Biofilm Formation and Antibiotic Resistance Profiles in Carbapenemase-Producing Gram-Negative Rods—A Comparative Analysis between Screening and Pathological Isolates
Abstract
1. Introduction
2. Results
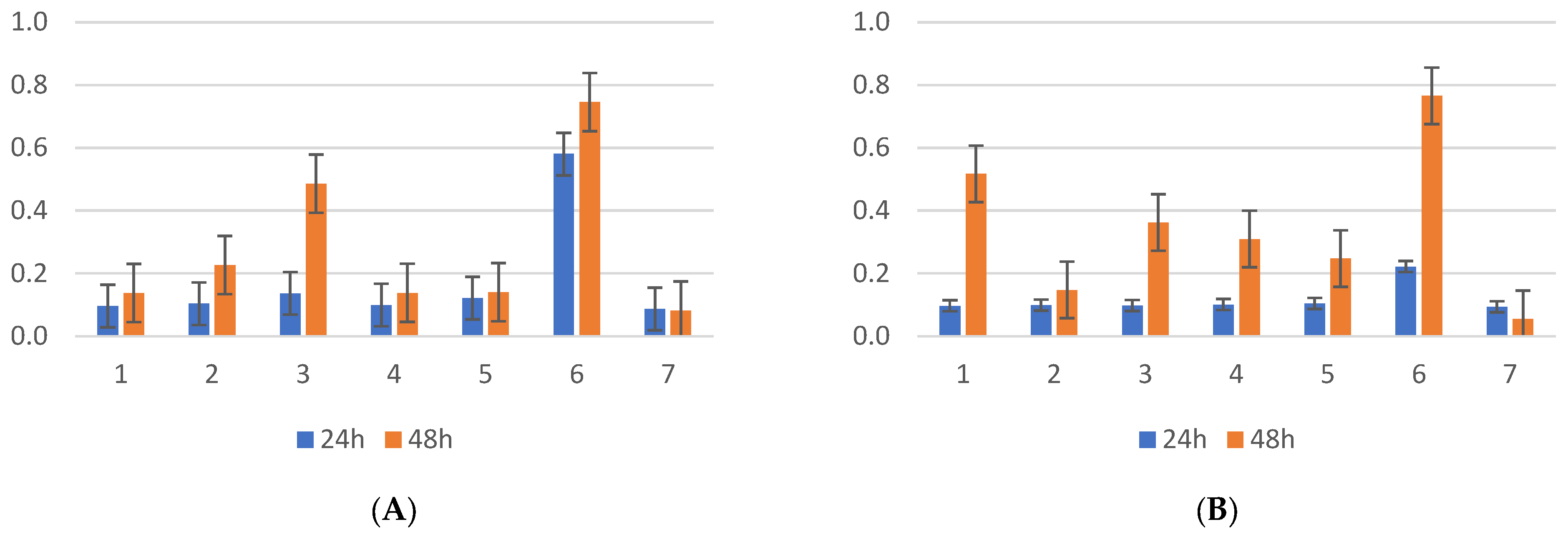
