Modification and Synergistic Studies of a Novel Frog Antimicrobial Peptide against Pseudomonas aeruginosa Biofilms
Abstract
:1. Introduction
2. Results
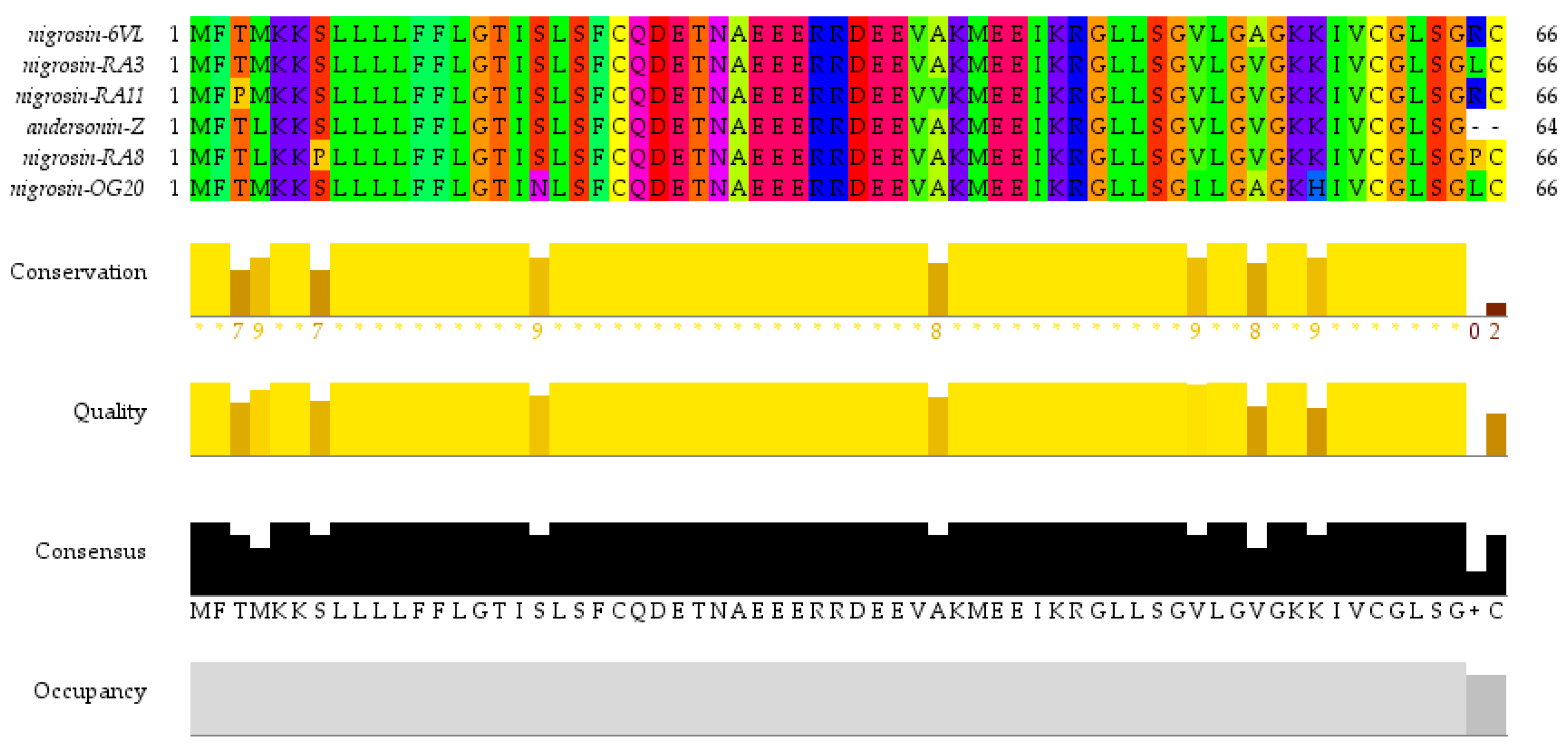
2.1. ‘Shotgun’ Cloning of Peptide Nigrosin-6VL Biosynthetic Precursor Complementary DNA (cDNA)
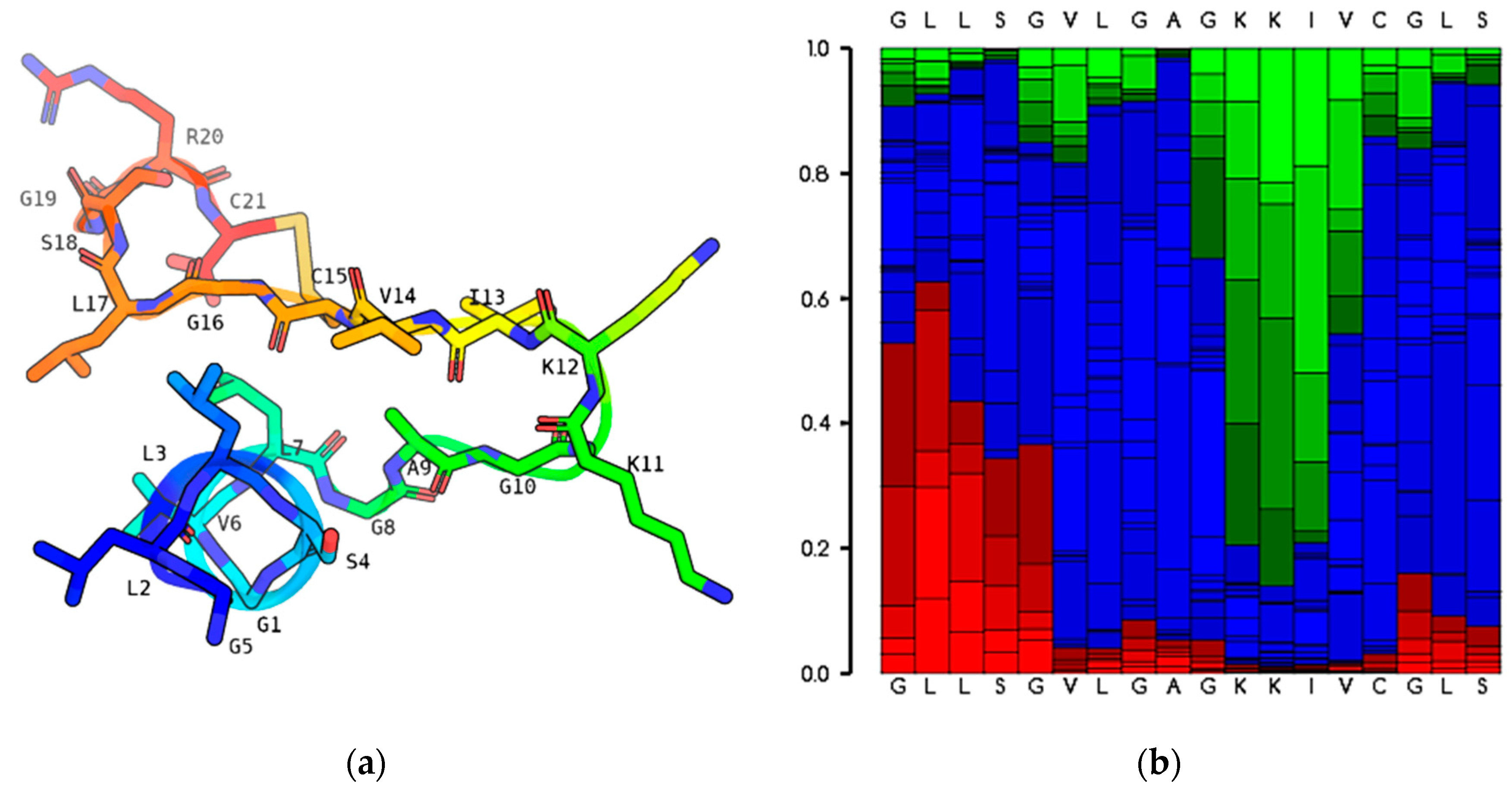
2.2. Predicted Secondary Structure and Physicochemical Characteristics of the Original Peptide Nigrosin-6VL
2.3. Physicochemical Properties and Structural Modification of Nigrosin-6VL
2.4. Anti-Planktonic Microorganism Activity Selection of Nigrosin-6VL and Its Analogues
2.5. Haemolysis Activity and TI Values of Nigrosin-6VL and Its Analogues
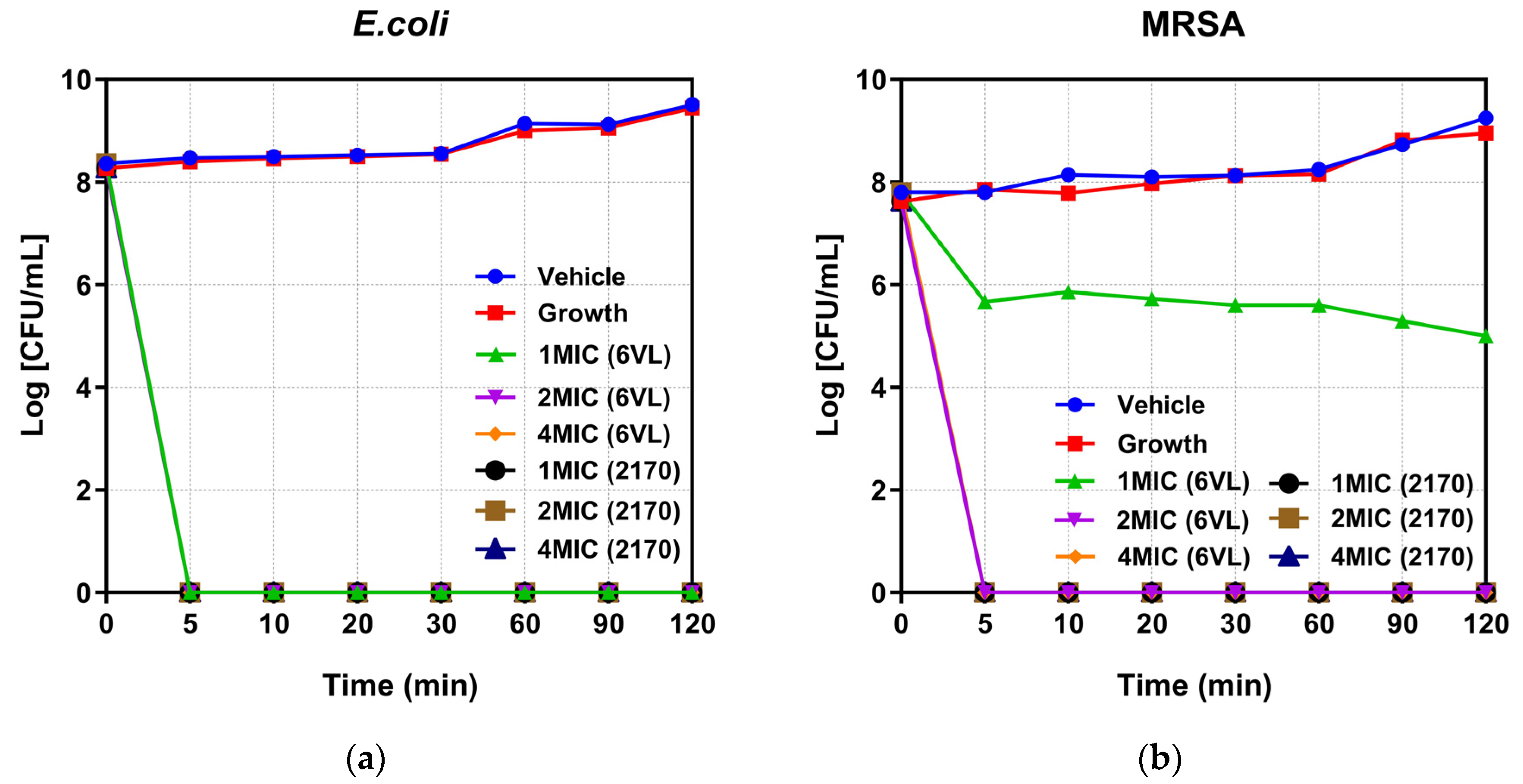
2.6. Time-Killing Kinetic Studies of Nigrosin-6VL and 2170-2R
2.7. Antimicrobial Mechanism Studies of Nigrosin-6VL and 2170-2R
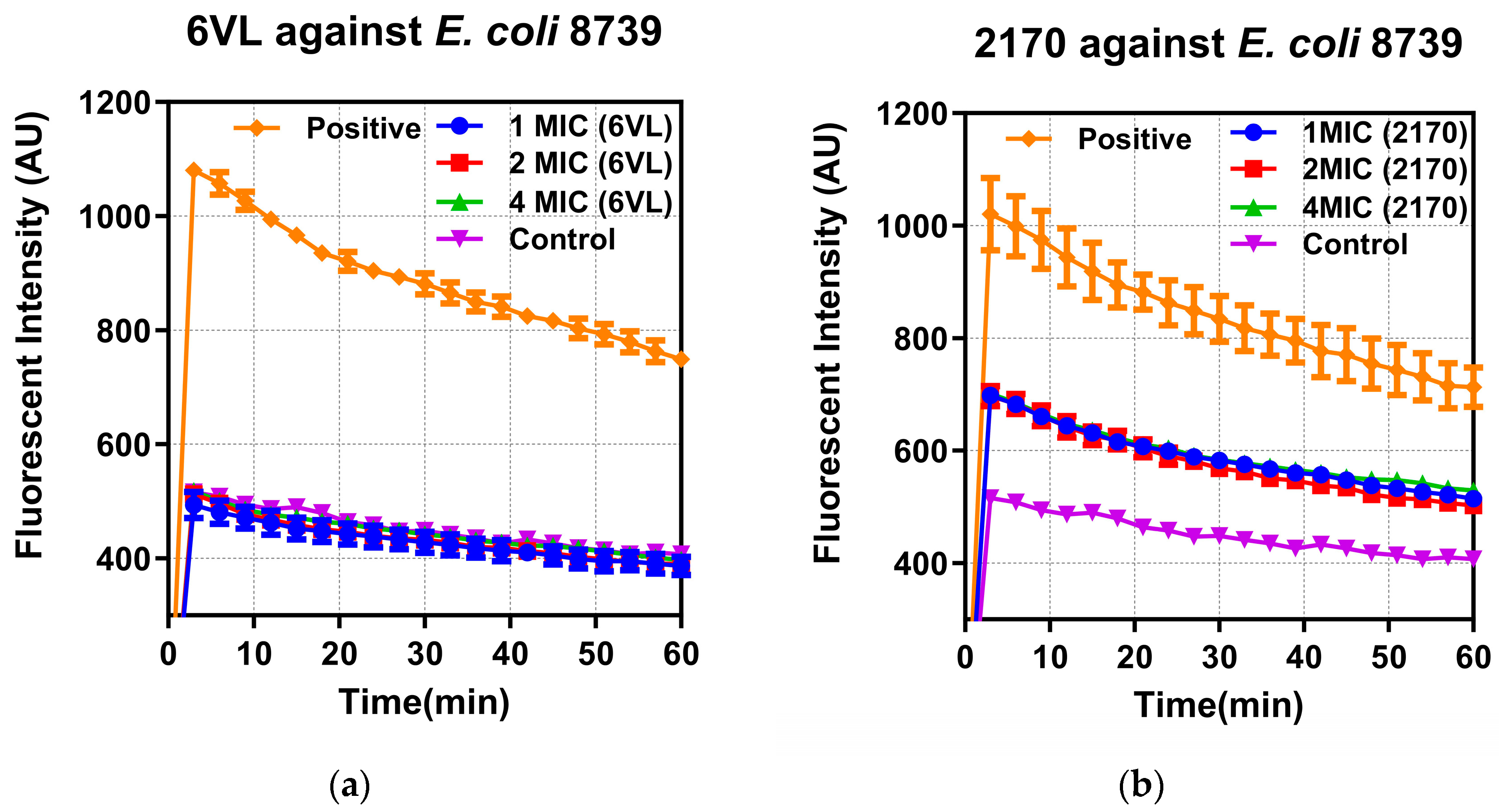
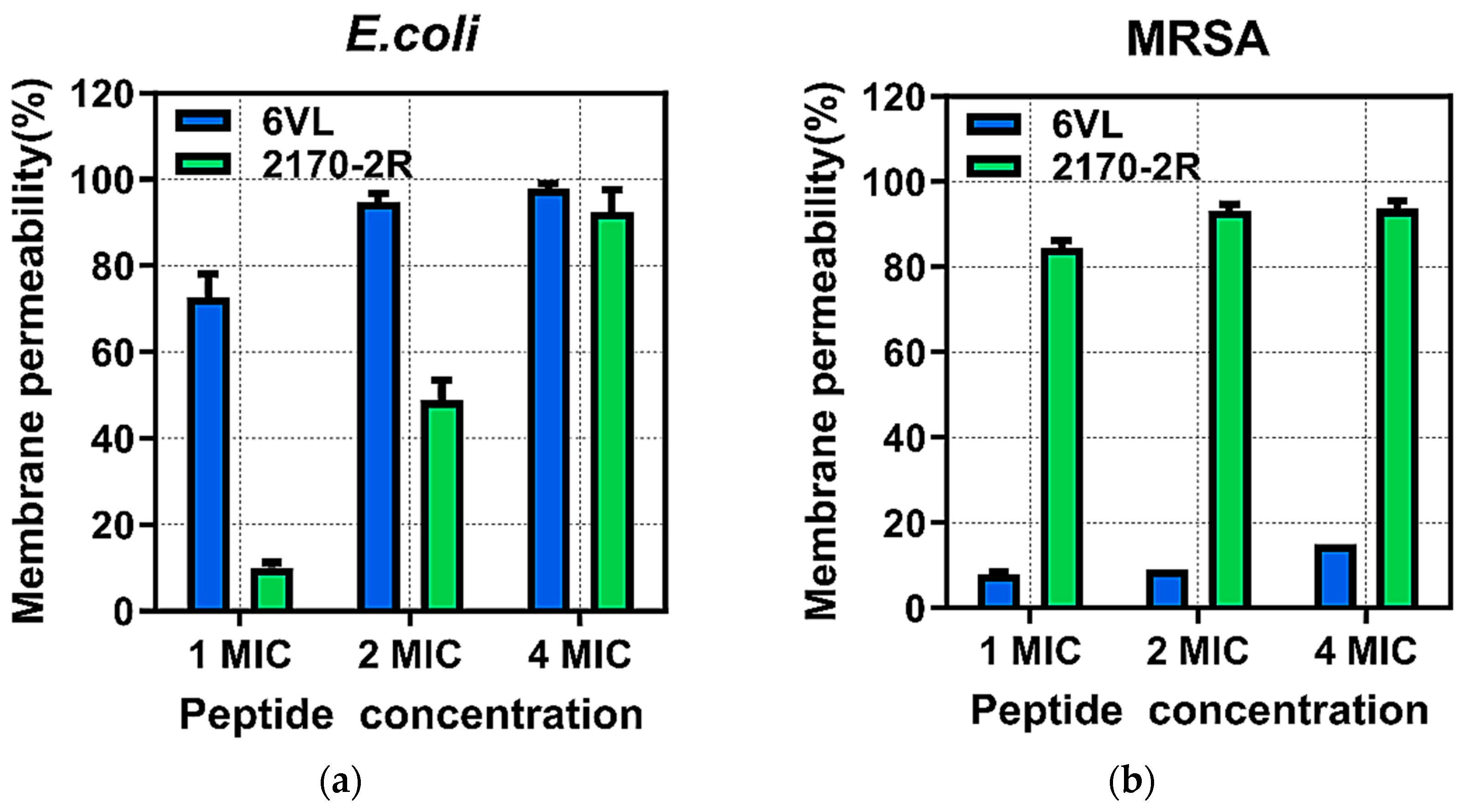
2.7.1. Effects on Bacterial Outer Membrane Permeability
2.7.2. Effects of Nigrosin-6VL and 2170-2R on Bacterial Intracellular Membrane Permeability
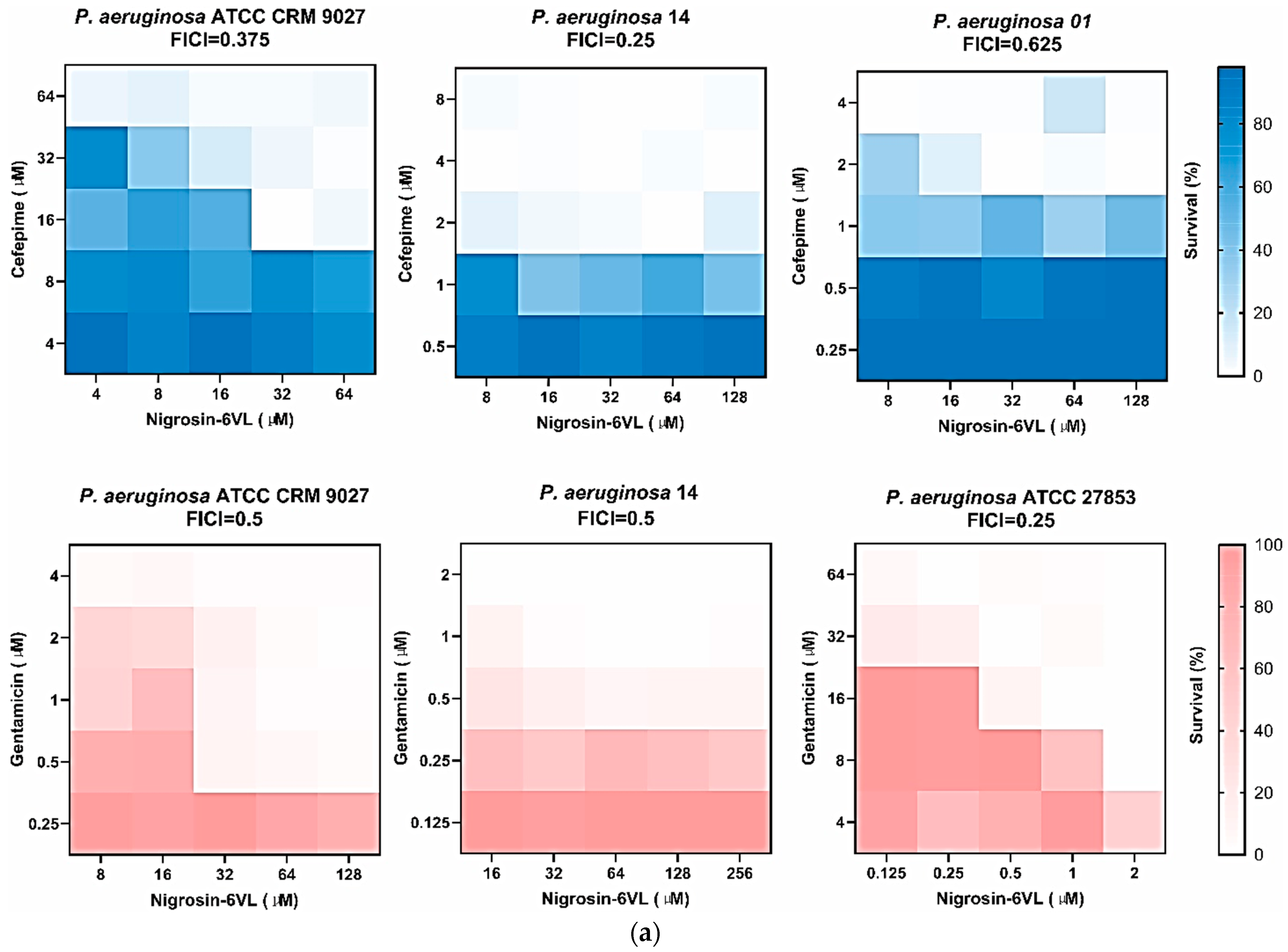
2.8. Peptide and Antibiotic Co-Administration Studies
2.9. Anti-Biofilm Studies
2.10. Peptide and Antibiotic Co-Administration Studies of Anti-Biofilm Activities
3. Discussion
4. Materials and Methods
4.1. Acquisition of Skin Secretions from Odorrana andersonii
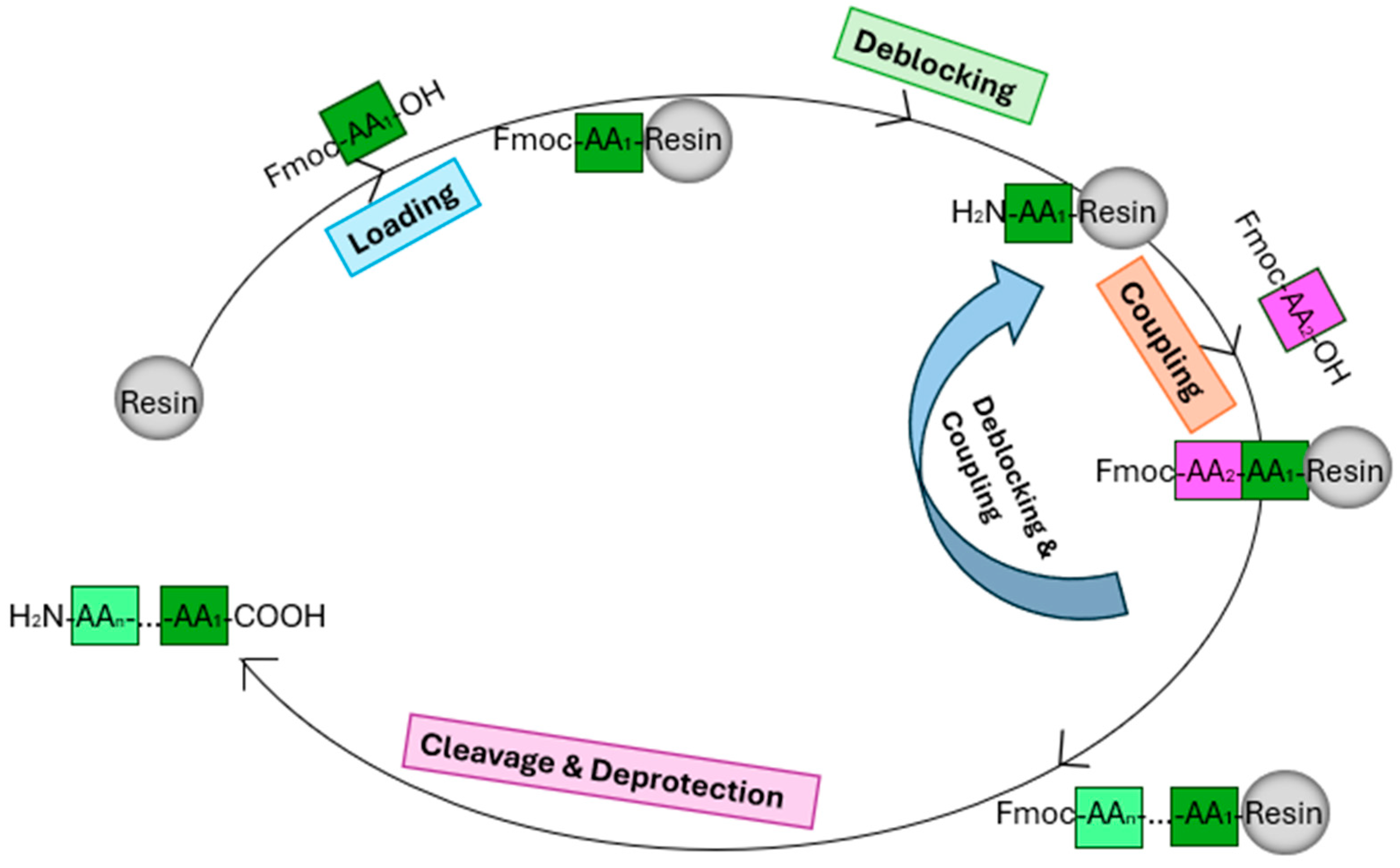
4.2. Molecular Cloning
4.3. Peptide Sequence Analysis, Secondary Structural Prediction, and Verification
4.4. Synthesis and Purification of Peptides
4.5. Anti-Planktonic Microorganism Activity Study
4.6. Haemolysis Assay and TI Value Calculation
4.7. Time-Killing Kinetic Assay
4.8. The SYTOX Green of the Bacterial Membrane Permeabilisation Assay
4.9. Outer Membrane Assay
4.10. Anti-Biofilm Assay
4.11. Peptide and Antibiotic Co-Administration Study
4.11.1. Peptide and Antibiotic Co-Administration of MIC Assay
4.11.2. Peptide and Antibiotic Co-Administration of MBIC Assay
4.12. Method of Measurement and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial Peptides for Therapeutic Use: Obstacles and Realistic Outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic Resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Mühlen, S.; Dersch, P. Anti-Virulence Strategies to Target Bacterial Infections. Curr. Top. Microbiol. Immunol. 2016, 398, 147–183. [Google Scholar] [CrossRef] [PubMed]
