Genotypic Shift and Diversification of MRSA Blood Stream Isolates in a University Hospital Setting: Evidence from a 12-Year Observational Study
Abstract
:1. Introduction
2. Results
2.1. Trends in SCCmec Types
2.2. Trends in Agr Types
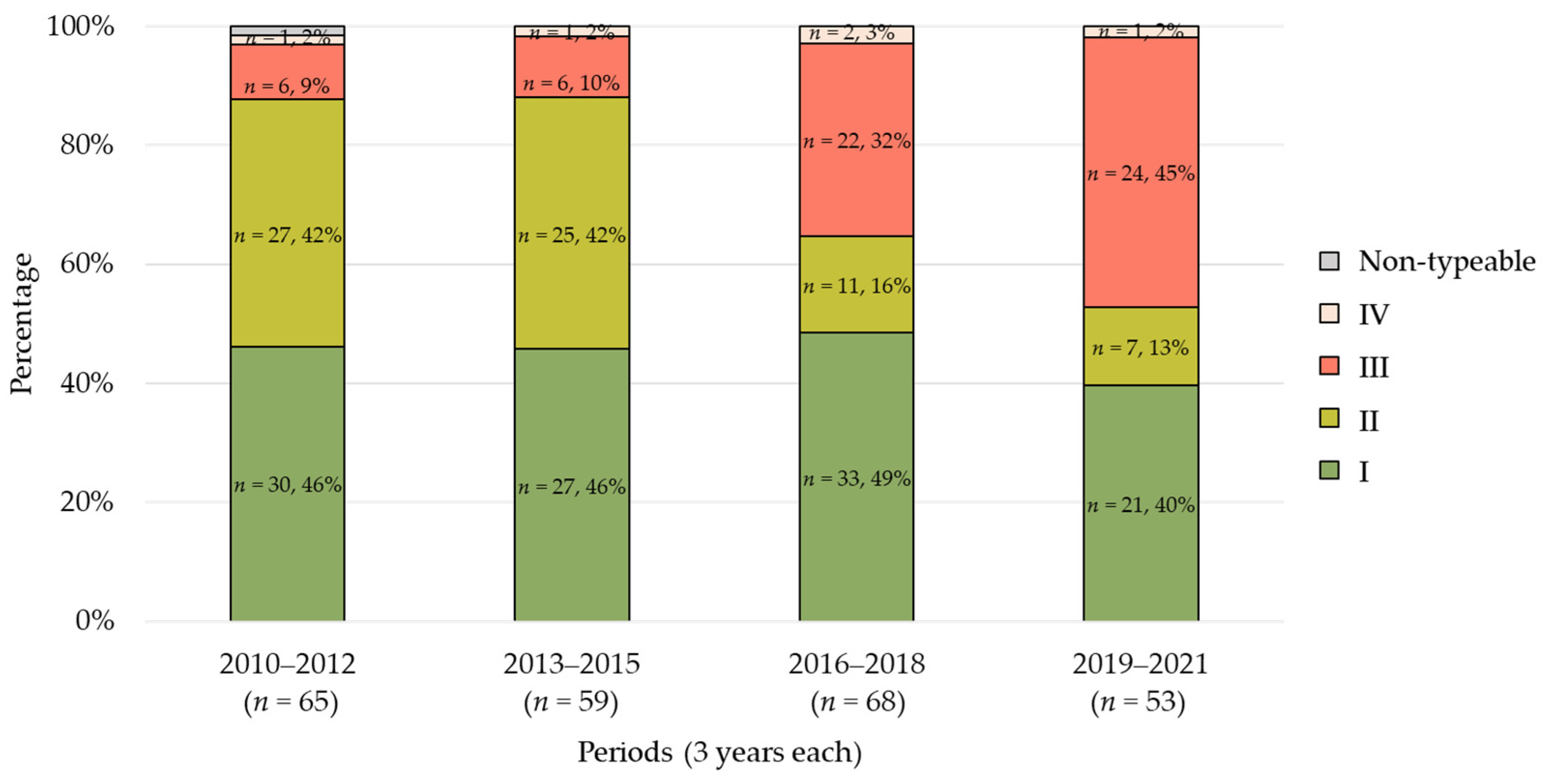
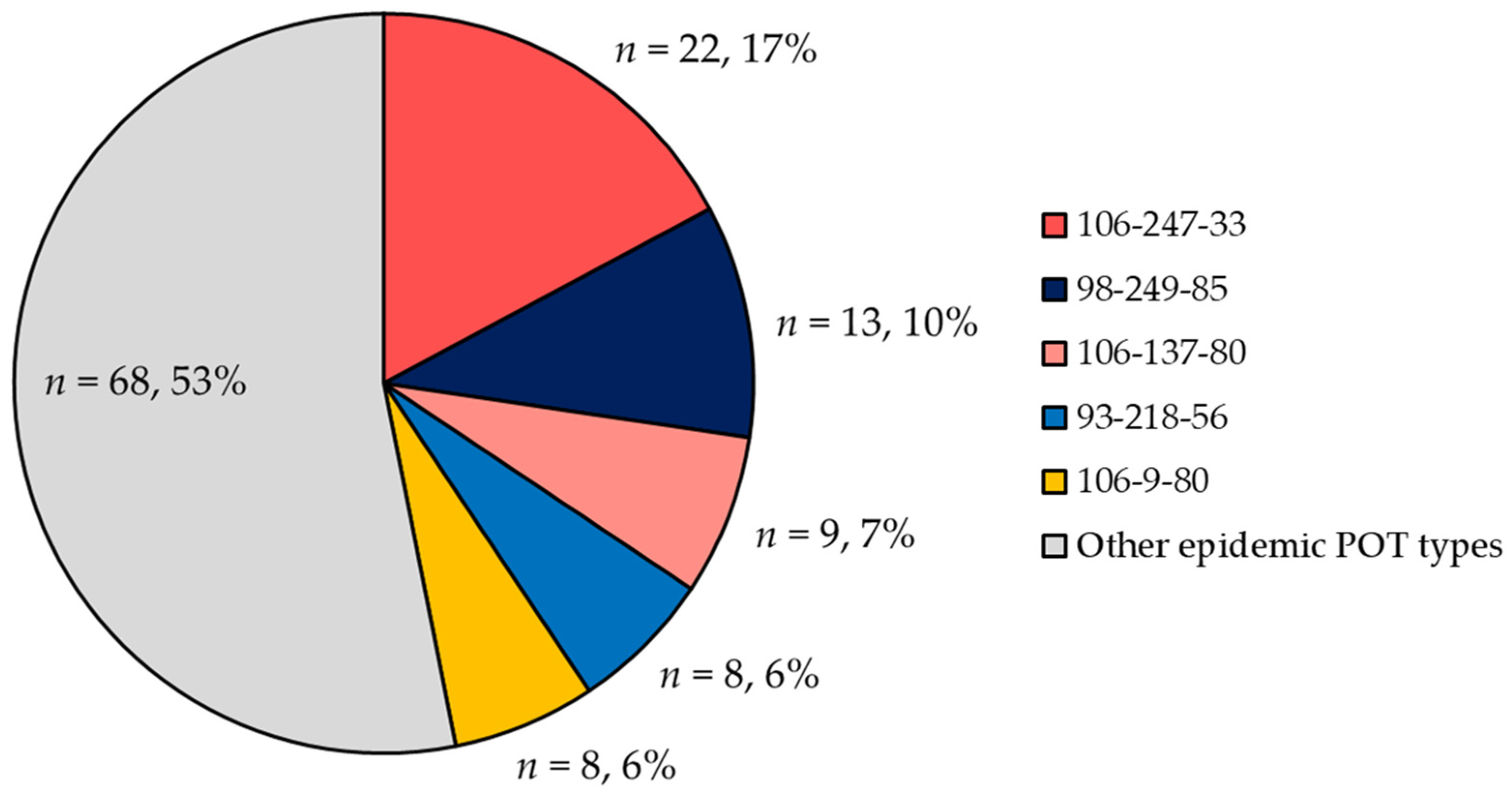
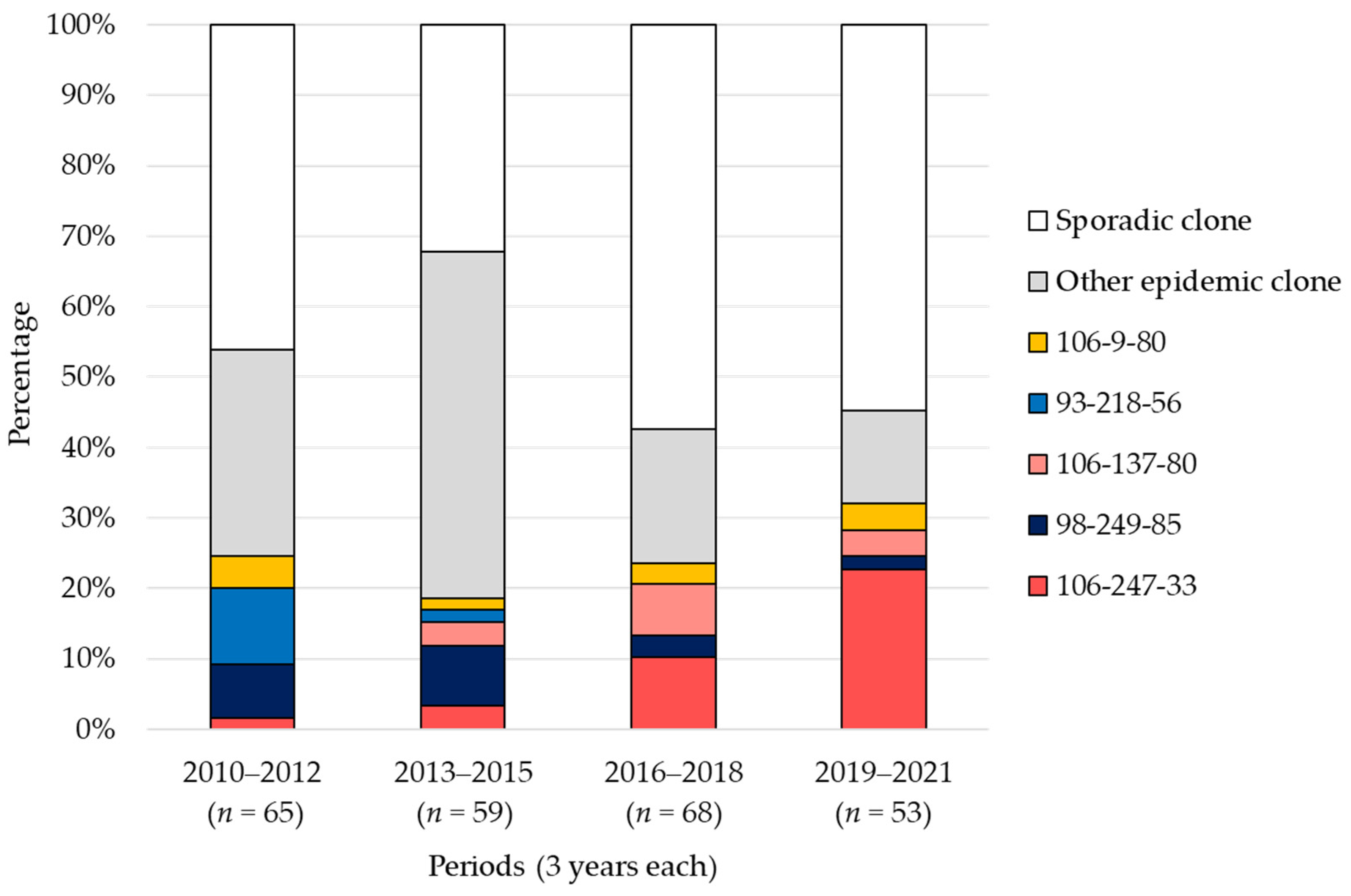
2.3. POT Type Distribution
2.4. Trends in POT Types
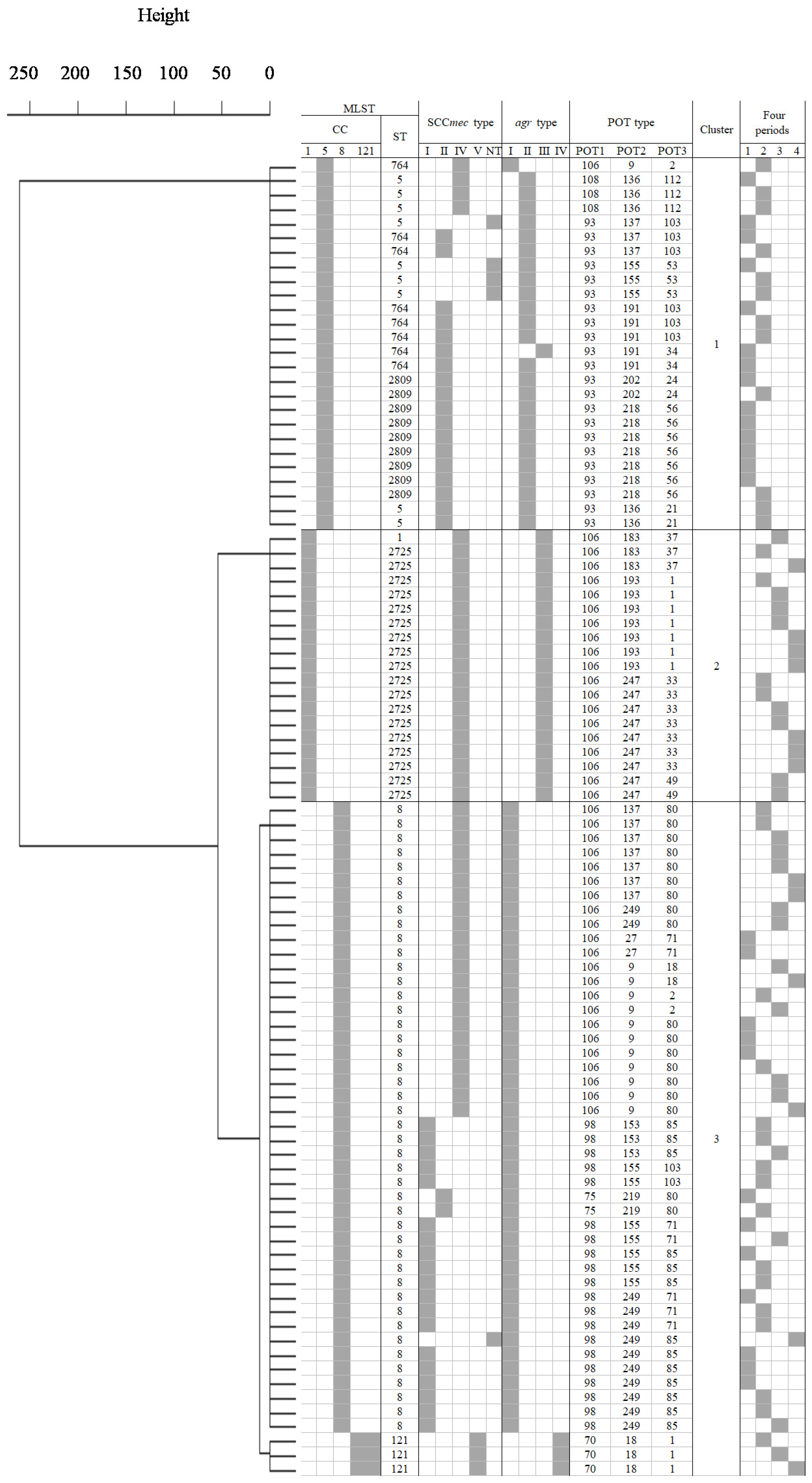
2.5. Cluster Analysis
2.6. Identification of Virulence Genes and Antimicrobial Resistance Profiles
2.7. Antimicrobial Resistance Profiles
2.8. Relations between Each Genotype (SCCmec and Agr Types) and VCM Susceptibility
3. Discussion
4. Materials and Methods
4.1. Clinical Isolates
4.2. SCCmec Typing, Agr Typing, and Identification of Virulence Genes
4.3. POT Type Detection
4.4. MLST Analysis
4.5. Antimicrobial Resistance Profile
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Hal, S.J.; Jensen, S.O.; Vaska, V.L.; Espedido, B.A.; Paterson, D.L.; Gosbell, I.B. Predictors of mortality in Staphylococcus aureus Bacteremia. Clin. Microbiol. Rev. 2012, 25, 362–386. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.X.; Ito, T.; Chongtrakool, P.; Hiramatsu, K. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J. Clin. Microbiol. 2006, 44, 4515–4527. [Google Scholar] [CrossRef] [PubMed]
- Taneike, I.; Otsuka, T.; Dohmae, S.; Saito, K.; Ozaki, K.; Takano, M.; Yamamoto, T. Molecular nature of methicillin-resistant Staphylococcus aureus derived from explosive nosocomial outbreaks of the 1980s in Japan. FEBS Lett. 2006, 580, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Yanagihara, K.; Araki, N.; Watanabe, S.; Kinebuchi, T.; Kaku, M.; Maesaki, S.; Yamaguchi, K.; Matsumoto, T.; Mikamo, H.; Takesue, Y.; et al. Antimicrobial susceptibility and molecular characteristics of 857 methicillin-resistant Staphylococcus aureus isolates from 16 medical centers in Japan (2008–2009): Nationwide survey of community-acquired and nosocomial MRSA. Diagn. Microbiol. Infect. Dis. 2012, 72, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Nishiyama, A.; Takano, T.; Yabe, S.; Higuchi, W.; Razvina, O.; Shi, D. Community-acquired methicillin-resistant Staphylococcus aureus: Community transmission, pathogenesis, and drug resistance. J. Infect. Chemother. 2010, 16, 225–254. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999. MMWR Morb. Mortal. Wkly. Rep. 1999, 48, 707–710. [Google Scholar]
