Screening of Klebsiella pneumoniae subsp. pneumoniae Strains with Multi-Drug Resistance and Virulence Profiles Isolated from an Italian Hospital between 2020 and 2023
Abstract
:1. Introduction
2. Results
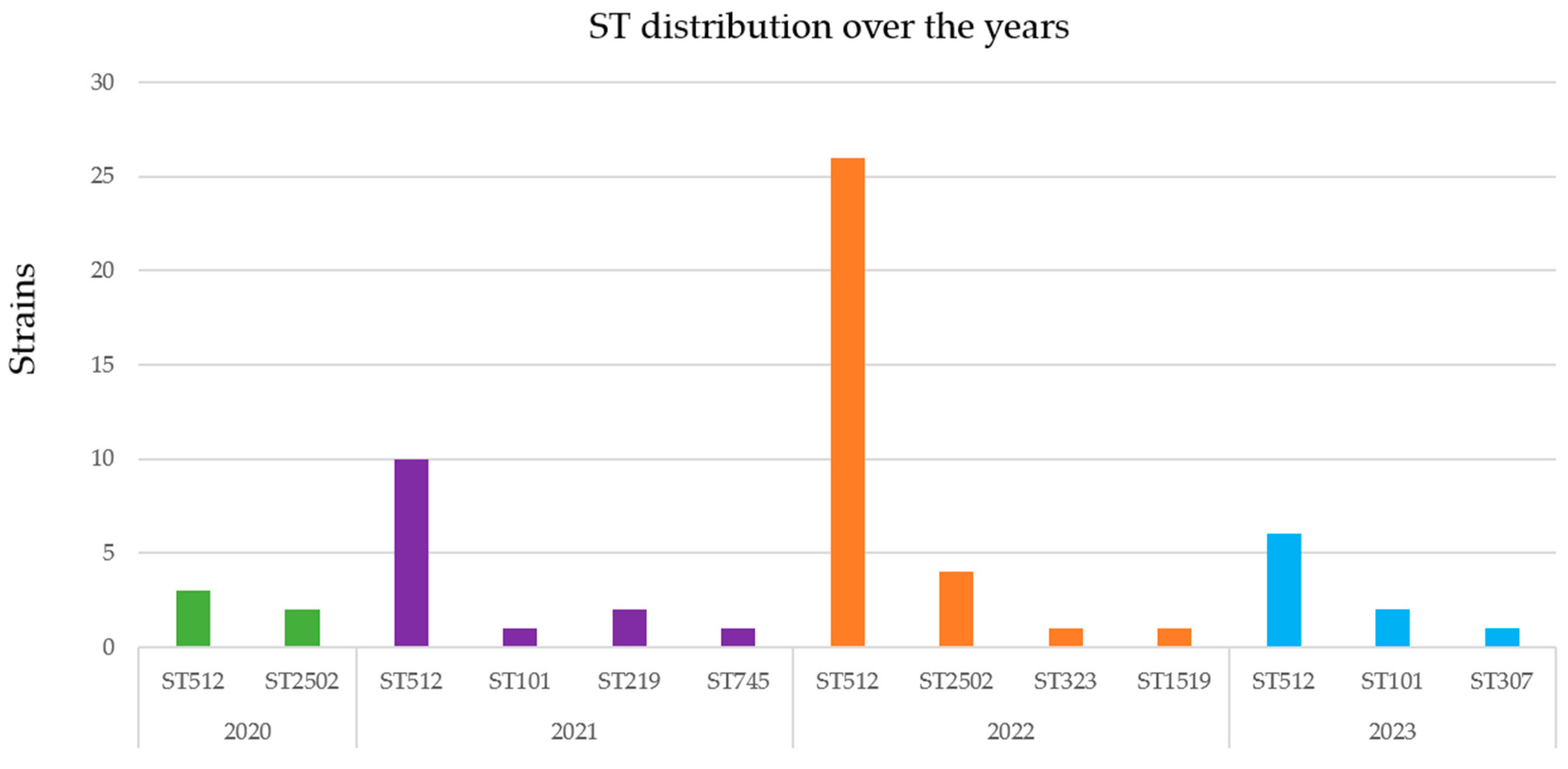
2.1. Phenotypic Characteristics
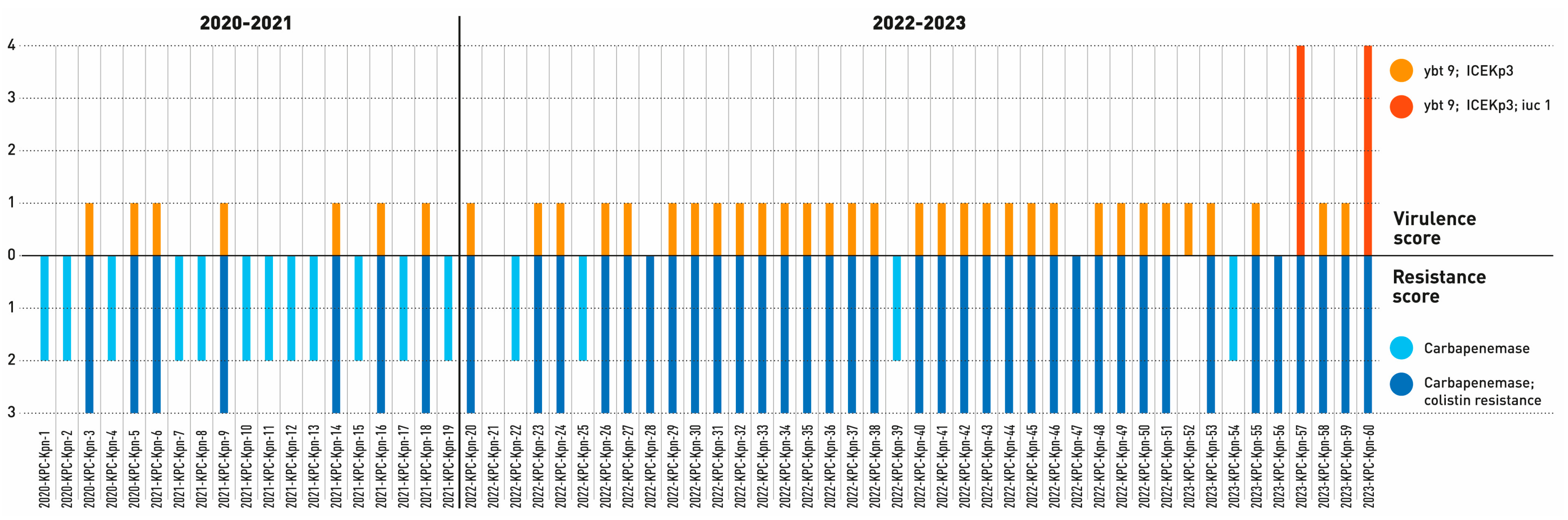
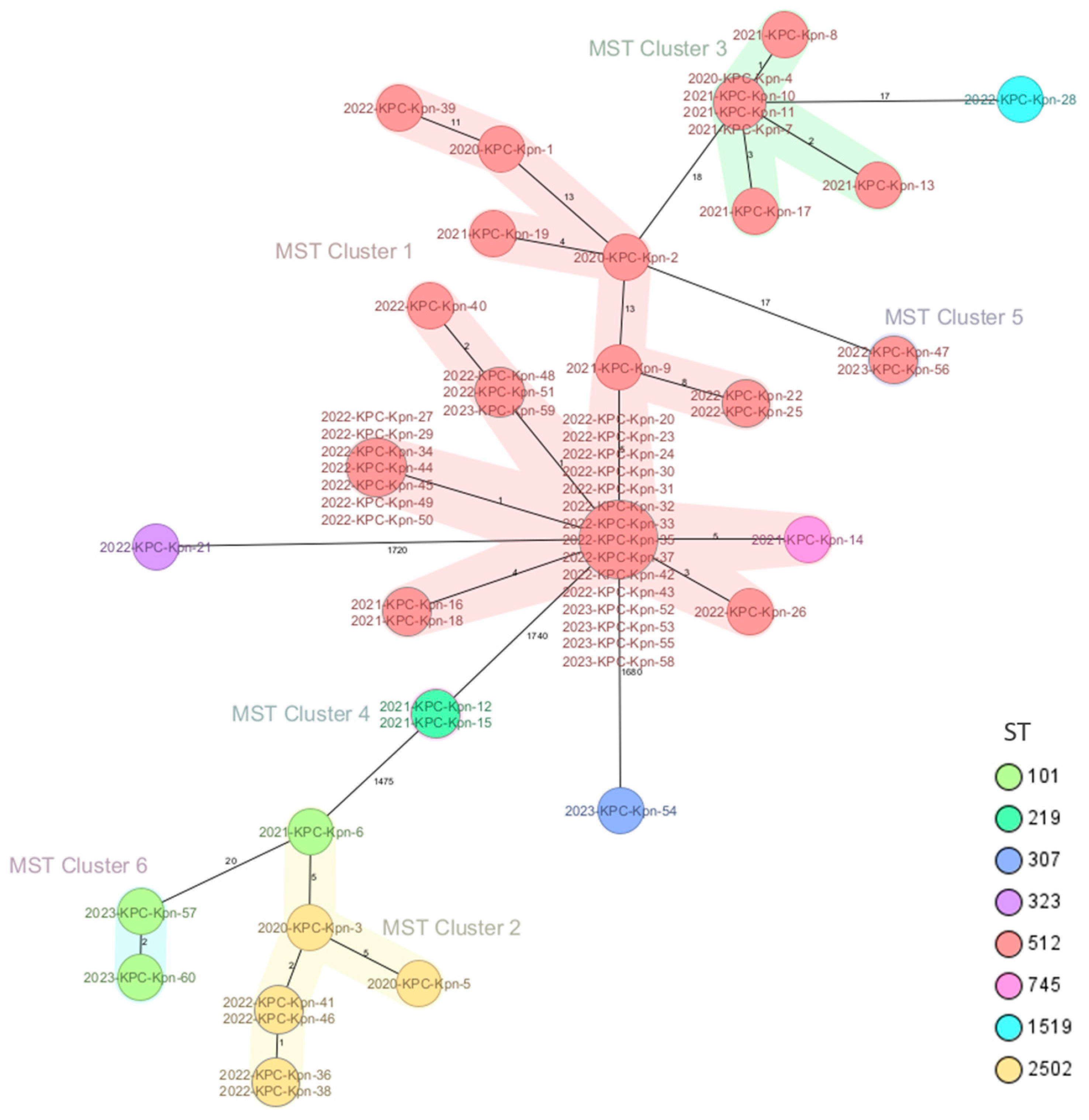
2.2. Genomic Characterization of Virulence and Resistance Profiles in KP Isolates
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Sanità, I.S.D. CRE: Sorveglianza Nazionale delle Batteriemie da Enterobatteri Resistenti ai Carbapenemi; ISS: Rome, Italy, 2022. [Google Scholar]
- Bartolini, A.; Basso, M.; Franchin, E.; Menegotto, N.; Ferrari, A.; De Canale, E.; Andreis, S.; Scaggiante, R.; Stefani, S.; Palu, G.; et al. Prevalence, molecular epidemiology and intra-hospital acquisition of Klebsiella pneumoniae strains producing carbapenemases in an Italian teaching hospital from January 2015 to September 2016. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2017, 59, 103–109. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net); ECDC: Stockolm, Sweden, 2022. [Google Scholar]
- Falcone, M.; Tiseo, G.; Giordano, C.; Leonildi, A.; Menichini, M.; Vecchione, A.; Pistello, M.; Guarracino, F.; Ghiadoni, L.; Forfori, F.; et al. Predictors of hospital-acquired bacterial and fungal superinfections in COVID-19: A prospective observational study. J. Antimicrob. Chemother. 2021, 76, 1078–1084. [Google Scholar] [CrossRef] [PubMed]
- Arcari, G.; Raponi, G.; Sacco, F.; Bibbolino, G.; Di Lella, F.M.; Alessandri, F.; Coletti, M.; Trancassini, M.; Deales, A.; Pugliese, F.; et al. Klebsiella pneumoniae infections in COVID-19 patients: A 2-month retrospective analysis in an Italian hospital. Int. J. Antimicrob. Agents 2021, 57, 106245. [Google Scholar] [CrossRef] [PubMed]
- Montrucchio, G.; Corcione, S.; Sales, G.; Curtoni, A.; De Rosa, F.G.; Brazzi, L. Carbapenem-resistant Klebsiella pneumoniae in ICU-admitted COVID-19 patients: Keep an eye on the ball. J. Glob. Antimicrob. Resist. 2020, 23, 398–400. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Reuter, S.; Harris, S.R.; Glasner, C.; Feltwell, T.; Argimon, S.; Abudahab, K.; Goater, R.; Giani, T.; Errico, G.; et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- WHO. The WHO Global Clinical Platform for COVID-19; WHO: Geneva, Switzerland, 2023. [Google Scholar]
- WHO. WHO reports widespread overuse of antibiotics in patients hospitalized with COVID-19. Saudi Med. J. 2024, 45, 547–548. [Google Scholar]
- ECDC. Emergence of Hypervirulent Klebsiella Pneumoniae ST23 Carrying Carbapenemase Genes in EU/EEA Countries; European Centre for Disease Prevention and Control: Stockolm, Sweden, 2021. [Google Scholar]
