Quorum Quenching Approaches against Bacterial-Biofilm-Induced Antibiotic Resistance
Abstract
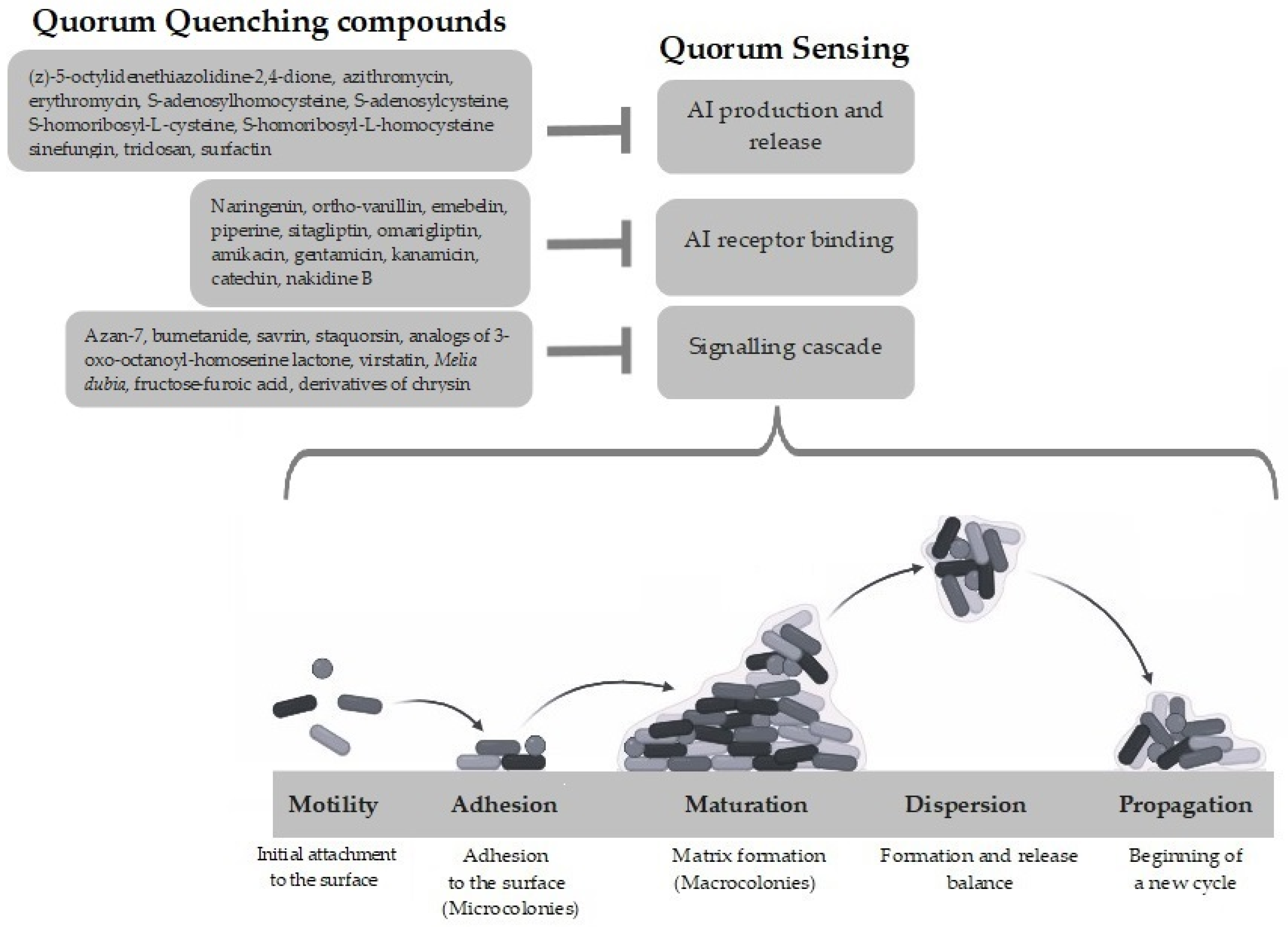
:1. Introduction
2. Biofilm and Antibiotic Resistance
3. Anti-QS Approaches to Overcome Biofilm Resistance
3.1. Inactivation of Signaling Molecules
3.2. Inactivation of Signaling Receptors
3.3. Inhibition of Signaling Cascade
4. Quorum Quenching Compounds
4.1. Plant-Derived Anti-Biofilm Compounds
4.2. Marine-Derived Anti-Biofilm Compounds
4.3. Microbial-Derived Anti-Biofilm Compounds
4.4. Antimicrobial Peptides
- (i)
- Inhibition of bacterial attachments to surfaces: AMPs can interfere in the early stages of biofilm formation to prevent the initial adhesion of bacteria to surfaces. Arslan and coll. reported that lactoferrin suppresses the initial attachment of Streptococcus gordonii coaggregates [187]. By downregulating genes encoding ABC transporters involved in cell-to-surface and cell-to-cell interactions, the peptide Nal-P-113 can inhibit Porphyromonas gingivalis biofilm formation [188].
- (ii)
- Disruption of the membrane potential of biofilm-embedded cells: Different kinds of bacteriocins, including nisin A and, to a lesser extent, lacticin Q, show antibiofilm activity on clinical isolates of methicillin-resistant S. aureus (MRSA), altering the membrane potential [189].
- (iii)
- (iv)
- Interruption of QS signaling and gene regulation: In P. aeruginosa, cathelicidic LL-37 and indolicidin prevent biofilm formation via the downregulation of the transcription of the Las and Rhl systems [192]. Additionally, peptide 1037 directly inhibits biofilms by reducing swimming and swarming motilities, stimulating twitching motility, and suppressing the expression of a variety of genes involved in biofilm formation [193]. The human β-defensin 3 (hBD-3) reduces the synthesis of polysaccharide intercellular adhesin (PIA) by downregulating the expression of the icaA, icaD, and icaR genes in S. epidermidis [194].
- (v)
- Degradation of the biofilm matrix: Peptide PI decrease S. mutans biofilm biomass by degrading the exopolysaccharide matrix [195]. In addition, an AMP complex produced by the maggots of blowfly Calliphora vicina effectively counteracts the formation of E. coli, S. aureus, and A. baumannii biofilms by destroying the biofilm matrix [196]. Similarly, the human antimicrobial peptide hepcidin 20 reduces the mass of the extracellular matrix and alters the architecture of the biofilm in S. epidermidis [197].
4.5. Synthetic Compounds
5. QQ Applications
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, W.J.; Bhatt, K.; Zhou, Z.; Huang, Y.; Zhang, L.H.; Chen, S.; Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Front. Plant Sci. 2023, 13, 1063393. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, C.R.; Parsek, M.R. New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 4317–4319. [Google Scholar] [CrossRef]
- McDougald, D.; Rice, S.A.; Barraud, N.; Steinberg, P.D.; Kjelleberg, S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2011, 10, 39–50. [Google Scholar] [CrossRef]
- Chen, L.; Wen, Y.M. The role of bacterial biofilm in persistent infections and control strategies. Int. J. Oral Sci. 2011, 3, 66–73. [Google Scholar] [CrossRef]
- Tseng, B.S.; Zhang, W.; Harrison, J.J.; Quach, T.P.; Song, J.L.; Penterman, J.; Singh, P.K.; Chopp, D.L.; Packman, A.I.; Parsek, M.R. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 2013, 15, 2865–2878. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, J.E.; Lam, H. Evolution of Bacterial Tolerance Under Antibiotic Treatment and Its Implications on the Development of Resistance. Front. Microbiol. 2021, 12, 617412. [Google Scholar] [CrossRef] [PubMed]
- Kosztołowicz, T.; Metzler, R. Diffusion of antibiotics through a biofilm in the presence of diffusion and absorption barriers. Phys. Rev. E 2020, 102, 032408. [Google Scholar] [CrossRef] [PubMed]
- Lewenza, S.; Johnson, L.; Charron-Mazenod, L.; Hong, M.; Mulcahy-O’Grady, H. Extracellular DNA controls expression of Pseudomonas aeruginosa genes involved in nutrient utilization, metal homeostasis, acid pH tolerance and virulence. J. Med. Microbiol. 2020, 69, 895–905. [Google Scholar] [CrossRef]
- André, A.C.; Laborde, M.; Marteyn, B.S. The battle for oxygen during bacterial and fungal infections. Trends Microbiol. 2022, 30, 643–653. [Google Scholar] [CrossRef]
- Lin, Q.; Pilewski, J.M.; Di, Y.P. Acidic Microenvironment Determines Antibiotic Susceptibility and Biofilm Formation of Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 747834. [Google Scholar] [CrossRef]
- Kouhsari, E.; Kaviar, V.H.; Asadi, A.; Ahmadi, A.; Sholeh, M.; Mirbalouchzehi, A.; Yaghoubi, S.; Abdi, M. Bacterial Persister Cells: Mechanisms of Formation, Control, and Eradication. Infect. Disord. Drug Targets 2023, 23, 17–28. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Zhou, Y.; Liao, H.; Pei, L.; Pu, Y. Combatting persister cells: The daunting task in post-antibiotics era. Cell Insight 2023, 2, 100104. [Google Scholar] [CrossRef]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef]
- He, X.; Lu, F.; Yuan, F.; Jiang, D.; Zhao, P.; Zhu, J.; Cheng, H.; Cao, J.; Lu, G. Biofilm Formation Caused by Clinical Acinetobacter baumannii Isolates Is Associated with Overexpression of the AdeFGH Efflux Pump. Antimicrob. Agents Chemother. 2015, 59, 4817–4825. [Google Scholar] [CrossRef] [PubMed]
- Buroni, S.; Matthijs, N.; Spadaro, F.; Van Acker, H.; Scoffone, V.C.; Pasca, M.R.; Riccardi, G.; Coenye, T. Differential roles of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob. Agents Chemother. 2014, 58, 7424–7429. [Google Scholar] [CrossRef]
- Liao, J.; Schurr, M.J.; Sauer, K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J. Bacteriol. 2013, 195, 3352–3363. [Google Scholar] [CrossRef] [PubMed]
- Hajiagha, M.N.; Kafil, H.S. Efflux pumps and microbial biofilm formation. Infect. Genet. Evol. 2023, 112, 105459. [Google Scholar] [CrossRef]
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef] [PubMed]
- Lidor, O.; Al-Quntar, A.; Pesci, E.C.; Steinberg, D. Mechanistic analysis of a synthetic inhibitor of the Pseudomonas aeruginosa LasI quorum-sensing signal synthase. Sci. Rep. 2015, 5, 16569. [Google Scholar] [CrossRef]
- Tateda, K.; Comte, R.; Pechere, J.C.; Köhler, T.; Yamaguchi, K.; Van Delden, C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2001, 45, 1930–1933. [Google Scholar] [CrossRef] [PubMed]
- Sofer, D.; Gilboa-Garber, N.; Belz, A.; Garber, N.C. ‘Subinhibitory’ erythromycin represses production of Pseudomonas aeruginosa lectins, autoinducer and virulence factors. Chemotherapy 1999, 45, 335–341. [Google Scholar] [CrossRef]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E., Jr.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365. [Google Scholar] [CrossRef]
- Kai, T.; Tateda, K.; Kimura, S.; Ishii, Y.; Ito, H.; Yoshida, H.; Kimura, T.; Yamaguchi, K. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 2009, 22, 483–486. [Google Scholar] [CrossRef]
- Hoang, T.T.; Schweizer, H.P. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): A target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 1999, 181, 5489–5497. [Google Scholar] [CrossRef]
- Yadav, M.K.; Park, S.W.; Chae, S.W.; Song, J.J. Sinefungin, a natural nucleoside analogue of S-adenosylmethionine, inhibits Streptococcus pneumoniae biofilm growth. BioMed Res. Int. 2014, 2014, 156987. [Google Scholar] [CrossRef]
- Han, X.; Lu, C. Biological activity and identification of a peptide inhibitor of LuxS from Streptococcus suis serotype 2. FEMS Microbiol. Lett. 2009, 294, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Zang, T.; Lee, B.W.; Cannon, L.M.; Ritter, K.A.; Dai, S.; Ren, D.; Wood, T.K.; Zhou, Z.S. A naturally occurring brominated furanone covalently modifies and inactivates LuxS. Bioorg. Med. Chem. Lett. 2009, 19, 6200–6204. [Google Scholar] [CrossRef] [PubMed]
- Benneche, T.; Hussain, Z.; Aamdal Scheieb, A.; Lönn-Stensrudb, J. Synthesis of 5-(bromomethylene) furan-2(5H)-ones and 3-(bromomethylene)isobenzofuran-1(3H)-ones as inhibitors of microbial quorum sensing. New J. Chem. 2008, 32, 1567–1572. [Google Scholar] [CrossRef]
- Shevate, S.N.; Shinde, S.S.; Bankar, A.V.; Patil, N.P. Identification of Quorum Quenching N-Acyl Homoserine Lactonases from Priestia aryabhattai J1D and Bacillus cereus G Isolated from the Rhizosphere. Curr. Microbiol. 2023, 80, 86. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, P.S.; Rai, V.R. Inhibition of QS-regulated virulence factors in Pseudomonas aeruginosa PAO1 and Pectobacterium carotovorum by AHL-lactonase of endophytic bacterium Bacillus cereus VT96. Biocatal. Agric. Biotechnol. 2016, 7, 154–163. [Google Scholar] [CrossRef]
- Carlier, A.; Uroz, S.; Smadja, B.; Fray, R.; Latour, X.; Dessaux, Y.; Faure, D. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-Acyl homoserine lactonase activity. Appl. Environ. Microbiol. 2003, 69, 4989–4993. [Google Scholar] [CrossRef]