- Raphael, E.; Riley, L.W. Infections Caused by Antimicrobial Drug-Resistant Saprophytic Gram-Negative Bacteria in the Environment. Front. Med. 2017, 4, 183. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, Ç.; Özcengiz, G. Antibiotics: Pharmacokinetics, Toxicity, Resistance and Multidrug Efflux Pumps. Biochem. Pharmacol. 2017, 133, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial Host Defence Peptides: Functions and Clinical Potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Dou, X.; Zhu, X.; Wang, J.; Dong, N.; Shan, A. Novel Design of Heptad Amphiphiles to Enhance Cell Selectivity, Salt Resistance, Antibiofilm Properties and Their Membrane-Disruptive Mechanism. J. Med. Chem. 2017, 60, 2257–2270. [Google Scholar] [CrossRef] [PubMed]
- Dehsorkhi, A.; Castelletto, V.; Hamley, I.W. Self-Assembling Amphiphilic Peptides. J. Pept. Sci. 2014, 20, 453–467. [Google Scholar] [CrossRef]
- Lee, D.L.; Hodges, R.S. Structure-Activity Relationships of de Novo Designed Cyclic Antimicrobial Peptides Based on Gramicidin S. Pept. Sci. 2003, 71, 28–48. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Yan, Y.; Li, Y.; Zhang, Z.; Wang, X.; Niu, Y.; Zhang, S.; Xu, W.; Ren, C. Advances of Peptides for Antibacterial Applications. Colloids Surf. B Biointerfaces 2021, 202, 111682. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung Infections Associated with Cystic Fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Activity of Antimicrobial Peptides and Ciprofloxacin against Pseudomonas aeruginosa Biofilms. Molecules 2020, 25, 3843. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.K.; Yeung, A.T.Y.; Hancock, R.E.W. Antibiotic Resistance in Pseudomonas aeruginosa Biofilms: Towards the Development of Novel Anti-Biofilm Therapies. J. Biotechnol. 2014, 191, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Govan, J.R.W.; Deretic, V. Microbial Pathogenesis in Cystic Fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia Cepacia. Microbiol. Rev. 1996, 60, 539–574. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Hancock, R.E.W. Pseudomonas aeruginosa: All Roads Lead to Resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Pizzolato-Cezar, L.R.; Okuda-Shinagawa, N.M.; Teresa Machini, M. Combinatory Therapy Antimicrobial Peptide-Antibiotic to Minimize the Ongoing Rise of Resistance. Front. Microbiol. 2019, 10, 1703. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, Z.; Li, X.; Tian, Y.; Fan, Y.; Yu, C.; Zhou, B.; Liu, Y.; Xiang, R.; Yang, L. Synergistic Effects of Antimicrobial Peptide DP7 Combined with Antibiotics against Multidrug-Resistant Bacteria. Drug Des. Devel Ther. 2017, 11, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Pollini, S.; Brunetti, J.; Sennati, S.; Rossolini, G.M.; Bracci, L.; Pini, A.; Falciani, C. Synergistic Activity Profile of an Antimicrobial Peptide against Multidrug-Resistant and Extensively Drug-Resistant Strains of Gram-Negative Bacterial Pathogens. J. Pept. Sci. 2017, 23, 329–333. [Google Scholar] [CrossRef]
- Rudilla, H.; Fusté, E.; Cajal, Y.; Rabanal, F.; Vinuesa, T.; Viñas, M. Synergistic Antipseudomonal Effects of Synthetic Peptide AMP38 and Carbapenems. Molecules 2016, 21, 1223. [Google Scholar] [CrossRef]
- Conlon, J.M.; Al-Ghaferi, N.; Abraham, B.; Jiansheng, H.; Cosette, P.; Leprince, J.; Jouenne, T.; Vaudry, H. Antimicrobial Peptides from Diverse Families Isolated from the Skin of the Asian Frog, Rana Grahami. Peptides 2006, 27, 2111–2117. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Park, S.H.; Ahn, H.C.; Kim, S.; Kim, S.S.; Lee, B.J.; Lee, B.J. Structural Study of Novel Antimicrobial Peptides, Nigrocins, Isolated from Rana Nigromaculata. FEBS Lett. 2001, 507, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, X.; Xu, C.; Zhou, W.; Zhang, K.; Yu, H.; Zhang, Y.; Zheng, Y.; Rees, H.H.; Lai, R.; et al. Anti-Infection Peptidomics of Amphibian Skin. Mol. Cell. Proteom. 2007, 6, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Iwakoshi-Ukena, E.; Ukena, K.; Okimoto, A.; Soga, M.; Okada, G.; Sano, N.; Fujii, T.; Sugawara, Y.; Sumida, M. Identification and Characterization of Antimicrobial Peptides from the Skin of the Endangered Frog Odorrana Ishikawae. Peptides 2011, 32, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Bao, K.; Yuan, W.; Ma, C.; Yu, X.; Wang, L.; Hong, M.; Xi, X.; Zhou, M.; Chen, T. Modification Targeting the “Rana Box” Motif of a Novel Nigrocin Peptide from Hylarana Latouchii Enhances and Broadens Its Potency against Multiple Bacteria. Front. Microbiol. 2018, 9, 2846. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Liu, L.; Ma, C.; Di, L.; Chen, T. A Novel Antimicrobial Peptide Found in Pelophylax Nigromaculatus. J. Genet. Eng. Biotechnol. 2022, 20, 76. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lai, R. The Chemistry and Biological Activities of Peptides from Amphibian Skin Secretions. Chem. Rev. 2015, 115, 1760–1846. [Google Scholar] [CrossRef] [PubMed]
- Durante-Mangoni, E.; Grammatikos, A.; Utili, R.; Falagas, M.E. Do We Still Need the Aminoglycosides? Int. J. Antimicrob. Agents 2009, 33, 201–205. [Google Scholar] [CrossRef]
