have been commonly used until now. These techniques, if performed before first-line therapy, could allow the identification of a subgroup of strains still sensitive to these drugs and, therefore, of patients who can benefit from old regimens (triple and sequential), whose administration has currently been discouraged by the current guidelines [15]. On the other hand, fecal molecular analysis has the advantage of improving patient compliance, reducing the time/cost ratio of the diagnostic procedure and improving the therapeutic outcome [16]. Finally, the potential risk of a future increase in resistance to quadruple regimens, suggested as first-line treatment by the guidelines, as a consequence of their use on a large scale and incomplete patient adherence could be avoided [37].
In the present study, we evaluated, with the use of molecular analysis of fecal specimens, the onset of secondary antibiotic resistance in subjects affected by H. pylori infection and subjected to one or more unsuccessful treatment regimens.
Despite the small sample, we observed that secondary resistance rates were almost low. Indeed, the percentage of secondary antibiotic resistance was 18% for metronidazole, 16% for clarithromycin, 14% for amoxicillin, 14% for levofloxacin, and 2% for tetracycline. Multi-resistance was detected in 18% and double resistance to clarithromycin and metronidazole was the most common resistance. Therefore, the overall secondary resistance rates, after the failure of one or more regimens, appears to be lower than expected. For example, in a Chinese series, the secondary resistances to clarithromycin, metronidazole, and levofloxacin were 96.7%, 90.7%, and 93.1%, respectively [38].
When secondary resistances were considered according to previous failing regimens, an unsuccessful clarithromycin-containing regimen was followed by the highest resistance (31.6%). Among patients with previous failure to an amoxicillin-containing regimen, amoxicillin resistance was found in 20% as well as among patients with previous failure in adhering to a metronidazole-containing regimen, resistance to this antibiotic was found in 14.3%. Patients with previous failure in adhering to tetracycline-containing therapy showed resistance to this antibiotic in 8.3% of cases. These results might suggest that clarithromycin use induces secondary resistance more easily than other antibiotics, even if metronidazole assumption also may be followed by secondary resistance onset in a substantial percentage. Previous exposure to clarithromycin is a known risk factor for secondary resistance, Karczewska found an increase in the resistance rate from 21% to 80% after one failed therapy course [39], and a mathematical model predicted that Clarithromycin resistance may originate from the transmission of resistant bacteria in 98.7% of cases, and derives from spontaneous mutations in the other 1.3% [40]. The rates of secondary resistance to this antibiotic are quite heterogeneous, ranging from 27.2% [41] to 82.9% [42], and this underlines how geographical factors may play a role [43]. In our experience, secondary levofloxacin resistance was 14%, close to the 16% of another study [39]. Interestingly, we found that failed levofloxacin-based therapy was not followed by induced resistances to this antibiotic. This result, even if needing to be confirmed in a large sample, could suggest that levofloxacin resistance is the result of a previous quinolone use for reasons other than H. pylori eradication. Indeed, it is well known that quinolone use induces cross-resistance. Finally, the low rate of tetracycline resistance is in agreement with most of the current literature [44]. Anyway, the quite low prevalence of secondary resistance may have many explanations. First, three out of fifty patients had previous early therapy termination due to side effects; therefore, in these patients, real secondary resistance may not have occurred. Another reason could be the molecular biology method. PCR with HRM detects every variation in the genetic sequence, but, differently from sequencing (NGS), it does not precisely define the point mutation; therefore, it is less accurate [45,46]. However, NGS is more expensive and not available in all laboratories. Based on this concern, we preferred HRM, which may be rapid and cost effective for a large-scale analysis. Finally, among the limitations, it should be acknowledged that not all genetic mutations may translate into phenotypic resistance [47].
4. Materials and Methods
4.1. Patients Selection
We enrolled consecutive dyspeptic patients [48] with at least one failure to a previous antibiotic course against H. pylori in the period June 2021–June 2023. Failure had to be demonstrated by persistent positivity to a non-invasive test (stool test or urea breath test) performed at least 4 weeks after stopping antibiotics. Patients were enrolled in two centers (the Gastroenterology Unit, University of Bari, and the National Institute of Gastroenterology “S. de Bellis” Research Hospital).