- Dong, W.; Cai, Y.; Xu, Z.; Fu, B.; Chen, Q.; Cui, Y.; Ruan, Z.; Liang, Y.; Peng, N.; Zhao, S. Heterologous expression of AHL lactonase AiiK by Lactobacillus casei MCJΔ1 with great quorum quenching ability against Aeromonas hydrophila AH-1 and AH-4. Microb. Cell Fact. 2020, 19, 191. [Google Scholar] [CrossRef]
- Dong, W.; Zhu, J.; Guo, X.; Kong, D.; Zhang, Q.; Zhou, Y.; Liu, X.; Zhao, S.; Ruan, Z. Characterization of AiiK, an AHL lactonase, from Kurthia huakui LAM0618T and its application in quorum quenching on Pseudomonas aeruginosa PAO1. Sci. Rep. 2018, 8, 6013. [Google Scholar] [CrossRef]
- Fan, X.; Liang, M.; Wang, L.; Chen, R.; Li, H.; Liu, X. Aii810, a Novel Cold-Adapted N-Acylhomoserine Lactonase Discovered in a Metagenome, Can Strongly Attenuate Pseudomonas aeruginosa Virulence Factors and Biofilm Formation. Front. Microbiol. 2017, 8, 1950. [Google Scholar] [CrossRef]
- Schipper, C.; Hornung, C.; Bijtenhoorn, P.; Quitschau, M.; Grond, S.; Streit, W.R. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 224–233. [Google Scholar] [CrossRef]
- Krysciak, D.; Schmeisser, C.; Preuss, S.; Riethausen, J.; Quitschau, M.; Grond, S.; Streit, W.R. Involvement of multiple loci in quorum quenching of autoinducer I molecules in the nitrogen-fixing symbiont Rhizobium (Sinorhizobium) sp. strain NGR234. Appl. Environ. Microbiol. 2011, 77, 5089–5099. [Google Scholar] [CrossRef]
- Chow, J.Y.; Yang, Y.; Tay, S.B.; Chua, K.L.; Yew, W.S. Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob. Agents Chemother. 2014, 58, 1802–1805. [Google Scholar] [CrossRef]
- Chow, J.Y.; Wu, L.; Yew, W.S. Directed evolution of a quorum-quenching lactonase from Mycobacterium avium subsp. paratuberculosis K-10 in the amidohydrolase superfamily. Biochemistry 2009, 48, 4344–4353. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.H. MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Bzdrenga, J.; Daudé, D.; Rémy, B.; Jacquet, P.; Plener, L.; Elias, M.; Chabrière, E. Biotechnological applications of quorum quenching enzymes. Chem. Biol. Interact. 2017, 267, 104–115. [Google Scholar] [CrossRef]
- Chun, C.K.; Ozer, E.A.; Welsh, M.J.; Zabner, J.; Greenberg, E.P. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl. Acad. Sci. USA 2004, 101, 3587–3590. [Google Scholar] [CrossRef] [PubMed]
- Curcic, J.; Dinic, M.; Novovic, K.; Vasiljevic, Z.; Kojic, M.; Jovcic, B.; Malesevic, M. A novel thermostable YtnP lactonase from Stenotrophomonas maltophilia inhibits Pseudomonas aeruginosa virulence in vitro and in vivo. Int. J. Biol. Macromol. 2024, 264, 130421. [Google Scholar] [CrossRef]
- Chen, C.N.; Chen, C.J.; Liao, C.T.; Lee, C.Y. A probable aculeacin A acylase from the Ralstonia solanacearum GMI1000 is N-acyl-homoserine lactone acylase with quorum-quenching activity. BMC Microbiol. 2009, 9, 89. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, S.J.; Oh, T.K.; Oh, J.W.; Koo, B.T.; Yum, D.Y.; Lee, J.K. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 2003, 149, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kang, H.O.; Jang, H.S.; Lee, J.K.; Koo, B.T.; Yum, D.Y. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 2005, 71, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- Terwagne, M.; Mirabella, A.; Lemaire, J.; Deschamps, C.; De Bolle, X.; Letesson, J.J. Quorum sensing and self-quorum quenching in the intracellular pathogen Brucellamelitensis. PLoS ONE 2013, 8, e82514. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Xu, J.L.; Hu, J.; Wang, L.H.; Ong, S.L.; Leadbetter, J.R.; Zhang, L.H. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 2003, 47, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Czajkowski, R.; Krzyżanowska, D.; Karczewska, J.; Atkinson, S.; Przysowa, J.; Lojkowska, E.; Williams, P.; Jafra, S. Inactivation of AHLs by Ochrobactrum sp. A44 depends on the activity of a novel class of AHL acylase. Environ. Microbiol. Rep. 2011, 3, 59–68. [Google Scholar] [CrossRef]
- Valera, M.J.; Mas, A.; Streit, W.R.; Mateo, E. GqqA, a novel protein in Komagataeibacter europaeus involved in bacterial quorum quenching and cellulose formation. Microb. Cell Fact. 2016, 15, 88. [Google Scholar] [CrossRef]
- Werner, N.; Petersen, K.; Vollstedt, C.; Garcia, P.P.; Chow, J.; Ferrer, M.; Fernandez-Lopez, L.; Falke, S.; Perbandt, M.; Hinrichs, W.; et al. The Komagataeibacter europaeus GqqA is the prototype of a novel bifunctional N-Acyl-homoserine lactone acylase with prephenate dehydratase activity. Sci. Rep. 2021, 11, 12255. [Google Scholar] [CrossRef]
- Shepherd, R.W.; Lindow, S.E. Two dissimilar N-acyl-homoserine lactone acylases of Pseudomonas syringae influence colony and biofilm morphology. Appl. Environ. Microbiol. 2009, 75, 45–53. [Google Scholar] [CrossRef]
- Kusada, H.; Tamaki, H.; Kamagata, Y.; Hanada, S.; Kimura, N. A Novel Quorum-Quenching N-Acylhomoserine Lactone Acylase from Acidovorax sp. Strain MR-S7 Mediates Antibiotic Resistance. Appl. Environ. Microbiol. 2017, 83, e00080-17. [Google Scholar] [CrossRef]
- Sio, C.F.; Otten, L.G.; Cool, R.H.; Diggle, S.P.; Braun, P.G.; Bos, R.; Daykin, M.; Cámara, M.; Williams, P.; Quax, W.J. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect. Immun. 2006, 74, 1673–1682. [Google Scholar] [CrossRef]
- Utari, P.D.; Setroikromo, R.; Melgert, B.N.; Quax, W.J. PvdQ Quorum Quenching Acylase Attenuates Pseudomonas aeruginosa Virulence in a Mouse Model of Pulmonary Infection. Front. Cell Infect. Microbiol. 2018, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Bokhove, M.; Nadal Jimenez, P.; Quax, W.J.; Dijkstra, B.W. The quorum-quenching N-acyl homoserine lactone acylase PvdQ is an Ntn-hydrolase with an unusual substrate-binding pocket. Proc. Natl. Acad. Sci. USA 2010, 107, 686–691. [Google Scholar] [CrossRef]
- Huang, J.J.; Petersen, A.; Whiteley, M.; Leadbetter, J.R. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 2006, 72, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Philem, P.D.; Yadav, Y.; Vellore Sunder, A.; Ghosh, D.; Prabhune, A.; Ramasamy, S. Structural and enzymatic analysis of a dimeric cholylglycine hydrolase like acylase active on N-acyl homoserine lactones. Biochimie 2020, 177, 108–116. [Google Scholar] [CrossRef]
- Lord, D.M.; Baran, A.U.; Wood, T.K.; Peti, W.; Page, R. BdcA, a protein important for Escherichia coli biofilm dispersal, is a short-chain dehydrogenase/reductase that binds specifically to NADPH. PLoS ONE 2014, 9, e105751. [Google Scholar] [CrossRef] [PubMed]
- Bijtenhoorn, P.; Schipper, C.; Hornung, C.; Quitschau, M.; Grond, S.; Weiland, N.; Streit, W.R. BpiB05, a novel metagenome-derived hydrolase acting on N-acylhomoserine lactones. J. Biotechnol. 2011, 155, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Bijtenhoorn, P.; Mayerhofer, H.; Müller-Dieckmann, J.; Utpatel, C.; Schipper, C.; Hornung, C.; Szesny, M.; Grond, S.; Thürmer, A.; Brzuszkiewicz, E.; et al. A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS ONE 2011, 6, e26278. [Google Scholar] [CrossRef]
- Chowdhary, P.K.; Keshavan, N.; Nguyen, H.Q.; Peterson, J.A.; González, J.E.; Haines, D.C. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry 2007, 46, 14429–14437. [Google Scholar] [CrossRef]
- Uroz, S.; Chhabra, S.R.; Cámara, M.; Williams, P.; Oger, P.; Dessaux, Y. N-Acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology 2005, 151, 3313–3322. [Google Scholar] [CrossRef]
- Weiland-Bräuer, N.; Kisch, M.J.; Pinnow, N.; Liese, A.; Schmitz, R.A. Highly Effective Inhibition of Biofilm Formation by the First Metagenome-Derived AI-2 Quenching Enzyme. Front. Microbiol. 2016, 7, 1098. [Google Scholar] [CrossRef]
- Dong, Y.H.; Wang, L.H.; Xu, J.L.; Zhang, H.B.; Zhang, X.F.; Zhang, L.H. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 2001, 411, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.H.; Xu, J.L.; Li, X.Z.; Zhang, L.H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 2000, 97, 3526–3531. [Google Scholar] [CrossRef] [PubMed]
- Bergonzi, C.; Schwab, M.; Naik, T.; Daudé, D.; Chabrière, E.; Elias, M. Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties. Sci. Rep. 2018, 8, 11262. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Raju, S.C.; Purohit, H.J. Genomic analysis reveals versatile organisms for quorum quenching enzymes: Acyl-homoserine lactone-acylase and -lactonase. Open Microbiol. J. 2011, 5, 1–13. [Google Scholar] [CrossRef]
- Leadbetter, J.R.; Greenberg, E.P. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 2000, 182, 6921–6926. [Google Scholar] [CrossRef]
- Yu, M.; Liu, N.; Zhao, Y.; Zhang, X. Quorum quenching enzymes of marine bacteria and implication in aquaculture. J. Ocean Univ. 2018, 48, 34–43. [Google Scholar]
- Marques, J.C.; Lamosa, P.; Russell, C.; Ventura, R.; Maycock, C.; Semmelhack, M.F.; Miller, S.T.; Xavier, K.B. Processing the interspecies quorum-sensing signal autoinducer-2 (AI-2): Characterization of phospho-(S)-4,5-dihydroxy-2,3-pentanedione isomerization by LsrG protein. J. Biol. Chem. 2011, 286, 18331–18343. [Google Scholar] [CrossRef]