- Mendes, G.; Santos, M.L.; Ramalho, J.F.; Duarte, A.; Caneiras, C. Virulence factors in carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2023, 14, 1325077. [Google Scholar] [CrossRef]
- Fasciana, T.; Gentile, B.; Aquilina, M.; Ciammaruconi, A.; Mascarella, C.; Anselmo, A.; Fortunato, A.; Fillo, S.; Petralito, G.; Lista, F.; et al. Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect. Dis. 2019, 19, 928. [Google Scholar] [CrossRef]
- Merla, C.; Kuka, A.; Mileto, I.; Petazzoni, G.; Gaiarsa, S.; De Vitis, D.; Ardizzone, M.; Corbella, M.; Baldanti, F.; Cambieri, P. One-year surveillance for hypervirulent Klebsiella pneumoniae detected carbapenem-resistant superbugs. Microbiol. Spectr. 2024, 12, e0329223. [Google Scholar] [CrossRef]
- Wahl, A.; Fischer, M.A.; Klaper, K.; Muller, A.; Borgmann, S.; Friesen, J.; Hunfeld, K.P.; Ilmberger, A.; Kolbe-Busch, S.; Kresken, M.; et al. Presence of hypervirulence-associated determinants in Klebsiella pneumoniae from hospitalised patients in Germany. Int. J. Med. Microbiol. IJMM 2024, 314, 151601. [Google Scholar] [CrossRef]
- de Sales, R.O.; Leaden, L.; Migliorini, L.B.; Severino, P. A Comprehensive Genomic Analysis of the Emergent Klebsiella pneumoniae ST16 Lineage: Virulence, Antimicrobial Resistance and a Comparison with the Clinically Relevant ST11 Strain. Pathogens 2022, 11, 1394. [Google Scholar] [CrossRef] [PubMed]
- Hallal Ferreira Raro, O.; Nordmann, P.; Dominguez Pino, M.; Findlay, J.; Poirel, L. Emergence of Carbapenemase-Producing Hypervirulent Klebsiella pneumoniae in Switzerland. Antimicrob. Agents Chemother. 2023, 67, e0142422. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Wyres, K.L.; Wick, R.R.; Judd, L.M.; Fostervold, A.; Holt, K.E.; Lohr, I.H. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 2019, 74, 1218–1222. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 15 January 2024).
- Hagiya, H.; Watanabe, N.; Maki, M.; Murase, T.; Otsuka, F. Clinical utility of string test as a screening method for hypermucoviscosity-phenotype Klebsiella pneumoniae. Acute Med. Surg. 2014, 1, 245–246. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; Soucy, J.R.; Leung, V.; So, M.; Kwan, A.T.H.; Portnoff, J.S.; Bertagnolio, S.; Raybardhan, S.; MacFadden, D.R.; Daneman, N. Antibiotic resistance associated with the COVID-19 pandemic: A systematic review and meta-analysis. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2023, 29, 302–309. [Google Scholar] [CrossRef]
- Sulayyim, H.J.A.; Ismail, R.; Hamid, A.A.; Ghafar, N.A. Antibiotic Resistance during COVID-19: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 11931. [Google Scholar] [CrossRef]