- Bian, W.; Meng, B.; Li, X.; Wang, S.; Cao, X.; Liu, N.; Yang, M.; Tang, J.; Wang, Y.; Yang, X. OA-GL21, a Novel Bioactive Peptide from Odorrana Andersonii, Accelerated the Healing of Skin Wounds. Biosci. Rep. 2018, 38, BSR20180215. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lee, W.H.; Zhang, Y. Extremely Abundant Antimicrobial Peptides Existed in the Skins of Nine Kinds of Chinese Odorous Frogs. J. Proteome Res. 2012, 11, 306–319. [Google Scholar] [CrossRef]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural Determinants of Host Defense Peptides for Antimicrobial Activity and Target Cell Selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Fu, H.; Ma, X. Design, Optimization, and Nanotechnology of Antimicrobial Peptides: From Exploration to Applications. Nano Today 2021, 39, 101229. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef] [PubMed]
- Onaizi, S.A.; Leong, S.S.J. Tethering Antimicrobial Peptides: Current Status and Potential Challenges. Biotechnol. Adv. 2011, 29, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chou, S.; Xu, L.; Zhu, X.; Dong, N.; Shan, A.; Chen, Z. High Specific Selectivity and Membrane-Active Mechanism of the Synthetic Centrosymmetric α-Helical Peptides with Gly-Gly Pairs. Sci. Rep. 2015, 5, 15963. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Hu, X.; Liao, M.; Fa, K.; Ciumac, D.; Clifton, L.A.; Sani, M.A.; King, S.M.; Maestro, A.; Separovic, F.; et al. Structural Disruptions of the Outer Membranes of Gram-Negative Bacteria by Rationally Designed Amphiphilic Antimicrobial Peptides. ACS Appl. Mater. Interfaces 2021, 13, 16062–16074. [Google Scholar] [CrossRef]
- Sun, L.; Wang, S.; Tian, F.; Zhu, H.; Dai, L. Organizations of Melittin Peptides after Spontaneous Penetration into Cell Membranes. Biophys. J. 2022, 121, 4368–4381. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Smith, D.H. Gentamicin: Adenine Mononucleotide Transferase: Partial Purification, Characterization, and Use in the Clinical Quantitation of Gentamicin. J. Infect. Dis. 1974, 129, 391–401. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, X.; Fu, Z.; Wang, S.; Li, X.; Liu, N.; Feng, Z.; Yang, M.; Tang, J.; Yang, X. Identification and Characterization of a Novel Gene-Encoded Antioxidant Peptide Obtained from Amphibian Skin Secretions. Nat. Prod. Res. 2020, 34, 754–758. [Google Scholar] [CrossRef]