- Xavier, K.B.; Miller, S.T.; Lu, W.; Kim, J.H.; Rabinowitz, J.; Pelczer, I.; Semmelhack, M.F.; Bassler, B.L. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem. Biol. 2007, 2, 128–136. [Google Scholar] [CrossRef]
- Gatta, V.; Ilina, P.; Porter, A.; McElroy, S.; Tammela, P. Targeting Quorum Sensing: High-Throughput Screening to Identify Novel LsrK Inhibitors. Int. J. Mol. Sci. 2019, 20, 3112. [Google Scholar] [CrossRef]
- Stotani, S.; Gatta, V.; Medarametla, P.; Padmanaban, M.; Karawajczyk, A.; Giordanetto, F.; Tammela, P.; Laitinen, T.; Poso, A.; Tzalis, D.; et al. DPD-Inspired Discovery of Novel LsrK Kinase Inhibitors: An Opportunity To Fight Antimicrobial Resistance. J. Med. Chem. 2019, 62, 2720–2737. [Google Scholar] [CrossRef]
- Medarametla, P.; Gatta, V.; Kajander, T.; Laitinen, T.; Tammela, P.; Poso, A. Structure-Based Virtual Screening of LsrK Kinase Inhibitors to Target Quorum Sensing. Chem. Med. Chem. 2018, 13, 2400–2407. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, W.; Zhang, M.; Cui, Y.; Chen, X.; Ni, J.; Yu, L.; Shang, F.; Xue, T. Imidazole decreases the ampicillin resistance of an Escherichia coli strain isolated from a cow with mastitis by inhibiting the function of autoinducer 2. J. Dairy Sci. 2018, 101, 3356–3362. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Alcalde-Rico, M.; Gil-Gil, T.; Valverde, J.R.; Martínez, J.L. Naringenin Inhibition of the Pseudomonas aeruginosa Quorum Sensing Response Is Based on Its Time-Dependent Competition with N-(3-Oxo-dodecanoyl)-L-homoserine Lactone for LasR Binding. Front. Mol. Biosci. 2020, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Paczkowski, J.E.; Mukherjee, S.; McCready, A.R.; Cong, J.P.; Aquino, C.J.; Kim, H.; Henke, B.R.; Smith, C.D.; Bassler, B.L. Flavonoids Suppress Pseudomonas aeruginosa Virulence through Allosteric Inhibition of Quorum-sensing Receptors. J. Biol. Chem. 2017, 292, 4064–4076. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.E.; Akhter, S.; Rodriguez, B.; Townsend, K.A.; Smith, N.; Smith, B.; Wambua, A.; Craddock, V.; Abisado-Duque, R.G.; Santa, E.E.; et al. Characterization of natural product inhibitors of quorum sensing in Pseudomonas aeruginosa reveals competitive inhibition of RhlR by ortho-vanillin. bioRxiv 2024. [Google Scholar] [CrossRef]
- Khayat, M.T.; Abbas, H.A.; Ibrahim, T.S.; Khayyat, A.N.; Alharbi, M.; Darwish, K.M.; Elhady, S.S.; Khafagy, E.S.; Safo, M.K.; Hegazy, W.A.H. Anti-Quorum Sensing Activities of Gliptins against Pseudomonas aeruginosa and Staphylococcus aureus. Biomedicines 2022, 10, 1169. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A.; Shaldam, M.A.; Eldamasi, D. Curtailing Quorum Sensing in Pseudomonas aeruginosa by Sitagliptin. Curr. Microbiol. 2020, 77, 1051–1060. [Google Scholar] [CrossRef]
- Dwivedi, D.; Singh, V. Effects of the natural compounds embelin and piperine on the biofilm-producing property of Streptococcus mutans. J. Tradit. Complement. Med. 2015, 6, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Lee, J.W.; Javaid, A.; Park, S.K.; Kim, Y.M. Inhibition of biofilm and virulence properties of Pseudomonas aeruginosa by sub-inhibitory concentrations of aminoglycosides. Microb. Pathog. 2020, 146, 104249. [Google Scholar] [CrossRef]
- Manson, D.E.; Ananiev, G.E.; Guo, S.; Ericksen, S.S.; Santa, E.E.; Blackwell, H.E. Abiotic Small Molecule Inhibitors and Activators of the LasR Quorum Sensing Receptor in Pseudomonas aeruginosa with Potencies Comparable or Surpassing N-Acyl Homoserine Lactones. ACS Infect. Dis. 2024, 10, 1212–1221. [Google Scholar] [CrossRef]
- Reverchon, S.; Chantegrel, B.; Deshayes, C.; Doutheau, A.; Cotte-Pattat, N. New synthetic analogues of N-acyl homoserine lactones as agonists or antagonists of transcriptional regulators involved in bacterial quorum sensing. Bioorg. Med. Chem. Lett. 2002, 12, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.L.; Hanzelka, B.L.; Eberhard, A.; Greenberg, E.P. Quorum sensing in Vibrio fischeri: Probing autoinducer-LuxR interactions with autoinducer analogs. J. Bacteriol. 1996, 178, 2897–2901. [Google Scholar] [CrossRef] [PubMed]
- Soltane, R.; Alasiri, A.; Taha, M.N.; Abd El-Aleam, R.H.; Alghamdi, K.S.; Ghareeb, M.A.; Keshek, D.E.; Cardoso, S.M.; Sayed, A.M. Norlobaridone Inhibits Quorum Sensing-Dependent Biofilm Formation and Some Virulence Factors in Pseudomonas aeruginosa by Disrupting Its Transcriptional Activator Protein LasR Dimerization. Biomolecules 2023, 13, 1573. [Google Scholar] [CrossRef]
- Chaieb, K.; Kouidhi, B.; Hosawi, S.B.; Baothman, O.A.S.; Zamzami, M.A.; Altayeb, H.N. Computational screening of natural compounds as putative quorum sensing inhibitors targeting drug resistance bacteria: Molecular docking and molecular dynamics simulations. Comput. Biol. Med. 2022, 145, 105517. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Aguirre, L.; Velasco-Bucheli, R.; García-Álvarez, B.; Saborido, A.; Arroyo, M.; de la Mata, I. Novel Bifunctional Acylase from Actinoplanes utahensis: A Versatile Enzyme to Synthesize Antimicrobial Compounds and Use in Quorum Quenching Processes. Antibiotics 2021, 10, 922. [Google Scholar] [CrossRef] [PubMed]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin Influences Quorum Sensing in Food Borne Bacteria: In-Vitro and In-Silico Evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef] [PubMed]
- Geske, G.D.; O’Neill, J.C.; Miller, D.M.; Wezeman, R.J.; Mattmann, M.E.; Lin, Q.; Blackwell, H.E. Comparative analyses of N-acylated homoserine lactones reveal unique structural features that dictate their ability to activate or inhibit quorum sensing. ChemBioChem 2008, 9, 389–400. [Google Scholar] [CrossRef]
- Garner, A.L.; Park, J.; Zakhari, J.S.; Lowery, C.A.; Struss, A.K.; Sawada, D.; Kaufmann, G.F.; Janda, K.D. A multivalent probe for AI-2 quorum-sensing receptors. J. Am. Chem. Soc. 2011, 133, 15934–15937. [Google Scholar] [CrossRef]
- Peng, H.; Cheng, Y.; Ni, N.; Li, M.; Choudhary, G.; Chou, H.T.; Lu, C.D.; Tai, P.C.; Wang, B. Synthesis and evaluation of new antagonists of bacterial quorum sensing in Vibrio harveyi. Chem. Med. Chem. 2009, 4, 1457–1468. [Google Scholar] [CrossRef]
- Bernabè, G.; Dal Pra, M.; Ronca, V.; Pauletto, A.; Marzaro, G.; Saluzzo, F.; Stefani, A.; Artusi, I.; De Filippis, V.; Ferlin, M.G.; et al. A Novel Aza-Derivative Inhibits agr Quorum Sensing Signaling and Synergizes Methicillin-Resistant Staphylococcus aureus to Clindamycin. Front. Microbiol. 2021, 12, 610859. [Google Scholar] [CrossRef]
- Mahdally, N.H.; George, R.F.; Kashef, M.T.; Al-Ghobashy, M.; Murad, F.E.; Attia, A.S. Staquorsin: A Novel Staphylococcus aureus Agr-Mediated Quorum Sensing Inhibitor Impairing Virulence in vivo without Notable Resistance Development. Front. Microbiol. 2021, 12, 700494. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, B.; Solomon, A.P. Targeting AgrA quorum sensing regulator by bumetanide attenuates virulence in Staphylococcus aureus—A drug repurposing approach. Life Sci. 2021, 273, 119306. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Beaber, J.W.; Moré, M.I.; Fuqua, C.; Eberhard, A.; Winans, S.C. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 1998, 180, 5398–5405. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Choi, C.H. Role of LuxIR Homologue AnoIR in Acinetobacter nosocomialis and the Effect of Virstatin on the Expression of anoR Gene. J. Microbiol. Biotechnol. 2015, 25, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- El-Shaer, S.; Shaaban, M.; Barwa, R.; Hassan, R. Control of quorum sensing and virulence factors of Pseudomonas aeruginosa using phenylalanine arginyl β-naphthylamide. J. Med. Microbiol. 2016, 65, 1194–1204. [Google Scholar] [CrossRef]
- Escobar-Muciño, E.; Arenas-Hernández, M.M.P.; Luna-Guevara, M.L. Mechanisms of Inhibition of Quorum Sensing as an Alternative for the Control of E. coli and Salmonella. Microorganisms 2022, 10, 884. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, J.M.; Wang, B.; Peng, A.Y.; Mao, Z.W.; Xia, W. Inhibition of Quorum-Sensing Regulator from Pseudomonas aeruginosa Using a Flavone Derivative. Molecules 2022, 27, 2439. [Google Scholar] [CrossRef] [PubMed]
- Atchan, A.P.N.; Monthe, O.C.; Tchamgoue, A.D.; Singh, Y.; Shivashankara, S.T.; Selvi, M.K.; Agbor, G.A.; Magni, P.; Piazza, S.; Manjappara, U.V.; et al. Anti-Inflammatory, Antioxidant Activities, and Phytochemical Characterization of Edible Plants Exerting Synergistic Effects in Human Gastric Epithelial Cells. Antioxidants 2023, 12, 591. [Google Scholar] [CrossRef]
- Dincheva, I.; Badjakov, I.; Galunska, B. New Insights into the Research of Bioactive Compounds from Plant Origins with Nutraceutical and Pharmaceutical Potential. Plants 2023, 12, 258. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Sureda, A.; Tenore, G.C.; Daglia, M.; Sharifi-Rad, M.; Valussi, M.; Tundis, R.; Sharifi-Rad, M.; Loizzo, M.R.; Ademiluyi, A.O.; et al. Biological Activities of Essential Oils: From Plant Chemoecology to Traditional Healing Systems. Molecules 2017, 22, 70. [Google Scholar] [CrossRef]