In detail, we enrolled subjects aged > 18 and who were able to express willingness to participate, presenting with dyspeptic symptoms, such as postprandial fullness, early satiation, epigastric pain, and epigastric burning. We excluded patients with history of gastric or extra-gastric cancer and those who were unable to express informed consent or refused to participate. Additional exclusion criteria were therapy with proton pump inhibitors or histamine receptor antagonists within two weeks from enrollment and use of antibiotics or bismuth salts in the previous four weeks. Furthermore, recent chronic diarrhea was another reason for exclusion because it could restrict the proper collection of fecal samples.
Details about eradication regimens, which were assumed by patients, before this study, were recorded.
The study was conducted in agreement with the indications of the Declaration of Helsinki, and the local Ethics Committee approved the protocol (AOU Consorziale Policlinico di Bari, protocol no. 74413, approved 16 November 2016). All patients signed informed consent.
4.2. Evaluation of Antibiotic Resistance
A stool sample was collected from all patients using the THD Fecal Test Device (THD s.p.a., Correggio, Reggio Emilia, Italy), which has shown a sensitivity of 90.2% and a specificity of 98.5% at detecting H. pylori genetic sequences [49,50]. This device has a filter blocking the real-time polymerase chain reaction (PCR), inhibiting substances like hemoglobin and its degradation products, polysaccharide complexes, heavy metals, and proteins. Furthermore, it eliminates large molecules such as fibers. The treated solution was finally taken from the reservoir and processed for DNA extraction using a QIAamp DNA Stool Minikit (Qiagen, Hilden, Germany). After this last phase, real-time PCR was performed to assess point mutations linked to H. pylori resistance to clarithromycin amoxicillin, metronidazole, levofloxacin, and tetracycline as previously described [16,50,51]. In particular, the following genes implied in antimicrobial resistance were amplified: pbp1 for amoxicillin, rdxA/frxA for metronidazole, 23S rRNA for clarithromycin, gyrA for levofloxacin, and 16S rRNA for tetracycline [52].
Real-time PCR followed by high-resolution melting (HRM) was set to detect mutations. Curves produced using HRM were compared to those derived for wild-type, nonmutated genes from susceptible strains of examined genes to detect resistances [53,54,55].
4.3. Statistics
Continuous data were expressed as the mean standard deviation, and categorical variables as proportions/percentages. Graphs were drawn using an Excel version for Windows, Microsoft (Microsoft Italia, Milan, Italy).
5. Conclusions
In conclusion, our study shows that, even though secondary resistances were not as high as expected, previous use of clarithromycin strongly increased the risk of novel resistance onset; therefore, its re-use should be discouraged. We did not observe new levofloxacin resistance after its use; therefore, it may be hypothesizable that such resistances have been acquired because of previous quinolones’ use, presumably even with the induction of cross resistance. The fact that the resistance rate to tetracycline is quite low may be a comforting finding, presumably due to its poor use in recent years. However, attention towards a careful use of such an antibiotic should be kept, in order to avoid future spread of its resistance [37]. Finally, it should be acknowledged that bismuth salts are an effective weapon against H. pylori, as they have an intrinsic antibacterial effect, and they do not elicit any antimicrobial resistance. It has been demonstrated that adding bismuth to an eradication regimen may result in an additional 30–40% of success in resistant infections [56]. Therefore, when available, adding bismuth should always be considered when facing difficult-to-eradicate H. pylori.
Author Contributions
Conceptualization, G.L., A.D.L., E.I. and A.I.; methodology, A.I. and G.L.; software, G.L., M.P. (Maria Pricci) and B.G.; validation, A.D.L. and F.C.; formal analysis, G.L.; investigation, M.M., M.P. (Maria Pricci), M.P. (Mariapaola Piazzolla), B.G., G.R., F.R. and G.G.; resources, M.M., I.D., M.P. (Mariapaola Piazzolla), B.G., G.R. and F.R.; data curation, M.P. (Mariapaola Piazzolla), M.M. and I.D.; writing—original draft preparation, G.L. and F.C.; writing—review and editing, A.I., E.I. and A.D.L.; supervision, A.D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in agreement with the indications of the Declaration of Helsinki, and the local Ethics Committee approved the protocol (AOU Consorziale Policlinico di Bari, protocol n. 74413).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
Alfredo Di Leo served as consultant for THD s.p.a.