- Vandenesch, F.; Naimi, T.; Enright, M.C.; Lina, G.; Nimmo, G.R.; Heffernan, H.; Liassine, N.; Bes, M.; Greenland, T.; Reverdy, M.E.; et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: Worldwide emergence. Emerg. Infect. Dis. 2003, 9, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Takeuchi, F.; Kuroda, M.; Yuzawa, H.; Aoki, K.; Oguchi, A.; Nagai, Y.; Iwama, N.; Asano, K.; Naimi, T.; et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 2002, 359, 1819–1827. [Google Scholar] [CrossRef]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.A.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006, 367, 731–739. [Google Scholar] [CrossRef]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.G.; Perdreau-Remington, F.; Rieg, G.; Mehdi, S.; Perlroth, J.; Bayer, A.S.; Tang, A.W.; Phung, T.O.; Spellberg, B. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N. Engl. J. Med. 2005, 352, 1445–1453. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Okamura, S.; Miura, Y.; Koyama, S.; Yanagisawa, H.; Matsumoto, T. Molecular Characterization of Community-Associated Methicillin-Resistant Staphylococcus aureus Isolated from Skin and Pus Samples of Outpatients in Japan. Microb. Drug Resist. 2015, 21, 441–447. [Google Scholar] [CrossRef]
- Nakaminami, H.; Takadama, S.; Ito, A.; Hasegawa, M.; Jono, C.; Noguchi, M.; Shoshi, M.; Wajima, T.; Fujii, T.; Maruyama, H.; et al. Characterization of SCCmec type IV methicillin-resistant Staphylococcus aureus clones increased in Japanese hospitals. J. Med. Microbiol. 2018, 67, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Yamaguchi, T.; Nakamura, I.; Koyama, S.; Tamai, K.; Okanda, T.; Matsumoto, T. Epidemiological Trends Observed from Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Blood Cultures at a Japanese University Hospital, 2012–2015. Microb. Drug Resist. 2018, 24, 70–75. [Google Scholar] [CrossRef]
- Kaku, N.; Yanagihara, K.; Morinaga, Y.; Yamada, K.; Harada, Y.; Migiyama, Y.; Nagaoka, K.; Matsuda, J.; Uno, N.; Hasegawa, H.; et al. Influence of antimicrobial regimen on decreased in-hospital mortality of patients with MRSA bacteremia. J. Infect. Chemother. 2014, 20, 350–355. [Google Scholar] [CrossRef]
- Aung, M.S.; Urushibara, N.; Kawaguchiya, M.; Ohashi, N.; Hirose, M.; Kimura, Y.; Kudo, K.; Ito, M.; Kobayashi, N. Molecular Epidemiological Characterization of Methicillin-Resistant Staphylococcus aureus from Bloodstream Infections in Northern Japan: Increasing Trend of CC1 and Identification of ST8-SCCmec IVa USA300-Like Isolate Lacking Arginine Catabolic Mobile Element. Microb. Drug Resist. 2023, 30, 63–72. [Google Scholar] [CrossRef]