- Huwyler, T.; Lenggenhager, L.; Abbas, M.; Ing Lorenzini, K.; Hughes, S.; Huttner, B.; Karmime, A.; Uçkay, I.; von Dach, E.; Lescuyer, P.; et al. Cefepime Plasma Concentrations and Clinical Toxicity: A Retrospective Cohort Study. Clin. Microbiol. Infect. 2017, 23, 454–459. [Google Scholar] [CrossRef]
- Yoshizawa, S.; Fourmy, D.; Puglisi, J.D. Structural Origins of Gentamicin Antibiotic Action. EMBO J. 1998, 17, 6437–6448. [Google Scholar] [CrossRef] [PubMed]
- Madurga, S.; Sánchez-Céspedes, J.; Belda, I.; Vila, J.; Giralt, E. Mechanism of Binding of Fluoroquinolones to the Quinolone Resistance-Determining Region of DNA Gyrase: Towards an Understanding of the Molecular Basis of Quinolone Resistance. ChemBioChem 2008, 9, 2081–2086. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the Regulation of Peptidoglycan Synthesis to Bacterial Growth and Morphology. Nat. Rev. Microbiol. 2012, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Savchenko, A. Molecular Mechanisms of Vancomycin Resistance. Protein Sci. 2020, 29, 654–669. [Google Scholar] [CrossRef] [PubMed]
- Dosler, S.; Karaaslan, E. Inhibition and Destruction of Pseudomonas aeruginosa Biofilms by Antibiotics and Antimicrobial Peptides. Peptides 2014, 62, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Speert, D.P. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Impact on Treatment. Drug Resist. Updates 2000, 3, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.F. Pseudomonas aeruginosa in Adults with Chronic Obstructive Pulmonary Disease. Curr. Opin. Pulm. Med. 2009, 15, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Magrini, N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; Volume 43. [Google Scholar]
- Page, M.G.; Heim, J. Prospects for the next Anti-Pseudomonas Drug. Curr. Opin. Pharmacol. 2009, 9, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Drenkard, E. Antimicrobial Resistance of Pseudomonas aeruginosa Biofilms. Microbes Infect. 2003, 5, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Burns, J.L.; Lory, S.; Lewis, K. Emergence of Pseudomonas aeruginosa Strains Producing High Levels of Persister Cells in Patients with Cystic Fibrosis. J. Bacteriol. 2010, 192, 6191–6199. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials 2018, 11, 2468. [Google Scholar] [CrossRef] [PubMed]
- Kurabayashi, A.; Sumida, M. PCR Primers for the Neobatrachian Mitochondrial Genome. Curr. Herpetol. 2009, 28, 1–11. [Google Scholar] [CrossRef]