- Aswathanarayan, J.B.; Vittal, R.R. Inhibition of biofilm formation and quorum sensing mediated phenotypes by berberine in Pseudomonas aeruginosa and Salmonella typhimurium. RSC Adv. 2018, 8, 36133–36141. [Google Scholar] [CrossRef]
- Qian, W.D.; Huang, J.; Zhang, J.N.; Li, X.C.; Kong, Y.; Wang, T.; Li, Y.D. Antimicrobial and Antibiofilm Activities and Mechanism of Action of Chelerythrine Against Carbapenem-Resistant Serratia marcescens In Vitro. Microb. Drug Resist. 2021, 27, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Tajani, A.S.; Amiri Tehranizadeh, Z.; Pourmohammad, A.; Pourmohammad, A.; Iranshahi, M.; Farhadi, F.; Soheili, V.; Fazly Bazzaz, B.S. Anti-quorum sensing and antibiofilm activity of coumarin derivatives against Pseudomonas aeruginosa PAO1: Insights from in vitro and in silico studies. Iran. J. Basic Med. Sci. 2023, 26, 445–452. [Google Scholar]
- He, Z.; Jiang, W.; Jiang, Y.; Dong, J.; Song, Z.; Xu, J.; Zhou, W. Anti-biofilm activities of coumarin as quorum sensing inhibitor for Porphyromonas gingivalis. J. Oral Microbiol. 2022, 14, 2055523. [Google Scholar] [CrossRef] [PubMed]
- Majik, M.S.; Naik, D.; Bhat, C.; Tilve, S.; Tilvi, S.; D’Souza, L. Synthesis of (R)-norbgugaine and its potential as quorum sensing inhibitor against Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2013, 23, 2353–2356. [Google Scholar] [CrossRef]
- Janeczko, M.; Masłyk, M.; Kubiński, K.; Golczyk, H. Emodin, a natural inhibitor of protein kinase CK2, suppresses growth, hyphal development, and biofilm formation of Candida albicans. Yeast 2017, 34, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Gu, S.; Shi, Y.; Cui, X.; Wen, S.; Ge, J. The effect of emodin on Staphylococcus aureus strains in planktonic form and biofilm formation in vitro. Arch. Microbiol. 2017, 199, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Jayaprakasha, G.K.; Uckoo, R.M.; Patil, B.S. Inhibition of Escherichia coli O157:H7 motility and biofilm by β-sitosterol glucoside. Biochim. Biophys. Acta 2013, 1830, 5219–5228. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Jesudhasan, P.R.; Jayaprakasha, G.K.; Pillai, S.D.; Patil, B.S. Citrus limonoids interfere with Vibrio harveyi cell-cell signalling and biofilm formation by modulating the response regulator LuxO. Microbiology 2011, 157, 99–110. [Google Scholar] [CrossRef]
- Quave, C.L.; Estévez-Carmona, M.; Compadre, C.M.; Hobby, G.; Hendrickson, H.; Beenken, K.E.; Smeltzer, M.S. Ellagic acid derivatives from Rubus ulmifolius inhibit Staphylococcus aureus biofilm formation and improve response to antibiotics. PLoS ONE 2012, 7, e28737. [Google Scholar] [CrossRef]
- Konozy, E.H.E.; Osman, M.E.M.; Dirar, A.I.; Ghartey-Kwansah, G. Plant lectins: A new antimicrobial frontier. Biomed. Pharmacother. 2022, 155, 113735. [Google Scholar] [CrossRef] [PubMed]
- Procópio, T.F.; de Siqueira Patriota, L.L.; de Moura, M.C.; da Silva, P.M.; de Oliveira, A.P.; do Nascimento Carvalho, L.V.; de Albuquerque Lima, T.; Soares, T.; da Silva, T.D.; Breitenbach Barroso Coelho, L.C.; et al. CasuL: A new lectin isolated from Calliandra surinamensis leaf pinnulae with cytotoxicity to cancer cells, antimicrobial activity and antibiofilm effect. Int. J. Biol. Macromol. 2017, 98, 419–429. [Google Scholar] [CrossRef]
- Jin, X.; Lee, Y.J.; Hong, S.H. Canavalia ensiformis-derived lectin inhibits biofilm formation of enterohemorrhagic Escherichia coli and Listeria monocytogenes. J. Appl. Microbiol. 2019, 126, 300–310. [Google Scholar] [CrossRef]
- Cavalcante, T.T.; Anderson Matias da Rocha, B.; Alves Carneiro, V.; Vassiliepe Sousa Arruda, F.; Fernandes do Nascimento, A.S.; Cardoso Sá, N.; do Nascimento, K.S.; Sousa Cavada, B.; Holanda Teixeira, E. Effect of lectins from Diocleinae subtribe against oral Streptococci. Molecules 2011, 16, 3530–3543. [Google Scholar] [CrossRef]
- Silva, P.M.; Napoleão, T.H.; Silva, L.C.; Fortes, D.T.; Lima, T.A.; Zingali, R.B.; Pontual, E.V.; Araújo, J.M.; Medeiros, P.L.; Rodrigues, C.G. The juicy sarcotesta of Punica granatum contains a lectin that affects growth, survival as well as adherence and invasive capacities of human pathogenic bacteria. J. Funct. Foods 2016, 27, 695–702. [Google Scholar] [CrossRef]
- Emad, F.; Khalafalah, A.K.; El Sayed, M.A.; Mohamed, A.H.; Stadler, M.; Helaly, S.E. Three new polyacetylene glycosides (PAGs) from the aerial part of Launaea capitata (Asteraceae) with anti-biofilm activity against Staphylococcus aureus. Fitoterapia 2020, 143, 104548. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, V.A.; Santos, H.S.; Arruda, F.V.; Bandeira, P.N.; Albuquerque, M.R.; Pereira, M.O.; Henriques, M.; Cavada, B.S.; Teixeira, E.H. Casbane diterpene as a promising natural antimicrobial agent against biofilm-associated infections. Molecules 2010, 16, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Murata, R.M.; Branco-de-Almeida, L.S.; Franco, E.M.; Yatsuda, R.; dos Santos, M.H.; de Alencar, S.M.; Koo, H.; Rosalen, P.L. Inhibition of Streptococcus mutans biofilm accumulation and development of dental caries in vivo by 7-epiclusianone and fluoride. Biofouling 2010, 26, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Raorane, C.J.; Lee, J.H.; Kim, Y.G.; Rajasekharan, S.K.; García-Contreras, R.; Lee, J. Antibiofilm and Antivirulence Efficacies of Flavonoids and Curcumin Against Acinetobacter baumannii. Front. Microbiol. 2019, 10, 990. [Google Scholar] [CrossRef]
- Luo, J.; Dong, B.; Wang, K.; Cai, S.; Liu, T.; Cheng, X.; Lei, D.; Chen, Y.; Li, Y.; Kong, J.; et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE 2017, 12, e0176883. [Google Scholar] [CrossRef]
- Das, M.C.; Sandhu, P.; Gupta, P.; Rudrapaul, P.; De, U.C.; Tribedi, P.; Akhter, Y.; Bhattacharjee, S. Attenuation of Pseudomonas aeruginosa biofilm formation by Vitexin: A combinatorial study with azithromycin and gentamicin. Sci. Rep. 2016, 6, 23347. [Google Scholar] [CrossRef] [PubMed]
- Rayan, M.; Abu Lafi, S.; Falah, M.; Kacergius, T.; Kirkliauskiene, A.; Gabe, V.; Rayan, A. Alkyl Gallates as Potential Antibiofilm Agents: A Review. Molecules 2023, 28, 1751. [Google Scholar] [CrossRef] [PubMed]
- Rudin, L.; Bornstein, M.M.; Shyp, V. Inhibition of biofilm formation and virulence factors of cariogenic oral pathogen Streptococcus mutans by natural flavonoid phloretin. J. Oral Microbiol. 2023, 15, 2230711. [Google Scholar] [CrossRef]
- Tapia-Rodriguez, M.R.; Bernal-Mercado, A.T.; Gutierrez-Pacheco, M.M.; Vazquez-Armenta, F.J.; Hernandez-Mendoza, A.; Gonzalez-Aguilar, G.A.; Martinez-Tellez, M.A.; Nazzaro, F.; Ayala-Zavala, J.F. Virulence of Pseudomonas aeruginosa exposed to carvacrol: Alterations of the Quorum sensing at enzymatic and gene levels. J. Cell Commun. Signal. 2019, 13, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Liu, C.C.; Li, Y.T.; Li, M.Q.; Fu, Y.T.; Li, X.C.; Qian, W.D. Antifungal and Antibiofilm In Vitro Activities of Ursolic Acid on Cryptococcus neoformans. Curr. Microbiol. 2022, 79, 293. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Sangwan, P.L.; Koul, S.; Pandey, A.; Bani, S.; Abdullah, S.T.; Sharma, P.R.; Kitchlu, S.; Khan, I.A. 4-epi-Pimaric acid: A phytomolecule as a potent antibacterial and anti-biofilm agent for oral cavity pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Ušjak, L.; Stojković, D.; Carević, T.; Milutinović, V.; Soković, M.; Niketić, M.; Petrović, S. Chemical Analysis and Investigation of Antimicrobial and Antibiofilm Activities of Prangos trifida (Apiaceae). Antibiotics 2024, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- D’Aquila, P.; Sena, G.; Crudo, M.; Passarino, G.; Bellizzi, D. Effect of Essential Oils of Apiaceae, Lamiaceae, Lauraceae, Myrtaceae, and Rutaceae Family Plants on Growth, Biofilm Formation, and Quorum Sensing in Chromobacterium violaceum, Pseudomonas aeruginosa, and Enterococcus faecalis. Microorganisms 2023, 11, 1150. [Google Scholar] [CrossRef]
- Alibi, S.; Ben Selma, W.; Ramos-Vivas, J.; Smach, M.A.; Touati, R.; Boukadida, J.; Navas, J.; Ben Mansour, H. Anti-oxidant, antibacterial, anti-biofilm, and anti-quorum sensing activities of four essential oils against multidrug-resistant bacterial clinical isolates. Curr. Res. Transl. Med. 2020, 68, 59–66. [Google Scholar] [CrossRef]
- Condò, C.; Anacarso, I.; Sabia, C.; Iseppi, R.; Anfelli, I.; Forti, L.; de Niederhäusern, S.; Bondi, M.; Messi, P. Antimicrobial activity of spices essential oils and its effectiveness on mature biofilms of human pathogens. Nat. Prod. Res. 2020, 34, 567–574. [Google Scholar] [CrossRef]
- Sharifi, A.; Mohammadzadeh, A.; Zahraei Salehi, T.; Mahmoodi, P. Antibacterial, antibiofilm and antiquorum sensing effects of Thymus daenensis and Satureja hortensis essential oils against Staphylococcus aureus isolates. J. Appl. Microbiol. 2018, 124, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Porfírio, E.M.; Melo, H.M.; Pereira, A.M.G.; Cavalcante, T.T.A.; Gomes, G.A.; de Carvalho, M.G.; Costa, R.A.; Júnior, F.E.A.C. In Vitro Antibacterial and Antibiofilm Activity of Lippia alba Essential Oil, Citral, and Carvone against Staphylococcus aureus. Sci. World J. 2017, 2017, 4962707. [Google Scholar] [CrossRef]