- Walia, K.; Mendelson, M.; Kang, G.; Venkatasubramanian, R.; Sinha, R.; Vijay, S.; Veeraraghavan, B.; Basnyat, B.; Rodrigues, C.; Bansal, N.; et al. How can lessons from the COVID-19 pandemic enhance antimicrobial resistance surveillance and stewardship? Lancet. Infect. Dis. 2023, 23, e301–e309. [Google Scholar] [CrossRef]
- Kariyawasam, R.M.; Julien, D.A.; Jelinski, D.C.; Larose, S.L.; Rennert-May, E.; Conly, J.M.; Dingle, T.C.; Chen, J.Z.; Tyrrell, G.J.; Ronksley, P.E.; et al. Antimicrobial resistance (AMR) in COVID-19 patients: A systematic review and meta-analysis (November 2019-June 2021). Antimicrob. Resist. Infect. Control 2022, 11, 45. [Google Scholar] [CrossRef]
- Karruli, A.; Boccia, F.; Gagliardi, M.; Patauner, F.; Ursi, M.P.; Sommese, P.; De Rosa, R.; Murino, P.; Ruocco, G.; Corcione, A.; et al. Multidrug-Resistant Infections and Outcome of Critically Ill Patients with Coronavirus Disease 2019: A Single Center Experience. Microb. Drug Resist. 2021, 27, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.; Favaro, M.; Campogiani, L.; Malagnino, V.; Minelli, S.; Bossa, M.C.; Altieri, A.; Andreoni, M.; Sarmati, L. Ceftazidime/Avibactam-Resistant Klebsiella pneumoniae subsp. pneumoniae Isolates in a Tertiary Italian Hospital: Identification of a New Mutation of the Carbapenemase Type 3 (KPC-3) Gene Conferring Ceftazidime/Avibactam Resistance. Microorganisms 2021, 9, 2356. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S. A parallel and silent emerging pandemic: Antimicrobial resistance (AMR) amid COVID-19 pandemic. J. Infect. Public Health 2023, 16, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam, M.M.C.; Duchene, S.; Jenney, A.; Holt, K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019, 15, e1008114. [Google Scholar] [CrossRef] [PubMed]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Pellegrino, M.; Giuzio, F.; Marra, M.; Rosano, C.; Saturnino, C.; Sinicropi, M.S.; Aquaro, S. Antibiotic-Resistant ESKAPE Pathogens and COVID-19: The Pandemic beyond the Pandemic. Viruses 2023, 15, 1843. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Jiang, Y.; Zhou, J.; Yu, Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2021, 25, 26–34. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Jeon, J.H.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 2017, 7, 483. [Google Scholar] [CrossRef]
- Venditti, C.; Butera, O.; Meledandri, M.; Balice, M.P.; Cocciolillo, G.C.; Fontana, C.; D’Arezzo, S.; De Giuli, C.; Antonini, M.; Capone, A.; et al. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2021, 27, 1040.e1–1040.e6. [Google Scholar] [CrossRef]
- Cristina, M.L.; Sartini, M.; Ottria, G.; Schinca, E.; Cenderello, N.; Crisalli, M.P.; Fabbri, P.; Lo Pinto, G.; Usiglio, D.; Spagnolo, A.M. Epidemiology and biomolecular characterization of carbapenem-resistant klebsiella pneumoniae in an Italian hospital. J. Prev. Med. Hyg. 2016, 57, E149–E156. [Google Scholar]