- Kaku, N.; Sasaki, D.; Ota, K.; Miyazaki, T.; Yanagihara, K. Changing molecular epidemiology and characteristics of MRSA isolated from bloodstream infections: Nationwide surveillance in Japan in 2019. J. Antimicrob. Chemother. 2022, 77, 2130–2141. [Google Scholar] [CrossRef]
- Kawasuji, H.; Ikezawa, Y.; Morita, M.; Sugie, K.; Somekawa, M.; Ezaki, M.; Koshiyama, Y.; Takegoshi, Y.; Murai, Y.; Kaneda, M.; et al. High Incidence of Metastatic Infections in Panton-Valentine Leucocidin-Negative, Community-Acquired Methicillin-Resistant Staphylococcus aureus Bacteremia: An 11-Year Retrospective Study in Japan. Antibiotics 2023, 12, 1516. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Mihara, T.; Ohara, J.; Inoue, K.; Kinoshita, M.; Sawa, T. Relationship between mortality and molecular epidemiology of methicillin-resistant Staphylococcus aureus bacteremia. PLoS ONE 2022, 17, e0271115. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Yamaguchi, T.; Sato, A.; Ono, D.; Aoki, K.; Kajiwara, C.; Kimura, S.; Maeda, T.; Sasaki, M.; Murakami, H.; et al. Increased Incidence and Plasma-Biofilm Formation Ability of SCCmec Type IV Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Patients With Bacteremia. Front. Cell. Infect. Microbiol. 2021, 11, 602833. [Google Scholar] [CrossRef] [PubMed]
- Nikolaras, G.P.; Papaparaskevas, J.; Samarkos, M.; Tzouvelekis, L.S.; Psychogiou, M.; Pavlopoulou, I.; Goukos, D.; Polonyfi, K.; Pantazatou, A.; Deliolanis, I.; et al. Changes in the rates and population structure of methicillin-resistant Staphylococcus aureus (MRSA) from bloodstream infections: A single-centre experience (2000–2015). J. Glob. Antimicrob. Resist. 2019, 17, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.J. Comparing the Phylogenetic Distribution of Multilocus Sequence Typing, Staphylococcal Protein A, and Staphylococcal Cassette Chromosome Mec Types in Methicillin-Resistant Staphylococcus aureus (MRSA) in Korea from 1994 to 2020. Antibiotics 2023, 12, 1397. [Google Scholar] [CrossRef]
- Niek, W.K.; Teh, C.S.J.; Idris, N.; Sit, P.S.; Lee, Y.Q.; Thong, K.L.; Sri La Sri Ponnampalavanar, S. Methicillin-resistant Staphylococcus aureus bacteraemia, 2003–2015: Comparative evaluation of changing trends in molecular epidemiology and clinical outcomes of infections. Infect. Genet. Evol. 2020, 85, 104567. [Google Scholar] [CrossRef]