- Yao, A.; Ma, Y.; Chen, X.; Zhou, M.; Xi, X.; Ma, C.; Ren, S.; Chen, T.; Shaw, C.; Wang, L. Modification Strategy of D-Leucine Residue Addition on a Novel Peptide from Odorrana Schmackeri, with Enhanced Bioactivity and in Vivo Efficacy. Toxins 2021, 13, 611. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wu, C.R.; Liu, W.; Zhang, J.W. Disulfide Bond Formation in Peptides by Dimethyl Sulfoxide. Scope and Applications. J. Am. Chem. Soc. 1991, 113, 6657–6662. [Google Scholar] [CrossRef]
- Chen, G.; Miao, Y.; Ma, C.; Zhou, M.; Shi, Z.; Chen, X.; Burrows, J.F.; Xi, X.; Chen, T.; Wang, L. Brevinin-2GHK from Sylvirana Guentheri and the Design of Truncated Analogs Exhibiting the Enhancement of Antimicrobial Activity. Antibiotics 2020, 9, 85. [Google Scholar] [CrossRef]


| Peptide | Sequence | Calculated Molecular Mass | Charge |
|---|---|---|---|
| Nigrosin-6VL | GLLSGVLGAGKKIVCGLSGRC | 1986.088 | +3 |
| 1885-C | GLLSGVLGAGKKIV-GLSGRC | 1885.099 | +3 |
| 1885-R | GLLSGVLGAGKKIVCGLSGR- | 1885.547 | +3 |
| 1924-A | GLLSGVLGAGKKIVAGLSGRA | 1924.037 | +3 |
| 1782-R | GLLSGVLGAGKKIV-GLSGR- | 1782.097 | +3 |
| 2057-C | GLLRGVLGAGKKIVCGLSGRC | 2057.665 | +4 |
| 2085-R | GLLRAVLAAGKKIVCGLSGRC | 2084.890 | +4 |
| 2170-2R | GLLRAVLRAGKKIVCGLSGRC | 2169.932 | +5 |
| Strains (Codes) | MICs/MBCs (µM) | |||||||
|---|---|---|---|---|---|---|---|---|
| Nigrosin-6VL | 1782-R | 1885-C | 1924-A | 1885-R | 2057-C | 2085-R | 2170-2R | |
| S. aureus (ATCC 6538) | 16/32 | >128/>128 | >128/>128 | 128/>128 | 128/>128 | 128/>128 | 16/16 | 4/8 |
| MRSA (NCTC 12493) | >128/>128 | >128/>128 | >128/>128 | >128/>128 | 64/64 | 64/128 | 16/32 | 16/16 |
| E. faecalis (NCTC 12697) | >128/>128 | >128/>128 | >128/>128 | >128/>128 | >128/>128 | >128/>128 | 64/256 | 128/128 |
| E. coli (ATCC 8739) | 64/128 | 128/>128 | 16/32 | 64/128 | 32/64 | 16/32 | 16/16 | 4/8 |
| K. pneumoniae (ATCC 43816) | 128/128 | >128/>128 | 64/128 | 64/128 | 32/64 | 64/128 | 32/64 | 32/32 |
| P. aeruginosa (ATCC CRM 9027) | 32/128 | >128/>128 | 64/128 | 128/>128 | 32/64 | 64/128 | 64/64 | 32/32 |
| A. baumannii (BAA 747) | >128/>128 | >128/>128 | >128/>128 | >128/>128 | 128/>128 | >128/>128 | 32/64 | 4/8 |
| C. albicans (ATCC 10231) | 128/>128 | >128/>128 | >128/>128 | 64/128 | >128/>128 | >128/>128 | 32/32 | 64/128 |
| Peptide | HC10 (μM) | GM (μM) | TI Value * | ||||
|---|---|---|---|---|---|---|---|
| G+ Bacteria | G− Bacteria | Yeast | G+ Bacteria | G− Bacteria | Yeast | ||
| Nigrosin-6VL | >256 | 101.59 | 90.51 | 128.00 | 5.04 | 5.66 | 4.00 |
| 1782-R | >256 | 256.00 | 215.27 | 256.00 | 2.00 | 2.38 | 2.00 |