- Zhu, J.; Wang, T.; Chen, L.; Du, H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 642484. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, T.; Harada, S.; Okamoto, K.; Ishino, S.; Kaneko, M.; Suzuki, M.; Ito, R.; Mizoguchi, M. COVID-19 and Fatal Sepsis Caused by Hypervirulent Klebsiella pneumoniae, Japan, 2020. Emerg. Infect. Dis. 2021, 27, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Falcone, M.; Tiseo, G.; Arcari, G.; Leonildi, A.; Giordano, C.; Tempini, S.; Bibbolino, G.; Mozzo, R.; Barnini, S.; Carattoli, A.; et al. Spread of hypervirulent multidrug-resistant ST147 Klebsiella pneumoniae in patients with severe COVID-19: An observational study from Italy, 2020–2021. J. Antimicrob. Chemother. 2022, 77, 1140–1145. [Google Scholar] [CrossRef] [PubMed]
- Morales-León, F.; Matus-Köhler, M.; Araya-Vega, P.; Aguilera, F.; Torres, I.; Vera, R.; Ibarra, C.; Venegas, S.; Bello-Toledo, H.; González-Rocha, G.; et al. Molecular Characterization of the Convergent Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae Strain K1-ST23, Collected in Chile during the COVID-19 Pandemic. Microbiol. Spectr. 2023, 11, e0054023. [Google Scholar] [CrossRef] [PubMed]
- Roe, C.C.; Vazquez, A.J.; Esposito, E.P.; Zarrilli, R.; Sahl, J.W. Diversity, Virulence, and Antimicrobial Resistance in Isolates From the Newly Emerging Klebsiella pneumoniae ST101 Lineage. Front. Microbiol. 2019, 10, 542. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Sun, S.; Zhao, Z.Y.; Sun, Y. The pathogenicity of rmpA or aerobactin-positive Klebsiella pneumoniae in infected mice. J. Int. Med. Res. 2019, 47, 4344–4352. [Google Scholar] [CrossRef] [PubMed]
- Parisi, S.G.; Bartolini, A.; Santacatterina, E.; Castellani, E.; Ghirardo, R.; Berto, A.; Franchin, E.; Menegotto, N.; De Canale, E.; Tommasini, T.; et al. Prevalence of Klebsiella pneumoniae strains producing carbapenemases and increase of resistance to colistin in an Italian teaching hospital from January 2012 to December 2014. BMC Infect. Dis. 2015, 15, 244. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.L.; Ko, W.C.; Cheng, K.C.; Lee, H.C.; Ke, D.S.; Lee, C.C.; Fung, C.P.; Chuang, Y.C. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2006, 42, 1351–1358. [Google Scholar] [CrossRef]
- Lee, A.H.Y.; Porto, W.F.; de Faria, C., Jr.; Dias, S.C.; Alencar, S.A.; Pickard, D.J.; Hancock, R.E.W.; Franco, O.L. Genomic insights into the diversity, virulence and resistance of Klebsiella pneumoniae extensively drug resistant clinical isolates. Microb. Genom. 2021, 7, 000613. [Google Scholar] [CrossRef]