References
- Jonaitis, L.; Pellicano, R.; Kupcinskas, L. Helicobacter pylori and nonmalignant upper gastrointestinal diseases. Helicobacter 2018, 23, e12522. [Google Scholar] [CrossRef]
- Ford, A.C.; Tsipotis, E.; Yuan, Y.; Leontiadis, G.I.; Moayyedi, P. Efficacy of Helicobacter pylori eradication therapy for functional dyspepsia: Updated systematic review and meta-analysis. Gut 2022, 71, 1967–1975. [Google Scholar] [CrossRef]
- Ford, A.C.; Mahadeva, S.; Carbone, M.F.; Lacy, B.E.; Talley, N.J. Functional dyspepsia. Lancet 2020, 396, 1689–1702. [Google Scholar] [CrossRef] [PubMed]
- Gravina, A.G.; Zagari, R.M.; De Musis, C.; Romano, L.; Loguercio, C.; Romano, M. Helicobacter pylori and extragastric diseases: A review. World J. Gastroenterol. 2018, 24, 3204–3221. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; He, F.; Clifford, G.M.; Li, M.; Fan, Z.; Li, X.; Wang, S.; Wei, W. A systematic review and meta-analysis on the relative and attributable risk of Helicobacter pylori infection and cardia and non-cardia gastric cancer. Expert Rev. Mol. Diagn. 2023, 23, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Fuccio, L.; Zagari, R.M.; Eusebi, L.H.; Laterza, L.; Cennamo, V.; Ceroni, L.; Grilli, D.; Bazzoli, F. Meta-analysis: Can Helicobacter pylori eradication treatment reduce the risk for gastric cancer? Ann. Intern. Med. 2009, 151, 121–128. [Google Scholar] [CrossRef]
- Fuccio, L.; Eusebi, L.H.; Bazzoli, F. Gastric cancer, Helicobacter pylori infection and other risk factors. World J. Gastrointest. Oncol. 2010, 2, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Venerito, M.; Vasapolli, R.; Rokkas, T.; Malfertheiner, P. Helicobacter pylori and Gastrointestinal Malignancies. Helicobacter 2015, 20 (Suppl. S1), 36–39. [Google Scholar] [CrossRef]
- Pellicano, R.; Ribaldone, D.G.; Fagoonee, S.; Astegiano, M.; Saracco, G.M.; Mégraud, F. A 2016 panorama of Helicobacter pylori infection: Key messages for clinicians. Panminerva Med. 2016, 58, 304–317. [Google Scholar]
- Nyssen, O.P.; McNicholl, A.G.; Megraud, F.; Savarino, V.; Oderda, G.; A Fallone, C.; Fischbach, L.; Bazzoli, F.; Gisbert, J.P. Sequential versus standard triple first-line therapy for Helicobacter pylori eradication. Cochrane Database Syst. Rev. 2016, 2016, CD009034. [Google Scholar] [CrossRef]
- Calvet, X. What is the best first-line therapy for Helicobacter pylori infection? Nat. Clin. Pract. Gastroenterol. Hepatol. 2008, 5, 606–607. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, V.; Margiotta, M.; Zullo, A.; Hassan, C.; Troiani, L.; Burattini, O.; Stella, F.; Di Leo, A.; Russo, F.; Marangi, S.; et al. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. Ann. Intern. Med. 2006, 144, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Xia, H.H.; Wang, J.D.; Wong, W.M.; Chan, A.O.; Lai, K.C.; Chan, C.K.; Yuen, M.F.; Fung, F.M.; Wong, K.W.; et al. Update on clarithromycin resistance in Helicobacter pylori in Hong Kong and its effect on clarithromycin-based triple therapy. Digestion 2006, 73, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Elitsur, Y.; Lawrence, Z.; Rüssmann, H.; Koletzko, S. Primary Clarithromycin Resistance to Helicobacter pylori and Therapy Failure in Children: The Experience in West Virginia. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 327–328. [Google Scholar] [CrossRef] [PubMed]
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. European Helicobacter and Microbiota Study group. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762. [Google Scholar] [CrossRef] [PubMed]