- Ellington, M.J.; Hope, R.; Livermore, D.M.; Kearns, A.M.; Henderson, K.; Cookson, B.D.; Pearson, A.; Johnson, A.P. Decline of EMRSA-16 amongst methicillin-resistant Staphylococcus aureus causing bacteraemias in the UK between 2001 and 2007. J. Antimicrob. Chemother. 2010, 65, 446–448. [Google Scholar] [CrossRef]
- Hsieh, Y.C.; Lin, Y.C.; Huang, Y.C. Vancomycin, teicoplanin, daptomycin, and linezolid MIC creep in methicillin-resistant Staphylococcus aureus is associated with clonality. Medicine 2016, 95, e5060. [Google Scholar] [CrossRef] [PubMed]
- Popovich, K.J.; Weinstein, R.A.; Hota, B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin. Infect. Dis. 2008, 46, 787–794. [Google Scholar] [CrossRef]
- Choi, S.H.; Lee, J.; Jung, J.; Kim, E.S.; Kim, M.J.; Chong, Y.P.; Kim, S.H.; Choi, S.H.; Lee, S.O.; Kim, Y.S. A Longitudinal Study of Adult Patients with Staphylococcus aureus Bacteremia over 11 Years in Korea. J. Korean Med. Sci. 2021, 36, e104. [Google Scholar] [CrossRef] [PubMed]
- Takata, T.; Miyazaki, M.; Li, H.; Healy, M.; Frye, S.; Tanaka, K.; Hara, S.; Kamimura, H.; Yoshimura, H.; Matsunaga, A.; et al. Methicillin-resistant Staphylococcus aureus bloodstream infections in a Japanese University Hospital between 1987 and 2004. J. Infect. Chemother. 2012, 18, 199–206. [Google Scholar] [CrossRef]
- Naimi, T.S.; LeDell, K.H.; Como-Sabetti, K.; Borchardt, S.M.; Boxrud, D.J.; Etienne, J.; Johnson, S.K.; Vandenesch, F.; Fridkin, S.; O’Boyle, C.; et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 2003, 290, 2976–2984. [Google Scholar] [CrossRef]
- Park, M.J.; Kim, H.S.; Kim, H.S.; Kim, J.S.; Song, W.; Kim, M.Y.; Lee, Y.K.; Kang, H.J. Accessory Gene Regulator Polymorphism and Vancomycin Minimum Inhibitory Concentration in Methicillin-Resistant Staphylococcus aureus. Ann. Lab. Med. 2015, 35, 399–403. [Google Scholar] [CrossRef]
- Moise, P.A.; Smyth, D.S.; Robinson, D.A.; El-Fawal, N.; McCalla, C.; Sakoulas, G. Genotypic and phenotypic relationships among methicillin-resistant Staphylococcus aureus from three multicentre bacteraemia studies. J. Antimicrob. Chemother. 2009, 63, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Cázares-Domínguez, V.; Cruz-Córdova, A.; Ochoa, S.A.; Escalona, G.; Arellano-Galindo, J.; Rodríguez-Leviz, A.; Hernández-Castro, R.; López-Villegas, E.O.; Xicohtencatl-Cortes, J. Vancomycin tolerant, methicillin-resistant Staphylococcus aureus reveals the effects of vancomycin on cell wall thickening. PLoS ONE 2015, 10, e0118791. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Chiu, C.F.; Lee, Y.T.; Hsueh, P.R.; Tsao, S.M. Molecular epidemiology and phenotypes of invasive methicillin-resistant vancomycin-intermediate Staphylococcus aureus in Taiwan. J. Microbiol. Immunol. Infect. 2022, 55 Pt 2, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Lee, S.Y.; Chiueh, T.S.; Lu, J.J. Molecular and phenotypic characteristics of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus isolates from patients with septic arthritis. J. Clin. Microbiol. 2009, 47, 3617–3623. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Zhou, K.; Wang, Y.; Song, J.; Liu, Y.; Zhou, Y. High prevalence of a globally disseminated hypervirulent clone, Staphylococcus aureus CC121, with reduced vancomycin susceptibility in community settings in China. J. Antimicrob. Chemother. 2019, 74, 2537–2543. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tawada, Y.; Kato, M.; Hori, H.; Mamiya, N.; Hayashi, Y.; Nakano, M.; Fukushima, R.; Katai, A.; Tanaka, T.; et al. Development of a rapid strain differentiation method for methicillin-resistant Staphylococcus aureus isolated in Japan by detecting phage-derived open-reading frames. J. Appl. Microbiol. 2006, 101, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Nada, T.; Yagi, T.; Ohkura, T.; Morishita, Y.; Baba, H.; Ohta, M.; Suzuki, M. Usefulness of phage open-reading frame typing method in an epidemiological study of an outbreak of methicillin-resistant Staphylococcus aureus infections. Jpn. J. Infect. Dis. 2009, 62, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, S.; Saito, R.; Sawabe, E.; Kozakai, T.; Shima, M.; Aiso, Y.; Fujie, T.; Nukui, Y.; Koike, R.; Hagihara, M.; et al. Molecular typing of methicillin-resistant Staphylococcus aureus: Comparison of PCR-based open reading frame typing, multilocus sequence typing, and Staphylococcus protein A gene typing. J. Infect. Chemother. 2018, 24, 312–314. [Google Scholar] [CrossRef]
- Osawa, K.; Shigemura, K.; Jikimoto, T.; Shirakawa, T.; Fujisawa, M.; Arakawa, S. Comparison between phage-open-reading frame typing and automated repetitive-sequence-based PCR for typing MRSA isolates. J. Antibiot. 2014, 67, 565–569. [Google Scholar] [CrossRef]