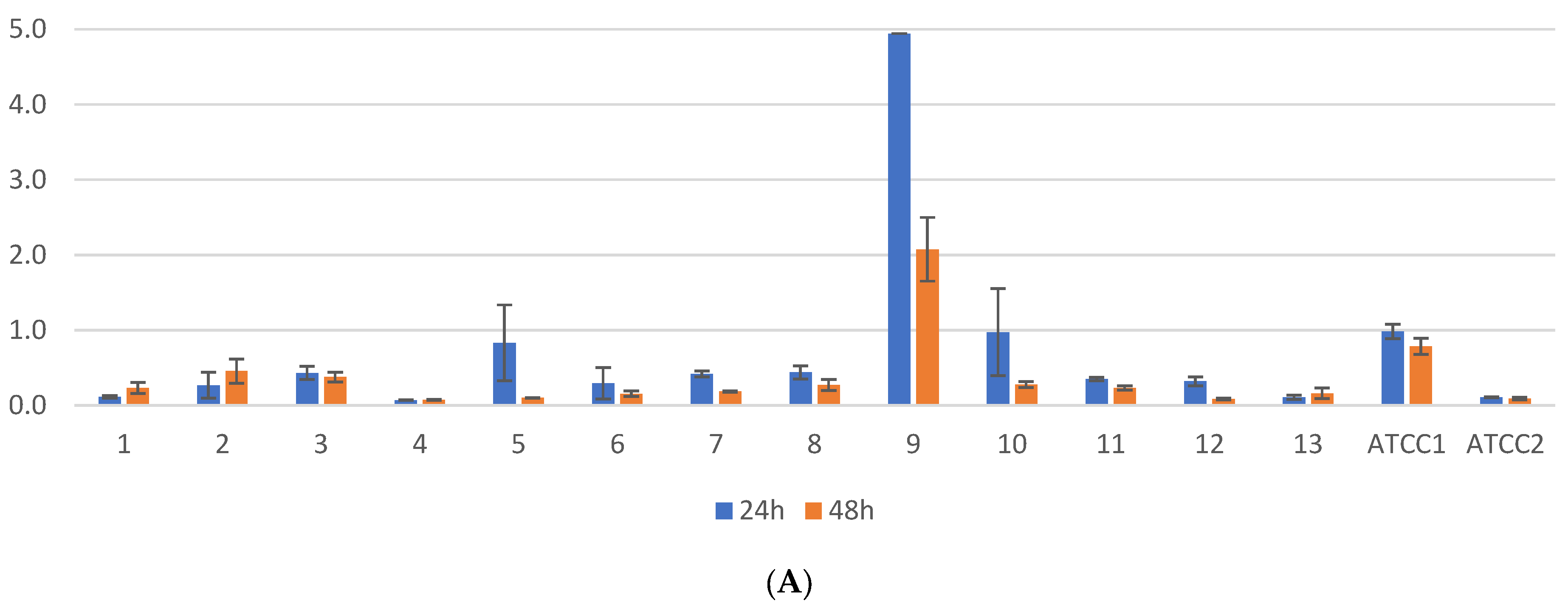
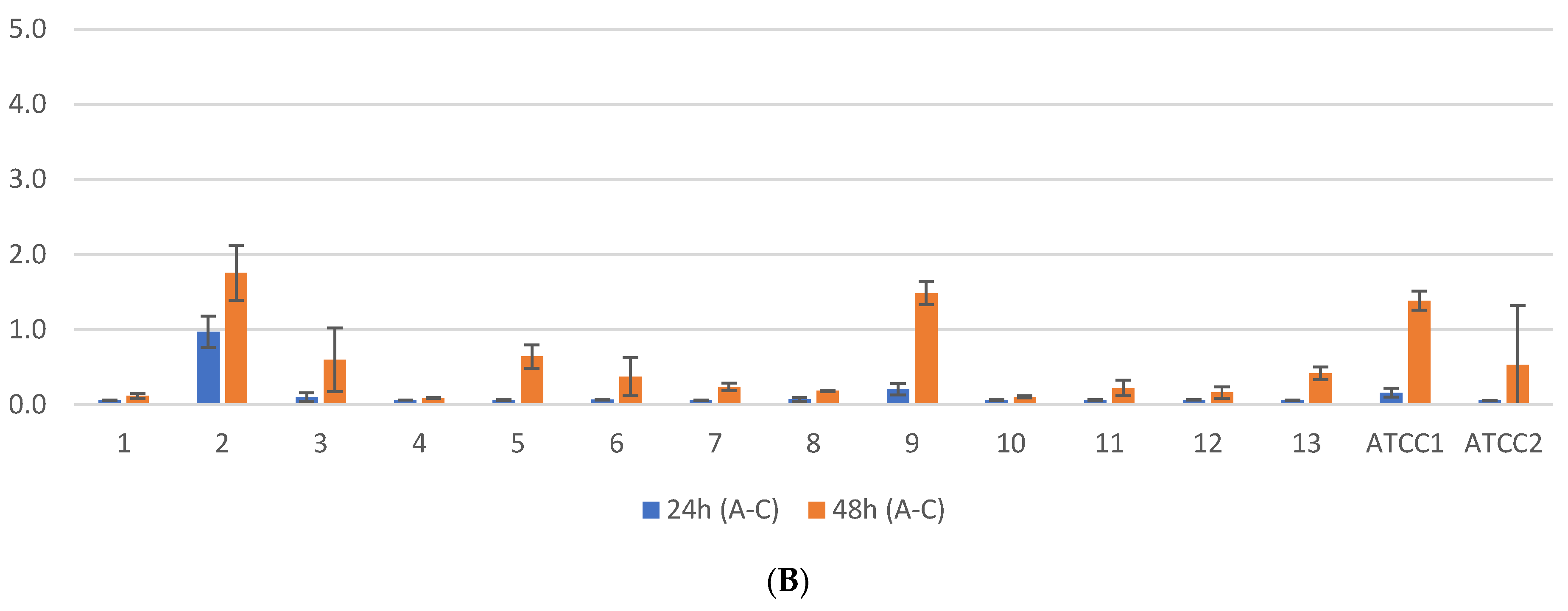