- Losurdo, G.; Giorgio, F.; Pricci, M.; Girardi, B.; Russo, F.; Riezzo, G.; Martulli, M.; Piazzolla, M.; Cocomazzi, F.; Abbruzzi, F.; et al. Helicobacter pylori Primary and Secondary Genotypic Resistance to Clarithromycin and Levofloxacin Detection in Stools: A 4-Year Scenario in Southern Italy. Antibiotics 2020, 9, 723. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Shokri-Shirvani, J.; Zamani, V. Letter: Levofloxacin resistance—A challenge for the treatment of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2017, 45, 572–573. [Google Scholar] [CrossRef]
- Trespalacios-Rangél, A.A.; Otero, W.; Arévalo-Galvis, A.; Poutou-Piñales, R.A.; Rimbara, E.; Graham, D.Y. Surveillance of Levofloxacin Resistance in Helicobacter pylori Isolates in Bogota-Colombia (2009–2014). PLoS ONE 2016, 11, e0160007. [Google Scholar] [CrossRef]
- Losurdo, G.; D’abramo, F.S.; Piazzolla, M.; Rima, R.; Continisio, A.; Pricci, M.; Ierardi, E.; Di Leo, A. Second-line Therapy for Helicobacter Pylori Eradication: State of the Art. Mini-Rev. Med. Chem. 2022, 22, 2430–2437. [Google Scholar] [CrossRef]
- Li, C.L.; Zhou, K.; Suo, B.J.; Tian, X.L.; Zhang, Y.X.; Ren, X.L.; Shi, Y.Y.; Zhou, L.Y.; Song, Z.Q. Tailored therapy guided by genotypic resistance of clarithromycin and levofloxacin detected by polymerase chain reaction in the first-line treatment of Helicobacter pylori infection. J. Dig. Dis. 2024, 25, 36–43. [Google Scholar] [CrossRef]
- Ierardi, E.; Giorgio, F.; Iannone, A.; Losurdo, G.; Principi, M.; Barone, M.; Pisani, A.; Di Leo, A. Noninvasive molecular analysis of Helicobacter pylori: Is it time for tailored first-line therapy? World J. Gastroenterol. 2017, 23, 2453–2458. [Google Scholar] [CrossRef] [PubMed]
- Mascellino, M.T.; Oliva, A.; Miele, M.C.; De Angelis, M.; Bruno, G.; Severi, C. Secondary Antibiotic Resistance, Correlation between Genotypic and Phenotypic Methods and Treatment in Helicobacter pylori Infected Patients: A Retrospective Study. Antibiotics 2020, 9, 549. [Google Scholar] [CrossRef]
- Thung, I.; Aramin, H.; Vavinskaya, V.; Gupta, S.; Park, J.Y.; Crowe, S.E.; Valasek, M.A. Review article: The global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharmacol. Ther. 2016, 43, 514–533. [Google Scholar] [CrossRef] [PubMed]
- Argueta, E.A.; Alsamman, M.A.; Moss, S.F.; D’agata, E.M. Impact of Antimicrobial Resistance Rates on Eradication of Helicobacter pylori in a US Population. Gastroenterology 2021, 160, 2181–2183.e1. [Google Scholar] [CrossRef] [PubMed]
- Godbole, G.; Mégraud, F.; Bessède, E. Review: Diagnosis of Helicobacter pylori infection. Helicobacter 2020, 25 (Suppl. S1), e12735. [Google Scholar] [CrossRef] [PubMed]
- Bénéjat, L.; Ducournau, A.; Lehours, P.; Mégraud, F. Real-time PCR for Helicobacter pylori diagnosis. The best tools available. Helicobacter 2018, 23, e12512. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.; Soares, J.-B.; Gonçalves, R. Efficacy and tolerability of culture-guided treatment for Helicobacter pylori infection. Eur. J. Gastroenterol. Hepatol. 2017, 29, 1258–1263. [Google Scholar] [CrossRef] [PubMed]
- Yuen, B.; Zbinden, R.; Fried, M.; Bauerfeind, P.; Bernardi, M. Cultural Recovery and Determination of Antimicrobial Susceptibility in Helicobacter pylori by Using Commercial Transport and Isolation Media. Infection 2005, 33, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.J.; Edwards-Jones, V.; Armitage, M. Metronidazole sensitivity testing of Helicobacter pylori: The importance of media. Br. J. Biomed. Sci. 1998, 55, 118–122. [Google Scholar]
- Nguyen, T.C.; Le, G.K.N.; Pham, D.T.H.; Van Pham, B.; Nguyen, L.T.H.; Che, T.H.; Nguyen, H.T.; Truong, D.Q.; Robert, A.; Bontems, P.; et al. Antibiotic resistance and heteroresistance in Helicobacter pylori isolates from symptomatic Vietnamese children: A prospective multicenter study. Helicobacter 2023, 28, e13009. [Google Scholar] [CrossRef]