- Nakaie, K.; Yamada, K.; Park, K.; Nakamura, Y.; Okada, Y.; Fujita, A.; Fujimoto, H.; Kaneko, Y.; Kakeya, H. Effectiveness of weekly polymerase chain reaction-based open reading frame typing analysis of all newly isolated methicillin-resistant Staphylococcus aureus strains for controlling nosocomial infections. J. Infect. Chemother. 2016, 22, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Kishita, M.; Matsumura, Y.; Yamamoto, M.; Nagao, M.; Takemura, M.; Sumi, M.; Osawa, M.; Nakano, Y.; Tanikawa, S.; Tsukaguchi, F.; et al. Increase in the frequency of community-acquired methicillin-resistant Staphylococcus aureus clones among inpatients of acute care hospitals in the Kyoto and Shiga regions, Japan. J. Infect. Chemother. 2023, 29, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Kitaoka, K.; Kimura, K.; Wachino, J.I.; Kondo, T.; Iinuma, Y.; Murakami, N.; Fujimoto, S.; Arakawa, Y. Spread of seb-Positive Methicillin-Resistant Staphylococcus aureus SCCmec Type II-ST764 Among Elderly Japanese in Nonacute Care Settings. Microb. Drug Resist. 2019, 25, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Ide, K.; Fukase, F.; Shimura, Y.; Yasuda, S.; Goto, H.; Fukuyama, A.; Nakajima, H. Polymerase chain reaction-based open reading frame typing (POT) method analysis for a methicillin-resistant Staphylococcus aureus (MRSA) outbreak through breast-feeding in the neonatal intensive care unit. IDCases 2018, 12, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, H.; Murakami, S.; Saito, M.; Tamaki, M.; Ochi, C.; Iyoda, M. Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus (MRSA) by POT Method, Antimicrobial Susceptibility Patterns, and Toxin-Producing Types. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 2020, 29, 45–52. (In Japanese) [Google Scholar]
- Sato, T.; Yamaguchi, T.; Aoki, K.; Kajiwara, C.; Kimura, S.; Maeda, T.; Yoshizawa, S.; Sasaki, M.; Murakami, H.; Hisatsune, J.; et al. Whole-genome sequencing analysis of molecular epidemiology and silent transmissions causing meticillin-resistant Staphylococcus aureus bloodstream infections in a university hospital. J. Hosp. Infect. 2023, 139, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Enright, M.C.; Robinson, D.A.; Randle, G.; Feil, E.J.; Grundmann, H.; Spratt, B.G. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 2002, 99, 7687–7692. [Google Scholar] [CrossRef] [PubMed]
- Konuma, T.; Takahashi, S.; Suzuki, M.; Tojo, A. Molecular characterization of healthcare and community-associated methicillin-resistant Staphylococcus aureus using phage open-reading frame typing. Iran. J. Microbiol. 2021, 13, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, S.; Inoue, O.; Yamagami, T.; Yanagimoto, K.; Uematsu, K.; Hisada, Y.; Uchida, T.; Ohta, M.; Suzuki-Inoue, K. Clinical characteristics and molecular analysis of USA300 and ST 764 methicillin-resistant Staphylococcus aureus isolates from outpatients in Japan by PCR-Based open reading frame typing. J. Infect. Chemother. 2021, 27, 466–472. [Google Scholar] [CrossRef]