- Kluytmans-van den Bergh, M.F.; Rossen, J.W.; Bruijning-Verhagen, P.C.; Bonten, M.J.; Friedrich, A.W.; Vandenbroucke-Grauls, C.M.; Willems, R.J.; Kluytmans, J.A. Whole-Genome Multilocus Sequence Typing of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2016, 54, 2919–2927. [Google Scholar] [CrossRef]

| Resistance Determinants | Virulence Setup | Typing | |||||
|---|---|---|---|---|---|---|---|
| Strain | β-Lactam | Additional Resistance Genes | Resistance Score | Virulence Genes | Virulence Score | ST 1 | CT 2 |
| 2020-KPC-Kpn-1 | blaKPC-3, blaOXA-9,blaSHV-11,blaTEM-1D | aac (6’) -Ib fosA_3 oqxA, oqxB | 2 | - | 0 | ST512 | 1 |
| 2020-KPC-Kpn-2 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | aac (6’) -Ib, aadA2, aph (3’) -Ia, sul1catA1, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A) | 2 | - | 0 | ST512 | 1 |
| 2020-KPC-Kpn-3 | blaKPC-3, blaSHV-11 | msr(E), mph(E), armA, fosA_3, fosA_6, oqxA, oqxB | 3 | ybt 9; ICEKp3 | 1 | ST2502 | 2 |
| 2020-KPC-Kpn-4 | blaKPC-3, blaOXA-9,blaSHV-11, blaTEM-1D | aac (6’) -Ib, aadA2, aph (3’) -Ia, sul1, catA1, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A) | 2 | - | 0 | ST512 | 3 |
| 2020-KPC-Kpn-5 | blaKPC-3, blaSHV-1 | fosA_3, fosA, oqxA, oqxB, armA, _6, msr(E), mph(E) | 3 | ybt 9; ICEKp3 | 1 | ST2502 | 2 |
| 2021-KPC-Kpn-6 | blaKPC-3, blaSHV-1,blaTEM-1D | aph(6)-Id, aph(3’)-Ib, aadA5, armA, sul1, sul2, tet(A), fosA_3, qacE, oqxA, oqxB, dfrA14, msr(E), mph(A), mph(E) | 3 | ybt 9; ICEKp3 | 1 | ST101 | 2 |
| 2021-KPC-Kpn-7 | blaKPC-3, blaOXA-9,blaSHV-11, blaTEM-1D | aac (6’) -Ib, aadA2, aph (3’) -Ia, sul1, catA1, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A) | 2 | - | 0 | ST512 | 3 |
| 2021-KPC-Kpn-8 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | fosA_3, aac (6’) -Ib, aadA2, aph (3’) -Ia, sul1, catA1, qacE, oqxA, oqxB, dfrA12, mph(A) | 2 | - | 0 | ST512 | 3 |
| 2021-KPC-Kpn-9 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | sul1, mph(A), aadA2, aph (3’) -Ia, catA1, fosA_3, qacE, oqxA, oqxB, dfrA12 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2021-KPC-Kpn-10 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | mph(A), aac (6’) -Ib, aadA2, aph(3’)-Ia, sul1, catA1, fosA_3, qacE, oqxA, oqxB, dfrA12 | 2 | - | 0 | ST512 | 3 |
| 2021-KPC-Kpn-11 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | aac (6’) -Ib, aadA2, aph (3’) -Ia, sul I, catA1, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A) | 2 | - | 0 | ST512 | 3 |
| 2021-KPC-Kpn-12 | blaKPC-3, blaCTX-M-15,blaSHV-1, blaTEM-1D | fosA_3, qacE, oqxA, oqxB, qnrS1, dfrA12, mph(A), aph (6) -Id, aph (3’) -Ib, aadA2, sul I, sul2 | 2 | - | 0 | ST219 | 4 |
| 2021-KPC-Kpn-13 | blaKPC-3, blaOXA-9,blaSHV-11, blaTEM-1D | mph(A), aac (6’) -Ib, aadA2, aph (3’) -Ia, sul1, catA1, fosA_3, qacE, oqxA, oqxB, dfrA12 | 2 | - | 0 | ST512 | 3 |