| 1885-C | >256 | 256.00 | 64.00 | 256.00 | 2.00 | 8.00 | 2.00 |
| 1924-A | >256 | 203.19 | 107.63 | 64.00 | 2.52 | 4.76 | 8.00 |
| 1885-R | >256 | 128.00 | 45.25 | 256.00 | 4.00 | 11.31 | 2.00 |
| 2057-C | >256 | 128.00 | 64.00 | 256.00 | 4.00 | 8.00 | 2.00 |
| 2085-R | >256 | 25.40 | 32.00 | 32.00 | 20.16 | 16.00 | 16.00 |
| 2170-2R | >256 | 25.16 | 11.31 | 64.00 | 20.35 | 45.27 | 8.00 |
| Strains | MICs/MBCs (µM) | |||
|---|---|---|---|---|
| Levofloxacin | Cefepime | Gentamicin | Vancomycin | |
| S. aureus ATCC 6538 | 0.5/0.5 | 32/64 | 2/2 | 8/64 |
| MRSA NCTC 12493 | 0.25/0.5 | 16/32 | 1/2 | 4/8 |
| E. faecalis NCTC 12697 | 2/8 | 128/>128 | 16/32 | 32/>128 |
| E. coli ATCC 8739 | 0.25/0.5 | 8/16 | 2/4 | 32/>128 |
| K. pneumoniae ATCC 43816 | >128/>128 | 128/>128 | 1/1 | >128/>128 |
| P. aeruginosa ATCC CRM 9027 | 2/2 | 2/2 | 0.25/0.5 | >128/>128 |
| A. baumannii BAA 747 | 1/1 | 1/2 | >128/>128 | >128/>128 |
| Strains | FICI Values | |||||||
|---|---|---|---|---|---|---|---|---|
| Vancomycin | Cefepime | Levofloxacin | Gentamicin | |||||
| Nigrosin-6VL | 2170-2R | Nigrosin-6VL | 2170-2R | Nigrosin-6VL | 2170-2R | Nigrosin-6VL | 2170-2R | |
| S. aureus ATCC 6538 | 2.25 | 0.625 | 0.625 | 1 | 0.625 | 0.625 | 1.5 | 1 |
| MRSA NCTC 12493 | 1 | 0.625 | 0.75 | 0.75 | 2 | 1.125 | 0.5 | 0.375 |
| E. faecalis NCTC 12697 | 0.625 | 0.625 | 0.125 | 0.15625 | 0.625 | 0.625 | 0.625 | 0.625 |
| E. coli ATCC 8739 | 1.125 | >4 | 0.375 | 0.53125 | 0.375 | 0.375 | 0.125 | 0.125 |
| K. pneumoniae ATCC 43816 | 1.125 | 1.125 | 0.1875 | 0.1875 | 0.625 | 1 | 0.1875 | 0.15625 |
| P. aeruginosa ATCC CRM 9027 | >4 | 1.125 | 0.5 | 0.5 | 0.5 | 0.625 | 0.5 | 0.5 |
| A. baumannii BAA 747 | 1.25 | 0.625 | 1 | 1.5 | >4 | >4 | 0.1875 | 0.25 |
| Strains | MBICs/MBECs (µM) | |
|---|---|---|
| Nigrosin-6VL | 2170-2R | |
| S. aureus ATCC 6538 | 128/>128 | 4/16 |
| MRSA NCTC 12493 | >128/>128 | 4/16 |
| E. faecalis NCTC 12697 | >128/>128 | 32/>128 |
| K. pneumoniae ATCC 43816 | >128/>128 | 32/>128 |
| P. aeruginosa ATCC CRM 9027 | >128/>128 | >128/>128 |
| A. baumannii BAA 747 | >128/>128 | 8/16 |
| Strains | MBICs/MBECs (µM) | |
|---|---|---|
| Cefepime | Gentamicin | |
| S. aureus ATCC 6538 | 4/8 | 8/16 |
| MRSA NCTC 12493 | >128/>128 | 2/>128 |
| E. faecalis NCTC 12697 | >128/>128 | >128/>128 |
| K. pneumoniae ATCC 43816 | >128/>128 | 32/32 |
| P. aeruginosa ATCC CRM 9027 | >128/>128 | 2/4 |
| A. baumannii BAA 747 | 128/>128 | 8/16 |
| Strains | Biofilm FICI Values | |||
|---|---|---|---|---|
| Cefepime | Gentamicin | |||