- Osaka, S.; Okuzumi, K.; Koide, S.; Tamai, K.; Sato, T.; Tanimoto, K.; Tomita, H.; Suzuki, M.; Nagano, Y.; Shibayama, K.; et al. Genetic shifts in methicillin-resistant Staphylococcus aureus epidemic clones and toxin gene profiles in Japan: Comparative analysis among pre-epidemic, epidemic and post-epidemic phases. J. Med. Microbiol. 2018, 67, 392–399. [Google Scholar] [CrossRef]
- Takadama, S.; Nakaminami, H.; Sato, A.; Shoshi, M.; Fujii, T.; Noguchi, N. Dissemination of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus USA300 clone in multiple hospitals in Tokyo, Japan. Clin. Microbiol. Infect. 2018, 24, 1211.e1–1211.e7. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Morinaga, Y.; Akamatsu, N.; Matsuda, J.; Yamaryo, T.; Kawakami, K.; Matsuo, H.; Kosai, K.; Uno, N.; Hasegawa, H.; et al. Antimicrobial susceptibility and molecular characteristics of methicillin-resistant Staphylococcus aureus in a Japanese secondary care facility. J. Infect. Chemother. 2016, 22, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.W.; Waddington, M.G.; Smith, D.H.; Manahan, J.M.; Kohner, P.C.; Highsmith, L.M.; Li, H.; Cockerill, F.R., 3rd; Thompson, R.L.; Montgomery, S.O.; et al. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1347–1351. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Boutite, F.; Tristan, A.; Bes, M.; Etienne, J.; Vandenesch, F. Bacterial competition for human nasal cavity colonization: Role of Staphylococcal agr alleles. Appl. Environ. Microbiol. 2003, 69, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Piémont, Y.; Godail-Gamot, F.; Bes, M.; Peter, M.O.; Gauduchon, V.; Vandenesch, F.; Etienne, J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 1999, 29, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhu, Y.; Song, Y.; Li, T.; Luo, T.; Sun, G.; Yang, C.; Cao, C.; Lu, Y.; Li, M. Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS ONE 2013, 8, e62742. [Google Scholar] [CrossRef]
- Suzuki, M.; Matsumoto, M.; Takahashi, M.; Hayakawa, Y.; Minagawa, H. Identification of the clonal complexes of Staphylococcus aureus strains by determination of the conservation patterns of small genomic islets. J. Appl. Microbiol. 2009, 107, 1367–1374. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, NY, USA, 2020. [Google Scholar]