| 2021-KPC-Kpn-14 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | fosA_3, oqxA, oqxB | 3 | ybt 9; ICEKp3 | 1 | ST745 | 1 |
| 2021-KPC-Kpn-15 | blaKPC-3, blaCTX-M-15, blaSHV-1, blaTEM-1D | fosA_3, qacE, oqxA, oqxB, qnrS1, dfrA12, mph(A), aph (6) -Id, aph(3’)-Ib, aadA2, sul1, sul2 | 2 | - | 0 | ST219 | 4 |
| 2021-KPC-Kpn-16 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | catA1, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A), aadA2, aph (3’) -Ia, sul1 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2021-KPC-Kpn-17 | blaKPC-3, blaOXA-9,blaSHV-11, blaTEM-1D | aac(6’)-Ib, aadA2, aph(3’)-Ia, sul1, catA1, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A) | 2 | - | 0 | ST512 | 3 |
| 2021-KPC-Kpn-18 | blaKPC-3, blaOXA-9,blaSHV-11, blaTEM-1D | catA1, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A), aac (6’) -Ib, aadA2, aph (3’) -Ia, sul1 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2021-KPC-Kpn-19 | blaKPC-3, blaOXA-9, blaSHV-11, blaTEM-1D | catAI, fosA_3, qacE, oqxA, oqxB, dfrA12, mph(A), aac (6’)-Ib, aadA2, aph (3’)-Ia, sul1 | 2 | 0 | ST512 | 1 | |
| 2022-KPC-Kpn-20 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA2, aph (3’)-VIb, aph3-Ia, sul1, sul2, tet(A), floR, catA1, cmlA5, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-21 | blaSHV-1 | aadA, sul1 | 0 | - | 0 | ST323 | - |
| 2022-KPC-Kpn-22 | blaKPC-3, blaSHV-11 | aac (6’)-Ib’, aadA2, aph3-Ia, sul I, catA1, mphA | 2 | - | 0 | ST512 | 1 |
| 2022-KPC-Kpn-23 | blaKPC-3, blaSHV-11, blaTEM-1D | mphA, aadA2, aph3-Ia, sul1, catA1 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-24 | blaKPC-3, blaSHV-11, blaTEM-1D | catA1, aadA2, aph3-Ia, sul1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-25 | blaKPC-3, blaSHV-1 | aac(6’)-Ib’, aadA2, aph3-Ia, sul1, catA1, mphA | 2 | - | 0 | ST512 | 1 |
| 2022-KPC-Kpn-26 | blaKPC-3, blaCTX-M-15, blaSHV-11, blaTEM-1D | aadA2, aph3-Ia, strA, strB, sul1, sul2, catA1, catII.2, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-27 | blaKPC-3, blaSHV-11, blaTEM-1D | aadA2, aph3-Ia, sul I, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-28 | blaKPC-3, blaSHV-11 | aac(6’)-Ib’, aadA2, aph3-Ia, sul1, catA1, mphA | 3 | - | 0 | ST1519 | - |
| 2022-KPC-Kpn-29 | blaKPC-19, blaSHV-11, blaTEM-1D | aadA2, aph3-Ia, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-30 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, strB, sul1, sul2, tet(A), floR, catA1, cmlA5, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-31 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, sul I, sul2, tet(A), floR, catA1, cmlA5 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-32 