| Nigrosin-6VL | 2170-2R | Nigrosin-6VL | 2170-2R | |
| S. aureus ATCC 6538 | 1 | 1 | 0.5 | 0.25 |
| MRSA NCTC 12493 | 0.75 | 0.5 | 0.375 | 1.125 |
| E. faecalis NCTC 12697 | 0.5 | 0.375 | 0.5 | 0.6125 |
| K. pneumoniae ATCC 43816 | 1.25 | 3 | 0.5 | 1.125 |
| P. aeruginosa ATCC CRM 9027 | 0.3125 | 0.5 | 0.5 | 1 |
| A. baumannii BAA 747 | 0.5 | 0.625 | 0.5 | 0.75 |
| Strains | MBICs/MBECs (µM) | |||
|---|---|---|---|---|
| Cefepime | Gentamicin | Nigrosin-6VL | 2170-2R | |
| P. aeruginosa ATCC CRM 9027 | >128/>128 | 2/4 | >128/>128 | >128/>128 |
| P. aeruginosa ATCC 27853 | 2/4 | 4/>128 | >128/>128 | 8/16 |
| P. aeruginosa ATCC BAA-2108 | 64/>128 | 16/128 | >128/>128 | >128/>128 |
| P. aeruginosa 01 | 4/>128 | 4/8 | >128/>128 | 32/64 |
| P. aeruginosa 14 | 8/>128 | 2/4 | >128/>128 | >128/>128 |
| Strains | Biofilm FICI Values | |||
|---|---|---|---|---|
| Cefepime | Gentamicin | |||
| Nigrosin-6VL | 2170-2R | Nigrosin-6VL | 2170-2R | |
| P. aeruginosa ATCC CRM 9027 | 0.3125 | 0.5 | 0.5 | 1 |
| P. aeruginosa ATCC 27853 | 1 | 0.5 | 0.25 | 0.25 |
| P. aeruginosa ATCC BAA-2108 | 0.625 | 1 | 1 | >3 |
| P. aeruginosa 01 | 0.625 | 0.5 | 0.625 | 1 |
| P. aeruginosa 14 | 0.25 | 0.375 | 0.5 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Shi, D.; Cheng, S.; Chen, X.; Ma, C.; Jiang, Y.; Wang, T.; Chen, T.; Shaw, C.; Wang, L.; et al. Modification and Synergistic Studies of a Novel Frog Antimicrobial Peptide against Pseudomonas aeruginosa Biofilms. Antibiotics 2024, 13, 574. https://doi.org/10.3390/antibiotics13070574
Liu X, Shi D, Cheng S, Chen X, Ma C, Jiang Y, Wang T, Chen T, Shaw C, Wang L, et al. Modification and Synergistic Studies of a Novel Frog Antimicrobial Peptide against Pseudomonas aeruginosa Biofilms. Antibiotics. 2024; 13(7):574. https://doi.org/10.3390/antibiotics13070574
Chicago/Turabian StyleLiu, Xinze, Daning Shi, Shiya Cheng, Xiaoling Chen, Chengbang Ma, Yangyang Jiang, Tao Wang, Tianbao Chen, Chris Shaw, Lei Wang, and et al. 2024. "Modification and Synergistic Studies of a Novel Frog Antimicrobial Peptide against Pseudomonas aeruginosa Biofilms" Antibiotics 13, no. 7: 574. https://doi.org/10.3390/antibiotics13070574
APA StyleLiu, X., Shi, D., Cheng, S., Chen, X., Ma, C., Jiang, Y., Wang, T., Chen, T., Shaw, C., Wang, L., & Zhou, M. (2024). Modification and Synergistic Studies of a Novel Frog Antimicrobial Peptide against Pseudomonas aeruginosa Biofilms. Antibiotics, 13(7), 574. https://doi.org/10.3390/antibiotics13070574