- Vieira, M.; Bessa, L.J.; Martins, M.R.; Arantes, S.; Teixeira, A.P.S.; Mendes, Â.; Martins da Costa, P.; Belo, A.D.F. Chemical Composition, Antibacterial, Antibiofilm and Synergistic Properties of Essential Oils from Eucalyptus globulus Labill. and Seven Mediterranean Aromatic Plants. Chem. Biodivers. 2017, 14, e1700006. [Google Scholar] [CrossRef] [PubMed]
- Lyu, X.; Lee, J.; Chen, W.N. Potential Natural Food Preservatives and Their Sustainable Production in Yeast: Terpenoids and Polyphenols. J. Agric. Food Chem. 2019, 67, 4397–4417. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Sun, Z.; Chen, W. Antimicrobial susceptibility and antibacterial mechanism of limonene against Listeria monocytogenes. Molecules 2020, 25, 33. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cai, J.; Chen, H.; Zhong, Q.; Hou, Y.; Chen, W.; Chen, W. Antibacterial activity and mechanism of linalool against Pseudomonas aeruginosa. Microb. Pathog. 2020, 141, 103980. [Google Scholar] [CrossRef]
- D’Aquila, P.; Paparazzo, E.; Crudo, M.; Bonacci, S.; Procopio, A.; Passarino, G.; Bellizzi, D. Antibacterial Activity and Epigenetic Remodeling of Essential Oils from Calabrian Aromatic Plants. Nutrients 2022, 14, 391. [Google Scholar] [CrossRef] [PubMed]
- Behzadnia, A.; Moosavi-Nasab, M.; Oliyaei, N. Anti-biofilm activity of marine algae-derived bioactive compounds. Front. Microbiol. 2024, 15, 1270174. [Google Scholar] [CrossRef]
- Al-Tmimi, S.L.; Al-Rrubaai, G.; Zaki, N.H. Antibiofilm activity of intracellular extracts of Westiellopsis prolifica isolated from local environment in Baghdad. JGPT 2018, 10, 281–288. [Google Scholar]
- El-Sapagh, S.; El-Shenody, R.; Pereira, L.; Elshobary, M. Unveiling the Potential of Algal Extracts as Promising Antibacterial and Antibiofilm Agents against Multidrug-Resistant Pseudomonas aeruginosa: In Vitro and In Silico Studies including Molecular Docking. Plants 2023, 12, 3324. [Google Scholar] [CrossRef]
- Nithya, C.; LewisOscar, F.; Kanaga, S.; Kavitha, R.; Bakkiyaraj, D.; Arunkumar, M.; Alharbi, N.S.; Chinnathambi, A.; Ali Alharbi, S.; Thajuddin, N. Biofilm inhibitory potential of Chlamydomonas sp. extract against Pseudomonas aeruginosa. J. Algal Biomass Utln. 2014, 5, 74–81. [Google Scholar]
- El Zawawy, N.A.; El-Shenody, R.A.; Ali, S.S.; El-Shetehy, M. A novel study on the inhibitory effect of marine macroalgal extracts on hyphal growth and biofilm formation of candidemia isolates. Sci. Rep. 2020, 10, 9339. [Google Scholar] [CrossRef]
- Cepas, V.; Gutiérrez-Del-Río, I.; López, Y.; Redondo-Blanco, S.; Gabasa, Y.; Iglesias, M.J.; Soengas, R.; Fernández-Lorenzo, A.; López-Ibáñez, S.; Villar, C.J.; et al. Microalgae and Cyanobacteria Strains as Producers of Lipids with Antibacterial and Antibiofilm Activity. Mar. Drugs 2021, 19, 675. [Google Scholar] [CrossRef]
- Besednova, N.N.; Andryukov, B.G.; Zaporozhets, T.S.; Kryzhanovsky, S.P.; Kuznetsova, T.A.; Fedyanina, L.N.; Makarenkova, I.D.; Zvyagintseva, T.N. Algae Polyphenolic Compounds and Modern Antibacterial Strategies: Current Achievements and Immediate Prospects. Biomedicines 2020, 8, 342. [Google Scholar] [CrossRef]
- Prasath, K.G.; Tharani, H.; Kumar, M.S.; Pandian, S.K. Palmitic Acid Inhibits the Virulence Factors of Candida tropicalis: Biofilms, Cell Surface Hydrophobicity, Ergosterol Biosynthesis, and Enzymatic Activity. Front. Microbiol. 2020, 11, 864. [Google Scholar] [CrossRef]
- Boutin, R.; Munnier, E.; Renaudeau, N.; Girardot, M.; Pinault, M.; Chevalier, S.; Chourpa, I.; Clément-Larosière, B.; Imbertet, C.; Boudesocque-Delaye, L. Spirulina platensis sustainable lipid extracts in alginate-based nanocarriers: An algal approach against biofilms. Algal Res. 2019, 37, 160–168. [Google Scholar] [CrossRef]
- Sampathkumar, S.J.; Srivastava, P.; Ramachandran, S.; Sivashanmugam, K.; Gothandam, K.M. Lutein: A potential antibiofilm and antiquorum sensing molecule from green microalga Chlorella pyrenoidosa. Microb. Pathog. 2019, 135, 103658. [Google Scholar] [CrossRef]
- Vishwakarma, J.; Vavilala, S.L. Evaluating the antibacterial and antibiofilm potential of sulphated polysaccharides extracted from green algae Chlamydomonas reinhardtii. J. Appl. Microbiol. 2019, 127, 1004–1017. [Google Scholar] [CrossRef]
- Jun, J.Y.; Jung, M.J.; Jeong, I.H.; Yamazaki, K.; Kawai, Y.; Kim, B.M. Antimicrobial and Antibiofilm Activities of Sulfated Polysaccharides from Marine Algae against Dental Plaque Bacteria. Mar. Drugs 2018, 16, 301. [Google Scholar] [CrossRef]
- Powell, L.C.; Pritchard, M.F.; Ferguson, E.L.; Powell, K.A.; Patel, S.U.; Rye, P.D.; Sakellakou, S.M.; Buurma, N.J.; Brilliant, C.D.; Copping, J.M.; et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. npj Biofilms Microbiomes 2018, 4, 13. [Google Scholar] [CrossRef]
- Gökalsın, B.; Aksoydan, B.; Erman, B.; Sesal, N.C. Reducing Virulence and Biofilm of Pseudomonas aeruginosa by Potential Quorum Sensing Inhibitor Carotenoid: Zeaxanthin. Microb. Ecol. 2017, 74, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Melander, R.J.; Basak, A.K.; Melander, C. Natural products as inspiration for the development of bacterial antibiofilm agents. Nat. Prod. Rep. 2020, 37, 1454–1477. [Google Scholar] [CrossRef]
- You, J.; Xue, X.; Cao, L.; Lu, X.; Wang, J.; Zhang, L.; Zhou, S. Inhibition of Vibrio biofilm formation by a marine actinomycete strain A66. Appl. Microbiol. Biotechnol. 2007, 76, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Oloke, J.K. Activity pattern of natural and synthetic antibacterial agents among hospital isolates. Microbios 2000, 102, 175–181. [Google Scholar] [PubMed]
- Eguchi, Y.; Kubo, N.; Matsunaga, H.; Igarashi, M.; Utsumi, R. Development of an antivirulence drug against Streptococcus mutans: Repression of biofilm formation, acid tolerance, and competence by a histidine kinase inhibitor, walkmycin C. Antimicrob. Agents Chemother. 2011, 55, 1475–1484. [Google Scholar] [CrossRef]
- Vasilchenko, A.S.; Julian, W.T.; Lapchinskaya, O.A.; Katrukha, G.S.; Sadykova, V.S.; Rogozhin, E.A. A Novel Peptide Antibiotic Produced by Streptomyces roseoflavus Strain INA-Ac-5812 With Directed Activity Against Gram-Positive Bacteria. Front. Microbiol. 2020, 11, 556063. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Pereira, F.; Grilo, I.R.; Godinho, C.C.; Paulino, M.; Almeida, V.; Gobbo-Neto, L.; Prieto-Davó, A.; Sobral, R.G.; Lopes, N.P.; et al. Intra-clade metabolomic profiling of MAR4 Streptomyces from the Macaronesia Atlantic region reveals a source of anti-biofilm metabolites. Environ. Microbiol. 2019, 21, 1099–1112. [Google Scholar] [CrossRef]
- Driche, E.H.; Sabaou, N.; Bijani, C.; Zitouni, A.; Pont, F.; Mathieu, F.; Badji, B. Streptomyces sp. AT37 isolated from a Saharan soil produces a furanone derivative active against multidrug-resistant Staphylococcus aureus. World J. Microbiol. Biotechnol. 2017, 33, 105. [Google Scholar] [CrossRef] [PubMed]
- Oja, T.; San Martin Galindo, P.; Taguchi, T.; Manner, S.; Vuorela, P.M.; Ichinose, K.; Metsä-Ketelä, M.; Fallarero, A. Effective Antibiofilm Polyketides against Staphylococcus aureus from the Pyranonaphthoquinone Biosynthetic Pathways of Streptomyces Species. Antimicrob. Agents Chemother. 2015, 59, 6046–6052. [Google Scholar] [CrossRef]
- Suzuki, N.; Ohtaguro, N.; Yoshida, Y.; Hirai, M.; Matsuo, H.; Yamada, Y.; Imamura, N.; Tsuchiya, T. A Compound Inhibits Biofilm Formation of Staphylococcus aureus from Streptomyces. Biol. Pharm. Bull. 2015, 38, 889–892. [Google Scholar] [CrossRef]
- Guillén-Navarro, D.; González-Vázquez, R.; León-Ávila, G.; Giono-Cerezo, S. Quorum Quenching with a Diffusible Signal Factor Analog in Stenotrophomonas maltophilia. Pathogens 2023, 12, 1448. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, L.H.; Cámara, M.; He, Y.W. The DSF Family of Quorum Sensing Signals: Diversity, Biosynthesis, and Turnover. Trends Microbiol. 2017, 25, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.N.; Davies, D.G.; Sauer, K. Control of Biofilms with the Fatty Acid Signaling Molecule cis-2-Decenoic Acid. Pharmaceuticals 2015, 8, 816–835. [Google Scholar] [CrossRef] [PubMed]
- Apel, C.; Barg, A.; Rheinberg, A.; Conrads, G.; Wagner-Döbler, I. Dental composite materials containing carolacton inhibit biofilm growth of Streptococcus mutans. Dent. Mater. 2013, 29, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Abouelhassan, Y.; Garrison, A.T.; Yang, H.; Chávez-Riveros, A.; Burch, G.M.; Huigens, R.W., 3rd. Recent Progress in Natural-Product-Inspired Programs Aimed to Address Antibiotic Resistance and Tolerance. J. Med. Chem. 2019, 62, 7618–7642. [Google Scholar] [CrossRef] [PubMed]
- Gayathiri, E.; Prakash, P.; Karmegam, N.; Varjani, S.; Awasthi, M.K.; Ravindran, B. Biosurfactants: Potential and eco-friendly material for sustainable agriculture and environmental safety—A review. Agronomy 2022, 12, 662. [Google Scholar] [CrossRef]