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, sul1, sul2, tet(A), floR, catA1, cmlA5, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-33 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, strB, sul1, sul2, tet(A), floR, catA1 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-34 | blaKPC-3, blaSHV-11, blaTEM-1D | aadA2, aph3-Ia, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-35 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, sul1, sul2, tet(A), floR, catA1, cmlA5, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-36 | blaKPC-3, blaSHV-1 | armA, mphE, msrE | 3 | ybt 9; ICEKp3 | 1 | ST2502 | 2 |
| 2022-KPC-Kpn-37 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, sul1, sul2, tet(A), floR, catA1, cmlA5, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-38 | blaKPC-3, blaSHV-1 | armA, mphE.v2, msrE | 3 | ybt 9; ICEKp3 | 1 | ST2502 | 2 |
| 2022-KPC-Kpn-39 | blaKPC-3, blaSHV-11, blaTEM-1D | aac(6’)-Ib’, aadA2, aph3-Ia, sul1, catA1, mphA | 2 | - | 0 | ST512 | 1 |
| 2022-KPC-Kpn-40 | blaKPC-3, blaSHV-11, blaTEM-1D | aadA2, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-41 | blaKPC-3, blaSHV-1 | armA, mphE.v2, msrE | 3 | ybt 9; ICEKp3 | 1 | ST2502 | 2 |
| 2022-KPC-Kpn-42 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, sul1, sul2, tet(A), floR, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-43 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11, blaTEM-1D | aadA2, aph (3’)-VIb, aph3-Ia, strA, strB, sul1, sul2, tet(A), floR, catA1 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-44 | blaKPC-3, blaSHV-11, blaTEM-1D | aadA2, aph3-Ia, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-45 | blaKPC-3, blaSHV-11, blaTEM-1D | aadA2, aph3-Ia, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-46 | blaKPC-3, blaSHV-1 | armA, mphE.v2, msrE | 3 | ybt 9; ICEKp3 | 1 | ST2502 | 2 |
| 2022-KPC-Kpn-47 | blaKPC-3, blaSHV-11, blaTEM-1D | mphA, aac (6’)-Ib’, aadA2, aph3-Ia, sul1, catA1 | 3 | - | 0 | ST512 | 5 |
| 2022-KPC-Kpn-48 | blaKPC-3, blaSHV-11, blaTEM-1D | sul1, aadA2, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-49 | blaKPC-3, blaSHV-11, blaTEM-1D | aadA2, aph3-Ia, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-50 | blaKPC-3, blaSHV-1, blaTEM-1D | aadA2, aph3-Ia, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2022-KPC-Kpn-51 | blaKPC-3, blaSHV-1, blaTEM-1D | aadA2, sul1, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2023-KPC-Kpn-52 | blaKPC-3,blaSHV-1, blaTEM-1D | aadA2, aph3-Ia, sul1, catA1, mphA | 0 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2023-KPC-Kpn-53 | blaKPC-3, blaCMY-16, blaOXA-10, blaTEM-1D, blaSHV-1 | aadA, aadA2, aph(3’)-VIb, aph3-Ia, strA, strB, sul1, sul2, tet(A), floR, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2023-KPC-Kpn-54 | blaKPC-3, blaOXA-10, blaCTX-M-15, blaSHV-28, blaTEM-1D | aac(3)-IIa, aac(6’)-Ib, strA, strB, sul2, CatB4 | 2 | - | 0 | ST307 | - |