- Sun, L.; Talarico, S.; Yao, L.; He, L.; Self, S.; You, Y.; Zhang, H.; Zhang, Y.; Guo, Y.; Liu, G.; et al. Droplet Digital PCR-Based Detection of Clarithromycin Resistance in Helicobacter pylori Isolates Reveals Frequent Heteroresistance. J. Clin. Microbiol. 2018, 56, e00019-18. [Google Scholar] [CrossRef]
- Fontana, C.; Favaro, M.; Pietroiusti, A.; Pistoia, E.S.; Galante, A.; Favalli, C. Detection of Clarithromycin-Resistant Helicobacter pylori in Stool Samples. J. Clin. Microbiol. 2003, 41, 3636–3640. [Google Scholar] [CrossRef]
- Brennan, D.E.; Omorogbe, J.; Hussey, M.; Tighe, D.; Holleran, G.; O’Morain, C.; Smith, S.M.; McNamara, D. Molecular detection of Helicobacter pylori antibiotic resistance in stool vs biopsy samples. World J. Gastroenterol. 2016, 22, 9214–9221. [Google Scholar] [CrossRef]
- Marrero Rolon, R.; Cunningham, S.A.; Mandrekar, J.N.; Polo, E.T.; Patel, R. Clinical evaluation of a Real_Time PCR assay for simultaneous detection of Helicobacter pylori and genotypic markers of Clarithromycin resistance directly from stool. J. Clin. Microbiol. 2021, 59, e03040-20. [Google Scholar] [CrossRef] [PubMed]
- Celiberto, F.; Losurdo, G.; Pricci, M.; Girardi, B.; Marotti, A.; Di Leo, A.; Ierardi, E. The State of the Art of Molecular Fecal Investigations for Helicobacter pylori (H. pylori) Antibiotic Resistances. Int. J. Mol. Sci. 2023, 24, 4361. [Google Scholar] [CrossRef]
- Noguchi, N.; Rimbara, E.; Kato, A.; Tanaka, A.; Tokunaga, K.; Kawai, T.; Takahashi, S.; Sasatsu, M. Detection of mixed clar-ithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J. Med. Microbiol. 2007, 56 Pt 9, 1174–1180. [Google Scholar] [CrossRef]
- Ng, H.Y.; Leung, W.K.; Cheung, K.S. Antibiotic Resistance, Susceptibility Testing and Stewardship in Helicobacter pylori Infection. Int. J. Mol. Sci. 2023, 24, 11708. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Dong, X.; Teng, G.; Zhang, W.; Cheng, H.; Gao, W.; Dai, Y.; Zhang, X.; Wang, W. The effect of previous eradication failure on antibiotic resistance of Helicobacter pylori: A retrospective study over 8 years in Beijing. Helicobacter 2021, 26, e12804. [Google Scholar] [CrossRef]
- Karczewska, E.; Wojtas-Bonior, I.; Sito, E.; Zwolińska-Wcisło, M.; Budak, A. Primary and secondary clarithromycin, metronidazole, amoxicillin and levofloxacin resistance to Helicobacter pylori in southern Poland. Pharmacol. Rep. 2011, 63, 799–807. [Google Scholar] [CrossRef]
- Kocsmár, É.; Buzás, G.M.; Szirtes, I.; Kocsmár, I.; Kramer, Z.; Szijártó, A.; Fadgyas-Freyler, P.; Szénás, K.; Rugge, M.; Fassan, M.; et al. Primary and secondary clarithromycin resistance in Helicobacter pylori and mathematical modeling of the role of macrolides. Nat. Commun. 2021, 12, 2255. [Google Scholar] [CrossRef]
- Tüzün, Y.; Bayan, K.; Yilmaz, S.; Dursun, M.; Ozekinci, T. The prevalence of primary and secondary Helicobacter pylori resistance to clarithromycin and probable contributing cofactors: Data from southeastern Anatolia. Hepatogastroenterology 2008, 55, 289–293. [Google Scholar] [PubMed]
- Wang, Y.-M.; Chen, M.-Y.; Chen, J.; Zhang, X.-H.; Feng, Y.; Han, Y.-X.; Li, Y.-L. Success of susceptibility-guided eradication of Helicobacter pylori in a region with high secondary clarithromycin and levofloxacin resistance rates. World J. Gastroenterol. 2024, 30, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Boyanova, L.; Mitov, I. Geographic map and evolution of primary Helicobacter pylori resistance to antibacterial agents. Expert Rev. Anti Infect. Ther. 2010, 8, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, N.; Nam, R.H.; Choi, S.I.; Lee, J.W.; Lee, D.H. Primary and secondary antibiotic resistance of Helicobacter pylori in Korea from 2003 to 2018. Helicobacter 2019, 24, e12660. [Google Scholar] [CrossRef] [PubMed]