- Paraszkiewicz, K.; Moryl, M.; Płaza, G.; Bhagat, D.K.; Satpute, S.; Bernat, P. Surfactants of microbial origin as antibiofilm agents. Int. J. Environ. Health Res. 2021, 31, 401–420. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, M.M.; Al-Omar, M.S.; Mohammed, H.A.; Emam, T.M. In Vitro and Ex Vivo Antibiofilm Activity of a Lipopeptide Biosurfactant Produced by the Entomopathogenic Beauveria bassiana Strain against Microsporum canis. Microorganisms 2020, 8, 232. [Google Scholar] [CrossRef]
- Abdollahi, S.; Tofighi, Z.; Babaee, T.; Shamsi, M.; Rahimzadeh, G.; Rezvanifar, H.; Saeidi, E.; Mohajeri Amiri, M.; Saffari Ashtiani, Y.; Samadi, N. Evaluation of Anti-oxidant and Anti-biofilm Activities of Biogenic Surfactants Derived from Bacillus amyloliquefaciens and Pseudomonas aeruginosa. Iran. J. Pharm. Res. 2020, 19, 115–126. [Google Scholar]
- Shakerifard, P.; Gancel, F.; Jacques, P.; Faille, C. Effect of different Bacillus subtilis lipopeptides on surface hydrophobicity and adhesion of Bacillus cereus 98/4 spores to stainless steel and Teflon. Biofouling 2009, 25, 533–541. [Google Scholar] [CrossRef]
- Pamp, S.J.; Tolker-Nielsen, T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 2531–2539. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta 2016, 1858, 1044–1060. [Google Scholar] [CrossRef] [PubMed]
- Pletzer, D.; Coleman, S.R.; Hancock, R.E. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr. Opin. Microbiol. 2016, 33, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Di Somma, A.; Moretta, A.; Canè, C.; Cirillo, A.; Duilio, A. Antimicrobial and Antibiofilm Peptides. Biomolecules 2020, 10, 652. [Google Scholar] [CrossRef]
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials 2018, 11, 2468. [Google Scholar] [CrossRef] [PubMed]
- Arslan, S.Y.; Leung, K.P.; Wu, C.D. The effect of lactoferrin on oral bacterial attachment. Oral Microbiol. Immunol. 2009, 24, 411–416. [Google Scholar] [CrossRef]
- Wang, H.Y.; Lin, L.; Tan, L.S.; Yu, H.Y.; Cheng, J.W.; Pan, Y.P. Molecular pathways underlying inhibitory effect of antimicrobial peptide Nal-P-113 on bacteria biofilms formation of Porphyromonas gingivalis W83 by DNA microarray. BMC Microbiol. 2017, 17, 37. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin(1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell Mol. Life Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef]
- Nagant, C.; Pitts, B.; Stewart, P.S.; Feng, Y.; Savage, P.B.; Dehaye, J.P. Study of the effect of antimicrobial peptide mimic, CSA-13, on an established biofilm formed by Pseudomonas aeruginosa. Microbiologyopen 2013, 2, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.S.; Jenssen, H. Human cathelicidin LL-37 prevents bacterial biofilm formation. Future Med. Chem. 2012, 4, 1587–1599. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef]
- Zhu, C.; Tan, H.; Cheng, T.; Shen, H.; Shao, J.; Guo, Y.; Shi, S.; Zhang, X. Human β-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation. J. Surg. Res. 2013, 183, 204–213. [Google Scholar] [CrossRef]
- Ansari, J.M.; Abraham, N.M.; Massaro, J.; Murphy, K.; Smith-Carpenter, J.; Fikrig, E. Anti-Biofilm Activity of a Self-Aggregating Peptide against Streptococcus mutans. Front. Microbiol. 2017, 8, 488. [Google Scholar] [CrossRef]
- Gordya, N.; Yakovlev, A.; Kruglikova, A.; Tulin, D.; Potolitsina, E.; Suborova, T.; Bordo, D.; Rosano, C.; Chernysh, S. Natural antimicrobial peptide complexes in the fighting of antibiotic resistant biofilms: Calliphora vicina medicinal maggots. PLoS ONE 2017, 12, e0173559. [Google Scholar] [CrossRef]
- Brancatisano, F.L.; Maisetta, G.; Di Luca, M.; Esin, S.; Bottai, D.; Bizzarri, R.; Campa, M.; Batoni, G. Inhibitory effect of the human liver-derived antimicrobial peptide hepcidin 20 on biofilms of polysaccharide intercellular adhesin (PIA)-positive and PIA-negative strains of Staphylococcus epidermidis. Biofouling 2014, 30, 435–446. [Google Scholar] [CrossRef]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Fighting biofilms with lantibiotics and other groups of bacteriocins. NPJ Biofilms Microbiomes 2018, 4, 9. [Google Scholar] [CrossRef]
- Chopra, L.; Singh, G.; Choudhary, V.; Sahoo, D.K. Sonorensin: An antimicrobial peptide, belonging to the heterocycloanthracin subfamily of bacteriocins, from a new marine isolate, Bacillus sonorensis MT93. Appl. Environ. Microbiol. 2014, 80, 2981–2990. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Xin, W.G.; Zhang, Q.L.; Lin, L.B.; Deng, X.Y. A Novel Bacteriocin against Shigella flexneri from Lactiplantibacillus plantarum Isolated from Tilapia Intestine: Purification, Antibacterial Properties and Antibiofilm Activity. Front. Microbiol. 2022, 12, 779315. [Google Scholar] [CrossRef]
- Yoon, J.W.; Kang, S.S. In Vitro Antibiofilm and Anti-Inflammatory Properties of Bacteriocins Produced by Pediococcus acidilactici Against Enterococcus faecalis. Foodborne Pathog. Dis. 2020, 17, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Ray Mohapatra, A.; Jeevaratnam, K. Inhibiting bacterial colonization on catheters: Antibacterial and antibiofilm activities of bacteriocins from Lactobacillus plantarum SJ33. J. Glob. Antimicrob. Resist. 2019, 19, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Vashistha, A.; Sharma, N.; Nanaji, Y.; Kumar, D.; Singh, G.; Barnwal, R.P.; Yadav, A.K. Quorum sensing inhibitors as Thera-peutics: Bacterial biofilm inhibition. Bioorg. Chem. 2023, 136, 106551. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Hu, H.; Yu, Y.; Chen, C.; Zhang, J.; Jarnda, K.V.; Ding, P. Therapeutic strategies for drug-resistant Pseudomonas aeruginosa: Metal and metal oxide nanoparticles. J. Biomed. Mater. Res. A 2024. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, A.; D’Aquila, P.; Palermo, G.; Dell’Aglio, M.; Passarino, G.; Strangi, G.; Bellizzi, D. Role of the Human Serum Albumin Protein Corona in the Antimicrobial and Photothermal Activity of Metallic Nanoparticles against Escherichia coli Bacteria. ACS Omega 2023, 8, 31333–31343. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Nazam, N.; Rizvi, S.M.D.; Ahmad, K.; Baig, M.H.; Lee, E.J.; Choi, I. Mechanistic Insights into the Antimicrobial Actions of Metallic Nanoparticles and Their Implications for Multidrug Resistance. Int. J. Mol. Sci. 2019, 20, 2468. [Google Scholar] [CrossRef] [PubMed]
- Niemirowicz, K.; Swiecicka, I.; Wilczewska, A.Z.; Misztalewska, I.; Kalska-Szostko, B.; Bienias, K.; Bucki, R.; Car, H. Gold-functionalized magnetic nanoparticles restrict growth of Pseudomonas aeruginosa. Int. J. Nanomed. 2014, 9, 2217–2224. [Google Scholar]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef]
- Robino, L.; Scavone, P. Nanotechnology in biofilm prevention. Future Microbiol. 2020, 15, 377–379. [Google Scholar] [CrossRef]
- Qayyum, S.; Khan, A.U. Nanoparticles: Vs. biofilms: A battle against another paradigm of antibiotic resistance. MedChemComm 2016, 7, 1479–1498. [Google Scholar] [CrossRef]
- Slomberg, D.L.; Lu, Y.; Broadnax, A.D.; Hunter, R.A.; Carpenter, A.W.; Schoenfisch, M.H. Role of size and shape on biofilm eradication for nitric oxide-releasing silica nanoparticles. ACS Appl. Mater. Interfaces 2013, 5, 9322–9329. [Google Scholar] [CrossRef] [PubMed]
- Lellouche, J.; Friedman, A.; Lellouche, J.P.; Gedanken, A.; Banin, E. Improved antibacterial and antibiofilm activity of magnesium fluoride nanoparticles obtained by water-based ultrasound chemistry. Nanomedicine 2012, 8, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, X.; Shen, Y.; Li, H.; Zou, Y.; Yuan, G.; Hu, P.; Hu, H. Mucus penetration enhanced lipid polymer nanoparticles improve the eradication rate of Helicobacter pylori biofilm. J. Control. Release 2019, 300, 52–63. [Google Scholar] [CrossRef]
- Li, X.; Yeh, Y.C.; Giri, K.; Mout, R.; Landis, R.F.; Prakash, Y.S.; Rotello, V.M. Control of nanoparticle penetration into biofilms through surface design. Chem. Commun. 2015, 51, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Fulaz, S.; Vitale, S.; Quinn, L.; Casey, E. Nanoparticle-Biofilm Interactions: The Role of the EPS Matrix. Trends Microbiol. 2019, 27, 915–926. [Google Scholar] [CrossRef]
- Omran, B.A.; Tseng, B.S.; Baek, K.H. Nanocomposites against Pseudomonas aeruginosa biofilms: Recent advances, challenges, and future prospects. Microbiol. Res. 2024, 282, 127656. [Google Scholar] [CrossRef]
- Kapoor, D.U.; Patel, R.J.; Gaur, M.; Parikh, S.; Prajapati, B.G. Metallic and metal oxide nanoparticles in treating Pseudomonas aeruginosa infections. J. Drug Deliv. Sci. Technol. 2024, 91, 105290. [Google Scholar] [CrossRef]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-Strategies to Fight Multidrug Resistant Bacteria—“A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef]
- El-Deeb, N.M.; Abo-Eleneen, M.A.; Al-Madboly, L.A.; Sharaf, M.M.; Othman, S.S.; Ibrahim, O.M.; Mubarak, M.S. Biogenically Synthesized Polysaccharides-Capped Silver Nanoparticles: Immunomodulatory and Antibacterial Potentialities Against Resistant Pseudomonas aeruginosa. Front. Bioeng. Biotechnol. 2020, 8, 643. [Google Scholar] [CrossRef]