| Antimicrobial Drug a | 2010–2012 (n = 65) | 2013–2015 (n = 59) | 2016–2018 (n = 68) | 2019–2021 (n = 52) | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mode MIC [μg/mL] b | R (%) c | Mode MIC [μg/mL] | R (%) | Mode MIC [μg/mL] | R (%) | Mode MIC [μg/mL] | R (%) | Mode MIC [μg/mL] | R (%) | |
| GM | ≥16 | 57% | ≥16 | 53% | ≥16 | 51% | ≤0.5 | 29% | ≥16 | 48% |
| EM | ≥8 | 83% | ≥8 | 81% | ≥8 | 65% | ≥8 | 55% | ≥8 | 72% |
| CLDM | ≥8 | 62% | ≥8 | 64% | ≤0.25 | 32% | ≤0.25 | 19% | ≤0.25 | 45% |
| MINO | ≥16 | 46% | ≥16 | 53% | ≤0.5 | 21% | ≤0.5 | 15% | ≤0.5 | 34% |
| LVFX | ≥8 | 77% | ≥8 | 85% | ≥8 | 76% | ≥8 | 75% | ≥8 | 79% |
| FOM | ≥8 | 23% | ≥8 | 34% | ≥8 | 12% | ≥8 | 6% | ≥8 | 19% |
| ABK | ≤1 | 2% | ≤1 | 0% | ≤1 | 1% | ≤1 | 0% | ≤1 | 1% |
| TEIC | ≤0.5 | 0% | ≤0.5 | 0% | ≤0.5 | 0% | ≤0.5 | 0% | ≤0.5 | 0% |
| VCM | 1 | 0% | 1 | 0% | 1 | 0% | 1 | 0% | 1 | 0% |
| LZD | 2 | 0% | 2 | 0% | 2 | 0% | 2 | 0% | 2 | 0% |
| DAP | N/D d | N/D | 1 | N/D | 0.25 | N/D | 0.25 | N/D | 0.25 | N/D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motomura, Y.; Miyazaki, M.; Kamada, M.; Morimoto, S.; Nakamura, Y.; Satho, T.; Takata, T.; Kashige, N. Genotypic Shift and Diversification of MRSA Blood Stream Isolates in a University Hospital Setting: Evidence from a 12-Year Observational Study. Antibiotics 2024, 13, 670. https://doi.org/10.3390/antibiotics13070670
Motomura Y, Miyazaki M, Kamada M, Morimoto S, Nakamura Y, Satho T, Takata T, Kashige N. Genotypic Shift and Diversification of MRSA Blood Stream Isolates in a University Hospital Setting: Evidence from a 12-Year Observational Study. Antibiotics. 2024; 13(7):670. https://doi.org/10.3390/antibiotics13070670
Chicago/Turabian StyleMotomura, Yuka, Motoyasu Miyazaki, Mitsuhiro Kamada, Shinichi Morimoto, Yoshihiko Nakamura, Tomomitsu Satho, Tohru Takata, and Nobuhiro Kashige. 2024. "Genotypic Shift and Diversification of MRSA Blood Stream Isolates in a University Hospital Setting: Evidence from a 12-Year Observational Study" Antibiotics 13, no. 7: 670. https://doi.org/10.3390/antibiotics13070670
APA StyleMotomura, Y., Miyazaki, M., Kamada, M., Morimoto, S., Nakamura, Y., Satho, T., Takata, T., & Kashige, N. (2024). Genotypic Shift and Diversification of MRSA Blood Stream Isolates in a University Hospital Setting: Evidence from a 12-Year Observational Study. Antibiotics, 13(7), 670. https://doi.org/10.3390/antibiotics13070670