| 2023-KPC-Kpn-55 | blaKPC-3, blaCMY-16, blaOXA-10, blaTEM-1D, blaSHV-1 | aadA, aadA2, aph (3’)-VIb, aph3-Ia, strA, strB, sul1, sul2, tet(A), floR, catA1, mphA | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2023-KPC-Kpn-56 | blaKPC-3, blaSHV-1 | aac(6’)-Ib’, aadA2, aph3-Ia, sul1, catA1, mphA | 3 | - | 0 | ST512 | 5 |
| 2023-KPC-Kpn-57 | blaKPC-3, blaSHV-1 | aadA, ant (2″)-Ia, sul1, catA1 | 3 | ybt 9; ICEKp3; iuc 1 | 4 | ST101 | 6 |
| 2023-KPC-Kpn-58 | blaKPC-3, blaCMY-16, blaOXA-10, blaSHV-11,blaTEM-1D | strA, strB, aadA, aadA2, aph (3’)-VIb, aph3Ia, sul1, sul2, tet(A), floR, catA1 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2023-KPC-Kpn-59 | blaKPC-3, blaSHV-11,blaTEM-1D | aadA2, sul1, catA1 | 3 | ybt 9; ICEKp3 | 1 | ST512 | 1 |
| 2023-KPC-Kpn-60 | blaKPC-3, blaSHV-1, | aadA, ant (2″)-Ia, sul1, catA1 | 3 | ybt 9; ICEKp3; iuc 1; rmpA2 | 4 | ST101 | 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimartino, V.; Venditti, C.; Messina, F.; D’Arezzo, S.; Selleri, M.; Butera, O.; Nisii, C.; Marani, A.; Arcangeli, A.; Gaziano, R.; et al. Screening of Klebsiella pneumoniae subsp. pneumoniae Strains with Multi-Drug Resistance and Virulence Profiles Isolated from an Italian Hospital between 2020 and 2023. Antibiotics 2024, 13, 561. https://doi.org/10.3390/antibiotics13060561
Dimartino V, Venditti C, Messina F, D’Arezzo S, Selleri M, Butera O, Nisii C, Marani A, Arcangeli A, Gaziano R, et al. Screening of Klebsiella pneumoniae subsp. pneumoniae Strains with Multi-Drug Resistance and Virulence Profiles Isolated from an Italian Hospital between 2020 and 2023. Antibiotics. 2024; 13(6):561. https://doi.org/10.3390/antibiotics13060561
Chicago/Turabian StyleDimartino, Valentina, Carolina Venditti, Francesco Messina, Silvia D’Arezzo, Marina Selleri, Ornella Butera, Carla Nisii, Alessandra Marani, Alessia Arcangeli, Roberta Gaziano, and et al. 2024. "Screening of Klebsiella pneumoniae subsp. pneumoniae Strains with Multi-Drug Resistance and Virulence Profiles Isolated from an Italian Hospital between 2020 and 2023" Antibiotics 13, no. 6: 561. https://doi.org/10.3390/antibiotics13060561
APA StyleDimartino, V., Venditti, C., Messina, F., D’Arezzo, S., Selleri, M., Butera, O., Nisii, C., Marani, A., Arcangeli, A., Gaziano, R., Cosio, T., Scanzano, P., & Fontana, C. (2024). Screening of Klebsiella pneumoniae subsp. pneumoniae Strains with Multi-Drug Resistance and Virulence Profiles Isolated from an Italian Hospital between 2020 and 2023. Antibiotics, 13(6), 561. https://doi.org/10.3390/antibiotics13060561