- LewisOscar, F.; Nithya, C.; Vismaya, S.; Arunkumar, M.; Pugazhendhi, A.; Nguyen-Tri, P.; Alharbi, S.A.; Alharbi, N.S.; Thajuddin, N. In vitro analysis of green fabricated silver nanoparticles (AgNPs) against Pseudomonas aeruginosa PA14 biofilm formation, their application on urinary catheter. Prog. Org. Coat. 2021, 151, 106058. [Google Scholar] [CrossRef]
- Hu, D.; Li, H.; Wang, B.; Ye, Z.; Lei, W.; Jia, F.; Jin, Q.; Ren, K.F.; Ji, J. Surface-Adaptive Gold Nanoparticles with Effective Adherence and Enhanced Photothermal Ablation of Methicillin-Resistant Staphylococcus aureus Biofilm. ACS Nano 2017, 11, 9330–9339. [Google Scholar] [CrossRef] [PubMed]
- Rajkumari, J.; Busi, S.; Vasu, A.C.; Reddy, P. Facile green synthesis of baicalein fabricated gold nanoparticles and their antibiofilm activity against Pseudomonas aeruginosa PAO1. Microb. Pathog. 2017, 107, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.G.; Cho, M.H.; Lee, J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol. Res. 2014, 169, 888–896. [Google Scholar] [CrossRef]
- Javanbakht, T.; Laurent, S.; Stanicki, D.; Wilkinson, K.J. Relating the Surface Properties of Superparamagnetic Iron Oxide Nanoparticles (SPIONs) to Their Bactericidal Effect towards a Biofilm of Streptococcus mutans. PLoS ONE 2016, 11, e0154445. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, W.; Wu, X.; Zhao, Y.; Dai, H. The antibacterial and antibiofilm activities of mesoporous hollow Fe3O4 nanoparticles in an alternating magnetic field. Biomater. Sci. 2020, 8, 4492–4507. [Google Scholar] [CrossRef]
- Subbiahdoss, G.; Sharifi, S.; Grijpma, D.W.; Laurent, S.; van der Mei, H.C.; Mahmoudi, M.; Busscher, H.J. Magnetic targeting of surface-modified superparamagnetic iron oxide nanoparticles yields antibacterial efficacy against biofilms of gentamicin-resistant staphylococci. Acta Biomater. 2012, 8, 2047–2055. [Google Scholar] [CrossRef] [PubMed]
- Auger, S.; Henry, C.; Péchoux, C.; Suman, S.; Lejal, N.; Bertho, N.; Larcher, T.; Stankic, S.; Vidic, J. Exploring multiple effects of Zn0.15Mg0.85O nanoparticles on Bacillus subtilis and macrophages. Sci. Rep. 2018, 8, 12276. [Google Scholar] [CrossRef]
- Eshed, M.; Lellouche, J.; Gedanken, A.; Banin, E. A Zn-doped CuO nanocomposite shows enhanced antibiofilm and antibacterial activities against Streptococcus mutans compared to nanosized CuO. Adv. Funct. Mater. 2014, 24, 1382–1390. [Google Scholar] [CrossRef]
- Ho, K.K.; Cole, N.; Chen, R.; Willcox, M.D.; Rice, S.A.; Kumar, N. Immobilization of antibacterial dihydropyrrol-2-ones on functional polymer supports to prevent bacterial infections in vivo. Antimicrob. Agents Chemother. 2012, 56, 1138–1141. [Google Scholar] [CrossRef]
- Roncari Rocha, G.; Sims, K.R., Jr.; Xiao, B.; Klein, M.I.; Benoit, D.S.W. Nanoparticle carrier co-delivery of complementary antibiofilm drugs abrogates dual species cariogenic biofilm formation in vitro. J. Oral Microbiol. 2021, 14, 1997230. [Google Scholar] [CrossRef]
- Vinoj, G.; Pati, R.; Sonawane, A.; Vaseeharan, B. In vitro cytotoxic effects of gold nanoparticles coated with functional acyl homoserine lactone lactonase protein from Bacillus licheniformis and their antibiofilm activity against Proteus species. Antimicrob. Agents Chemother. 2015, 59, 763–771. [Google Scholar] [CrossRef]
- Lee, H.W.; Kharel, S.; Loo, S.C.J. Lipid-Coated Hybrid Nanoparticles for Enhanced Bacterial Biofilm Penetration and Antibiofilm Efficacy. ACS Omega 2022, 7, 35814–35824. [Google Scholar] [CrossRef]
- Maurizi, L.; Forte, J.; Ammendolia, M.G.; Hanieh, P.N.; Conte, A.L.; Relucenti, M.; Donfrancesco, O.; Ricci, C.; Rinaldi, F.; Marianecci, C.; et al. Effect of Ciprofloxacin-Loaded Niosomes on Escherichia coli and Staphylococcus aureus Biofilm Formation. Pharmaceutics 2022, 14, 2662. [Google Scholar] [CrossRef]
- Lambadi, P.R.; Sharma, T.K.; Kumar, P.; Vasnani, P.; Thalluri, S.M.; Bisht, N.; Pathania, R.; Navani, N.K. Facile biofunctionalization of silver nanoparticles for enhanced antibacterial properties, endotoxin removal, and biofilm control. Int. J. Nanomed. 2015, 10, 2155–2171. [Google Scholar]
- Ho, D.K.; Murgia, X.; De Rossi, C.; Christmann, R.; Hüfner de Mello Martins, A.G.; Koch, M.; Andreas, A.; Herrmann, J.; Müller, R.; Empting, M.; et al. Squalenyl Hydrogen Sulfate Nanoparticles for Simultaneous Delivery of Tobramycin and an Alkylquinolone Quorum Sensing Inhibitor Enable the Eradication of P. aeruginosa Biofilm Infections. Angew. Chem. Int. Ed. Engl. 2020, 59, 10292–10296. [Google Scholar] [CrossRef]
- Ivanova, K.; Ivanova, A.; Hoyo, J.; Pérez-Rafael, S.; Tzanov, T. Nano-Formulation Endows Quorum Quenching Enzyme-Antibiotic Hybrids with Improved Antibacterial and Antibiofilm Activities against Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022, 23, 7632. [Google Scholar] [CrossRef]
- Charkowski, A.O. The Changing Face of Bacterial Soft-Rot Diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef]
- Rodríguez, M.; Torres, M.; Blanco, L.; Béjar, V.; Sampedro, I.; Llamas, I. Plant growth-promoting activity and quorum quenching-mediated biocontrol of bacterial phytopathogens by Pseudomonas segetis strain P6. Sci. Rep. 2020, 10, 4121. [Google Scholar] [CrossRef]
- Dong, Y.H.; Zhang, X.F.; Xu, J.L.; Zhang, L.H. Insecticidal Bacillus thuringiensis silences Erwinia carotovora virulence by a new form of microbial antagonism, signal interference. Appl. Environ. Microbiol. 2004, 70, 954–960. [Google Scholar] [CrossRef]
- Zhang, W.; Fan, X.; Li, J.; Ye, T.; Mishra, S.; Zhang, L.; Chen, S. Exploration of the Quorum-Quenching Mechanism in Pseudomonas nitroreducens W-7 and Its Potential to Attenuate the Virulence of Dickeya zeae EC1. Front. Microbiol. 2021, 12, 694161. [Google Scholar] [CrossRef]
- Yu, H.; Chen, W.J.; Bhatt, K.; Zhou, Z.; Zhu, X.; Liu, S.; He, J.; Zhang, L.H.; Chen, S.; Wang, H.; et al. A novel bacterial strain Burkholderia sp. F25 capable of degrading diffusible signal factor signal shows strong biocontrol potential. Front. Plant Sci. 2022, 13, 1071693. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Zhang, W.; Feng, Z.; Fan, X.; Xu, X.; Mishra, S.; Zhang, L.; Chen, S. Characterization of a Novel Quorum-Quenching Bacterial Strain, Burkholderia anthina HN-8, and Its Biocontrol Potential against Black Rot Disease Caused by Xanthomonas campestris pv. campestris. Microorganisms 2020, 8, 1485. [Google Scholar] [CrossRef]
- Ye, T.; Zhou, T.; Fan, X.; Bhatt, P.; Zhang, L.; Chen, S. Acinetobacter lactucae Strain QL-1, a Novel Quorum Quenching Candidate Against Bacterial Pathogen Xanthomonas campestris pv. campestris. Front. Microbiol. 2019, 10, 2867. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo-Picard, C.; Chapelle, E.; Ratet, P.; Faure, D.; Dessaux, Y. Transgenic plants expressing the quorum quenching lactonase AttM do not significantly alter root-associated bacterial populations. Res. Microbiol. 2011, 162, 951–958. [Google Scholar] [CrossRef]
- Chu, W.; Zhou, S.; Zhu, W.; Zhuang, X. Quorum quenching bacteria Bacillus sp. QSI-1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Sci. Rep. 2014, 4, 5446. [Google Scholar] [CrossRef] [PubMed]
- Vinoj, G.; Vaseeharan, B.; Thomas, S.; Spiers, A.J.; Shanthi, S. Quorum-quenching activity of the AHL-lactonase from Bacillus licheniformis DAHB1 inhibits Vibrio biofilm formation in vitro and reduces shrimp intestinal colonisation and mortality. Mar. Biotechnol. 2014, 16, 707–715. [Google Scholar] [CrossRef]
- Monzón-Atienza, L.; Bravo, J.; Torrecillas, S.; Gómez-Mercader, A.; Montero, D.; Ramos-Vivas, J.; Galindo-Villegas, J.; Acosta, F. An In-Depth Study on the Inhibition of Quorum Sensing by Bacillus velezensis D-18: Its Significant Impact on Vibrio Biofilm Formation in Aquaculture. Microorganisms 2024, 12, 890. [Google Scholar] [CrossRef]
- Chen, B.; Peng, M.; Tong, W.; Zhang, Q.; Song, Z. The Quorum Quenching Bacterium Bacillus licheniformis T-1 Protects Zebrafish against Aeromonas hydrophila Infection. Probiotics Antimicrob. Proteins 2020, 12, 160–171. [Google Scholar] [CrossRef]
- Ghanei-Motlagh, R.; Mohammadian, T.; Gharibi, D.; Menanteau-Ledouble, S.; Mahmoudi, E.; Khosravi, M.; Zarea, M.; El-Matbouli, M. Quorum Quenching Properties and Probiotic Potentials of Intestinal Associated Bacteria in Asian Sea Bass Lates calcarifer. Mar. Drugs 2019, 18, 23. [Google Scholar] [CrossRef]
- Zhou, S.; Yu, Z.; Chu, W. Effect of quorum-quenching bacterium Bacillus sp. QSI-1 on protein profiles and extracellular enzymatic activities of Aeromonas hydrophila YJ-1. BMC Microbiol. 2019, 19, 135. [Google Scholar] [CrossRef]
- Maddela, N.R.; Abiodun, A.S.; Zhang, S.; Prasad, R. Biofouling in Membrane Bioreactors-Mitigation and Current Status: A Review. Appl. Biochem. Biotechnol. 2023, 195, 5643–5668. [Google Scholar] [CrossRef] [PubMed]
- Ergön-Can, T.; Köse-Mutlu, B.; Koyuncu, I.; Lee, C.H. Biofouling control based on bacterial quorum quenching with a new application: Rotary microbial carrier frame. J. Membr. Sci. 2017, 525, 116–124. [Google Scholar] [CrossRef]