- Nezami, B.G.; Jani, M.; Alouani, D.; Rhoads, D.D.; Sadri, N. Helicobacter pylori Mutations Detected by Next-Generation Sequencing in Formalin-Fixed, Paraffin-Embedded Gastric Biopsy Specimens Are Associated with Treatment Failure. J. Clin. Microbiol. 2019, 57, e01834-18. [Google Scholar] [CrossRef] [PubMed]
- Egli, K.; Wagner, K.; Keller, P.M.; Risch, L.; Risch, M.; Bodmer, T. Comparison of the Diagnostic Performance of qPCR, Sanger Sequencing, and Whole-Genome Sequencing in Determining Clarithromycin and Levofloxacin Resistance in Helicobacter pylori. Front. Cell Infect. Microbiol. 2020, 10, 596371. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.; Aljaberi, H.S.M.; Ansari, N.K.; Sun, Y.; Yin, S.; Nasifu, L.; Sun, H.; Xu, T.; Pan, Y.; Nie, Z.; et al. Phenotype and genotype analysis for Helicobacter pylori antibiotic resistance in outpatients: A retrospective study. Microbiol. Spectr. 2023, 11, e0055023. [Google Scholar] [CrossRef] [PubMed]
- Ebell, M.H. H. pylori Eradication: Effective for Cure or Improvement of Functional Dyspepsia, Especially if Eradication Is Confirmed. Am. Fam. Physician 2023, 107, Online. [Google Scholar] [PubMed]
- Giorgio, F.; Ierardi, E.; Sorrentino, C.; Principi, M.; Barone, M.; Losurdo, G.; Iannone, A.; Giangaspero, A.; Monno, R.; Di Leo, A. Helicobacter pylori DNA isolation in the stool: An essential pre-requisite for bacterial noninvasive molecular analysis. Scand. J. Gastroenterol. 2016, 51, 1429–1432. [Google Scholar] [CrossRef]
- Iannone, A.; Giorgio, F.; Russo, F.; Riezzo, G.; Girardi, B.; Pricci, M.; Palmer, S.C.; Barone, M.; Principi, M.; Strippoli, G.F.; et al. New fecal test for non-invasive Helicobacter pylori detection: A diagnostic accuracy study. World J. Gastroenterol. 2018, 24, 3021–3029. [Google Scholar] [CrossRef]
- Losurdo, G.; Pricci, M.; De Bellis, M.; Celiberto, F.; Russo, F.; Riezzo, G.; D’Attoma, B.; Iannone, A.; Rendina, M.; Ierardi, E.; et al. Effect of metronidazole resistance on Helicobacter pylori eradication regimens. J. Dig. Dis. 2022, 23, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Mégraud, F. H pylori antibiotic resistance: Prevalence, importance, and advances in testing. Gut 2004, 53, 1374–1384. [Google Scholar] [CrossRef] [PubMed]
- Kaya, A.D.; Öztürk, C.E.; Akcan, Y.; Behçet, M.; Karakoç, A.E.; Yücel, M.; Mısırlıoglu, M.; Tuncer, S. Prevalence of Helicobacter pylori in symptomatic patients and detection of clarithromycin resistance using melting curve analysis. Curr. Ther. Res. 2007, 68, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Oleastro, M.; Ménard, A.; Santos, A.; Lamouliatte, H.; Monteiro, L.; Barthélémy, P.; Mégraud, F. Real-Time PCR Assay for Rapid and Accurate Detection of Point Mutations Conferring Resistance to Clarithromycin in Helicobacter pylori. J. Clin. Microbiol. 2003, 41, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Puz, S.; Innerhofer, A.; Ramharter, M.; Haefner, M.; Hirschl, A.M.; Kovách, Z.; Rotter, M.; Makristathis, A. A novel noninvasive genotyping method of Helicobacter pylori using stool specimens. Gastroenterology 2008, 135, 1543–1551. [Google Scholar] [CrossRef]
- Dore, M.P.; Lu, H.; Graham, D.Y. Role of bismuth in improving Helicobacter pylori eradication with triple therapy. Gut 2016, 65, 870–878. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).