- Lade, H.; Paul, D.; Kweon, J.H. Quorum quenching mediated approaches for control of membrane biofouling. Int. J. Biol. Sci. 2014, 10, 550–565. [Google Scholar] [CrossRef] [PubMed]
| Enzyme Category | Name | Source | Substrates | Effect | Reference |
|---|---|---|---|---|---|
| Lactonases | AiiA | Bacillus sp. Priestia aryabhattai | C4-, C6-, C8-, C10-HSL; 3OC4-, 3OC6-, 3OC8-, 3OC12-HSL; 3-OH-C4-HSL | Inhibition of biofilm and production of pyocyanin, rhamnolipid, and exopolysaccarides in P. aeruginosa, Vibrio cholerae, and S. aureus. | [36,37] |
| AiiB | Agrobacterium tumefaciens | C4-, C6-, C7-, C8-, C10-HSL; 3OC6-, 3OC8-HSL | Reduction in the bacterial virulence in Erwinia carotovora. | [38] | |
| AiiK | Lactobacillus casei, Kurthia huakui | C10-HSL | Attenuation of swimming motility, virulence factor production, and biofilm formation in Aeromonas hydrophila and P. aeruginosa | [39,40] | |
| Aii810 | Bacteria from Mao-tofu | 3OC12-HSL | Inhibition of virulence and biofilm in P. aeruginosa. | [41] | |
| bpiB01, B04, B07 | Bacteria from soil samples | 3OC8-HSL; 3OC6-, 3OC8-, 3OC12-HSL | Biofilm inhibition in P. aeruginosa. | [42] | |
| DlhR, QsdR1 | Rhizobium sp. | 3OC8-HSL | Inhibition of biofilm and other QS-dependent processes in P. aeruginosa, Chromobacterium violaceum, and A. tumefaciens. | [43] | |
| ND | Geobacillus kaustophilus | 3-OH-C10-HSL, 3-OH-C12-HSL | Distruption of biofilm on multiple strains of A. baumannii. | [44] | |
| MCP | Mycobacterium avium | C6-, C7-, C8-, C10-, C12-HSL | The enzyme is a metal-dependent N-acylhomoserine lactonase | [45] | |
| MomL | Muricauda olearia | C6-HSL | Attenuation of the virulence of P. aeruginosa. | [46] | |
| SsoPox-1 | Sulfolobus solfataricus | C4-, C6-, C8-, C12-HSL; 3OC6-, 3OC8-, 3OC10-, 3OC12-HSL | Attenuation of QS signaling, virulence factor production, and biofilm formation in vitro. | [47] | |
| PON1-3 | Homo sapiens | C7-, C12-, C14-HSL; 3OC6-, 3OC10-, 3OC12-HSL | Inhibition of biofilm formation and extracellular virulence factors in P. aeruginosa. | [48] | |
| YtnP | Stenotrophomonas maltophilia | 3O-C12-HSL | Inhibition of biofilm formation, induction of biofilm decomposition, and reducetion of extracellular virulence factors production. | [49] | |
| Acylases | Aac | Ralstonia solanacearum | C7-, C8-, C10-HSL; 3OC8-HSL | Inhibition of QS mechanism in C. violaceum. | [50] |
| AhlD | Arthrobacter sp. | OHHL, OHL and OdDH | Reduction in the AHL amount and pectate lyase activity in Erwinia carotovora. | [51] | |
| AhlM | Streptomyces sp. | C6-, C8-, C10-HSL; 3OC6-, 3OC8-, 3OC12-HSL | Decrease of the production of virulence factors, including elastase, total protease, and LasA, in P. aeruginosa. | [52] | |
| aibP | Brucella melitensis | 3OC8-, 3OC12-HSL | Decrease of endogenous AHL accumulation within Brucella melitensis. | [53] | |
| AiiD | Ralstonia sp. | OC6-, OC8-, OC10-, OC12-HSL | QQ activity in P. aeruginosa achieved through an Acyl-homoserine lactone acylase hydrolyses the AHL amide, releasing homoserine lactone and the corresponding fatty acid. | [54] | |
| AiiO | Ochrobactrum sp. A44 | 3OC4-14-HSL | Inhibition of the virulence in Pectobacterium carotovorum. | [55] | |
| GqqA | Komagataeibacter europaeus | C8-, C10-, C12-HS | Inhibiton of biofilm formation. | [56,57] | |
| HacA, B | Pseudomonas syringae | C6-, C8-, C10-, C12-HSL; OC8-, OC10-, OC12-, OC14-HSL | Inhibition of biofilm formation, and colony morphology. | [58] | |
| MacQ | Acidovorax sp. | C6-, C8-, C10-, C12-HSL; 3OC8-HSL; OC6-, OC8-, OC10-, OC12-, OC14-HSL | Interference with the QS system. | [59] | |
| PA2385 | P. aeruginosa | 3COC12-HSL | QQ activity achieved through an N-acyl-homoserine lactone acylase | [60] | |
| PvdQ | P. aeruginosa | C8-, C10-, C12-HSL; OC12-HSL | Attenuation of P. aeruginosa virulence. | [61,62] | |
| QuiP | P. aeruginosa | C8-, C10-, C12-HSL; OC12-HSL | Gene expression regulation of QS systems in P. aeruginosa. | [63] | |
| Slac1, 2 | Shewanella loihica-PV4 | 3OC10-HSL | Exhibition of in vitro QQ activity. | [64] | |
| Oxidoreductases | BdcA | E. coli | 3OC6-HSL | Dehydrogenase/reductase activity. | [65] |
| BpiB05 | Bacteria from soil sample | 3-oxo-C8-HSL | Exhibition of in vitro QQ activity. | [66] | |
| BpiB09 | Bacteria from soil sample | 3-oxo-C8-HSL | Reduction in pyocyanin production, decreased motility, poor biofilm formation in P. aeruginosa. | [67] | |
| CYP102A1 | Bacillus megaterium | C12-20-HSL (ω-1, ω-2, ω-3 hydroxylated) | In vitro oxidation of AHLs and their lactonolysis products acyl homoserines. | [68] | |
| ND | Rhodococcus erythropolis | 3-oxo-AHL, N-(3-oxo-6-phenylhexanoyl) homoserine lactone, 3-oxododecanamide, n-(3-oxododecanoyl)-L-homoserine lactone | Exhibition of AHL oxidoreductase and amidolytic activities. | [69] | |
| QQ-2 | Bacteria from water sample | 3-oxo-C6-HSL; 3,4,4-trihydroxy-2-pentanone-5-phosphate | Reduction of AHL and AI-2 in QS-inactive hydroxy-derivatives and inhibits biofilm formation in Klebsiella oxytoca and Klebsiella pneumoniae. | [70] |
| Phytochemical Class | Compounds | Natural Source | Molecular Effect | Reference |
|---|---|---|---|---|
| Alkaloids | Berberine | Coptis, Hydrastis and Berberis genus | Downregulation of QS-related genes. | [111] |
| Chelerythrine | Chelidonium majus L. | Disruption of membrane integrity, inhibition of biofilm components. | [112] | |
| Furocoumarins | Grapefruits | Inhibition of QS signalling; Biofilm dispersion. | [113,114] | |
| Norbgugaine | Arisarum vulgare | Inhibition of cell motility and of adhesion. | [115] | |
| Anthraquinones | Emodin | Rheum palmatum L. | Suppression growth, hyphal development, and biofilm of Candida albicans by targeting cellular kinase signalling. | [116] |
| Polygonum cuspidatum, Rheum palmatum | Downregulation of biofilm-forming related genes in S. aureus. | [117] | ||
| Glycosides | β-sitosterol glucoside; Isolimonic acid; Ichangin | Citrus species | Inhibition of the biofilm and motility through the repression of flagellar master operon flhDC Interferece with cell–cell signalling and biofilm formation by the modulation of luxO expression. | [118,119] |
| Ellagic acid | Rubus ulmifolius | Inhibition of the biofilm and increase antibiotic susceptibility. | [120] | |
| Lectins | AGL | Amaranthus gangeticus | Inhibition of biofilm formation. | [121] |
| CasuL | Calliandra surinamensi | Inhibition of biofilm formation. | [122] | |
| ConA | Canavalia ensiformis | Reduction in bacterial adhesion. | [123] | |
| ConBol | Canavalia boliviana | Reduction in bacterial adhesion. | [124] | |
| ConM | Canavalia maritima | Reduction in bacterial adhesion. | [124] | |
| PgTeL | Punica granatum | Reduction in bacterial adhesion. | [125] | |
| Polyacetylenes | Glicosides (PAGs) | Launaea capitata | [126] | |
| Polyphenols | Casbane diterpene | Croton nepetaefolius | Inhibition of biofilm and reduction of bacterial adhesion. | [127] |
| 7-epiclusianone | Rheedia brasiliensis | Inhibition of biofilm and reduction of bacterial adhesion. | [128] | |
| Flavonoids (baicalin, catechin, curcumin, kaempherol, luteolin, proaminophyllin, quercetin, vitexin) | Scutellaria baicalensis and Vitex species | Decrease in the QS signaling molecules 3-oxo-C12-HSL and C4-HSL; Attenuation of LasA, LasB, and LuxR. | [129,130,131] | |
| Methyl-gallate | Suppression of the synthesis of the extracellular polymeric matrix, inhibition of QS signaling, and alteration of the microbial cell membrane. | [132] | ||
| Phloretin | Apple, pear and strawberry fruits | Downregulation of virulence genes essential for surface attachment, biofilm formation, and QS. | [133] | |
| Terpenoids | Carvacrol | Organum vulgare | Reduction in QS signalling. | [134] |
| Ursolic acid | Many medical plants | Inhibition of biofilm formation and disruption of cell membrane. | [135] | |
| 4-epi-primaric acid | Aralia cachemirica L. | Inhibition of biofilm formation and disruption of cell membrane. | [136] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Aquila, P.; De Rose, E.; Sena, G.; Scorza, A.; Cretella, B.; Passarino, G.; Bellizzi, D. Quorum Quenching Approaches against Bacterial-Biofilm-Induced Antibiotic Resistance. Antibiotics 2024, 13, 619. https://doi.org/10.3390/antibiotics13070619
D’Aquila P, De Rose E, Sena G, Scorza A, Cretella B, Passarino G, Bellizzi D. Quorum Quenching Approaches against Bacterial-Biofilm-Induced Antibiotic Resistance. Antibiotics. 2024; 13(7):619. https://doi.org/10.3390/antibiotics13070619
Chicago/Turabian StyleD’Aquila, Patrizia, Elisabetta De Rose, Giada Sena, Angelo Scorza, Bonaventura Cretella, Giuseppe Passarino, and Dina Bellizzi. 2024. "Quorum Quenching Approaches against Bacterial-Biofilm-Induced Antibiotic Resistance" Antibiotics 13, no. 7: 619. https://doi.org/10.3390/antibiotics13070619
APA StyleD’Aquila, P., De Rose, E., Sena, G., Scorza, A., Cretella, B., Passarino, G., & Bellizzi, D. (2024). Quorum Quenching Approaches against Bacterial-Biofilm-Induced Antibiotic Resistance. Antibiotics, 13(7), 619. https://doi.org/10.3390/antibiotics13070619