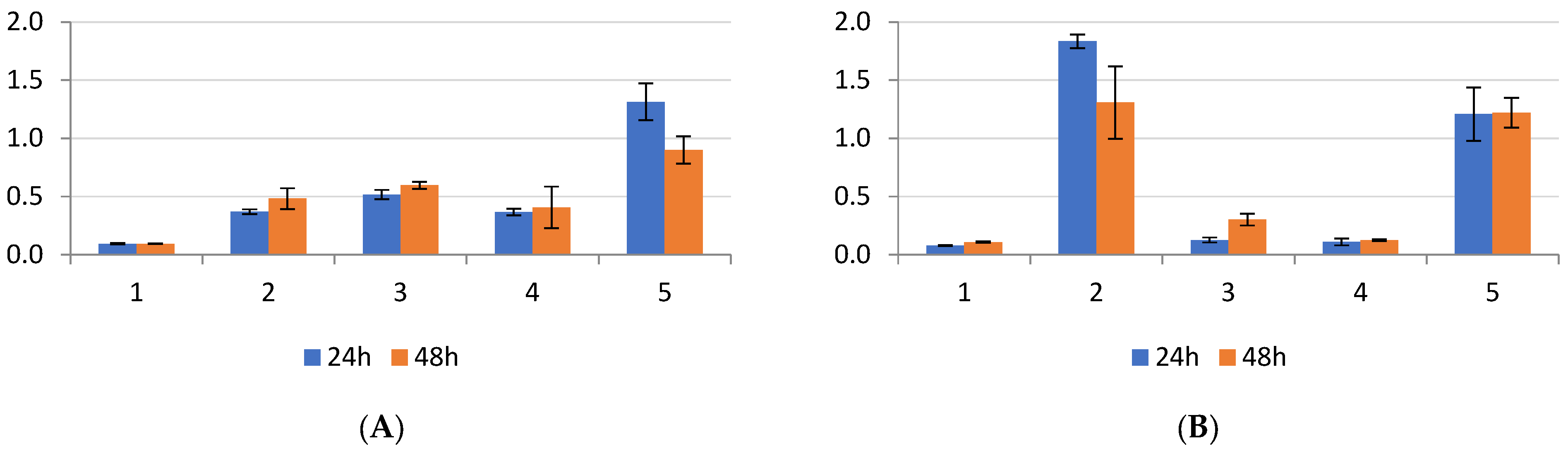
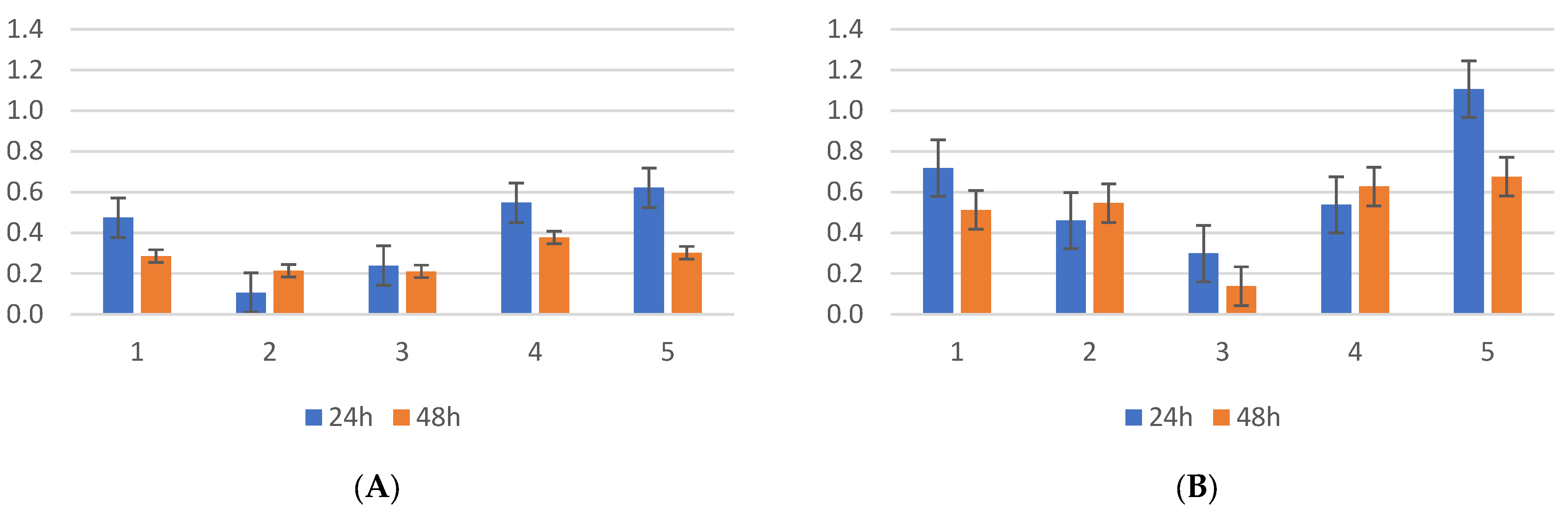
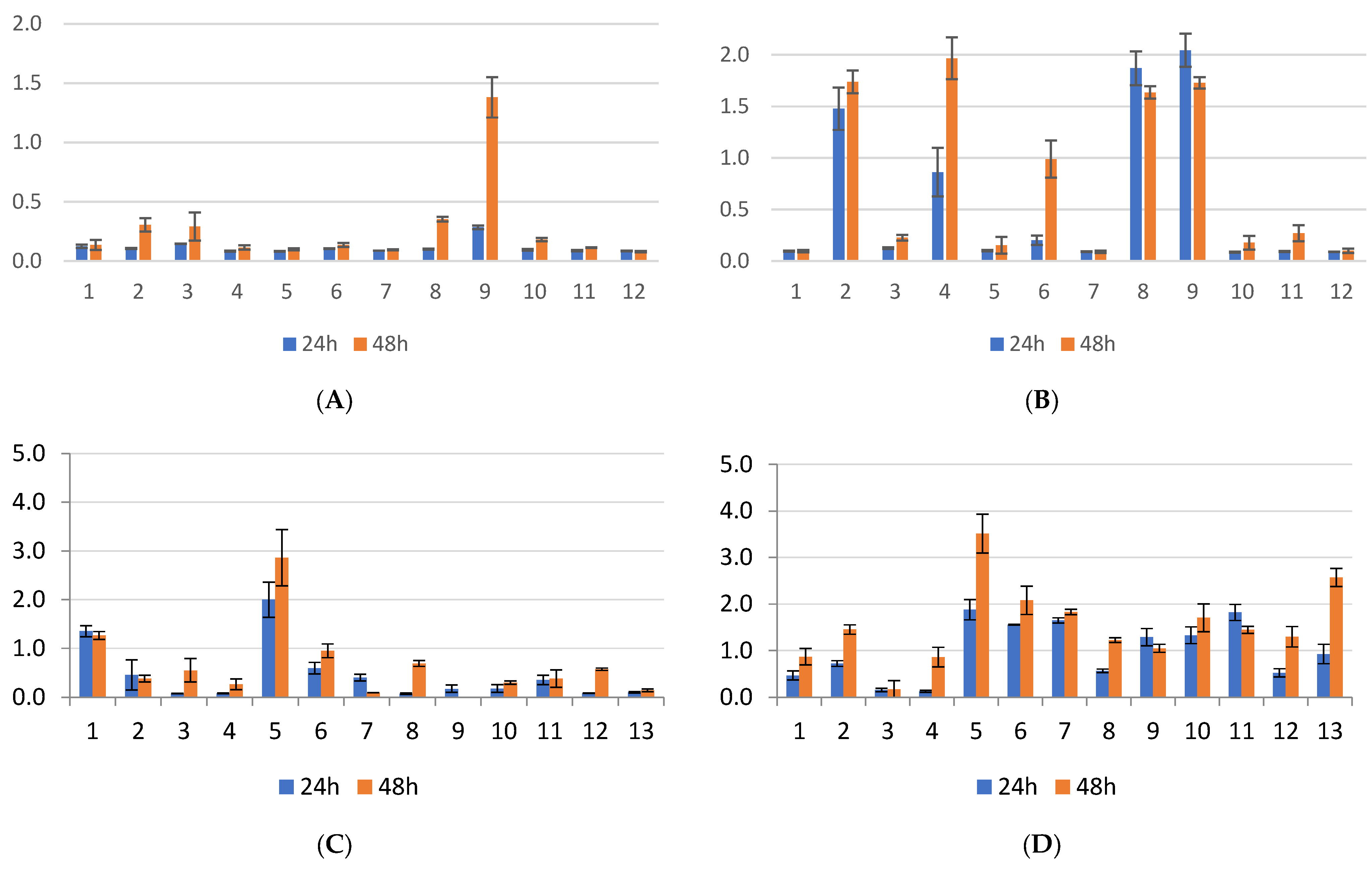
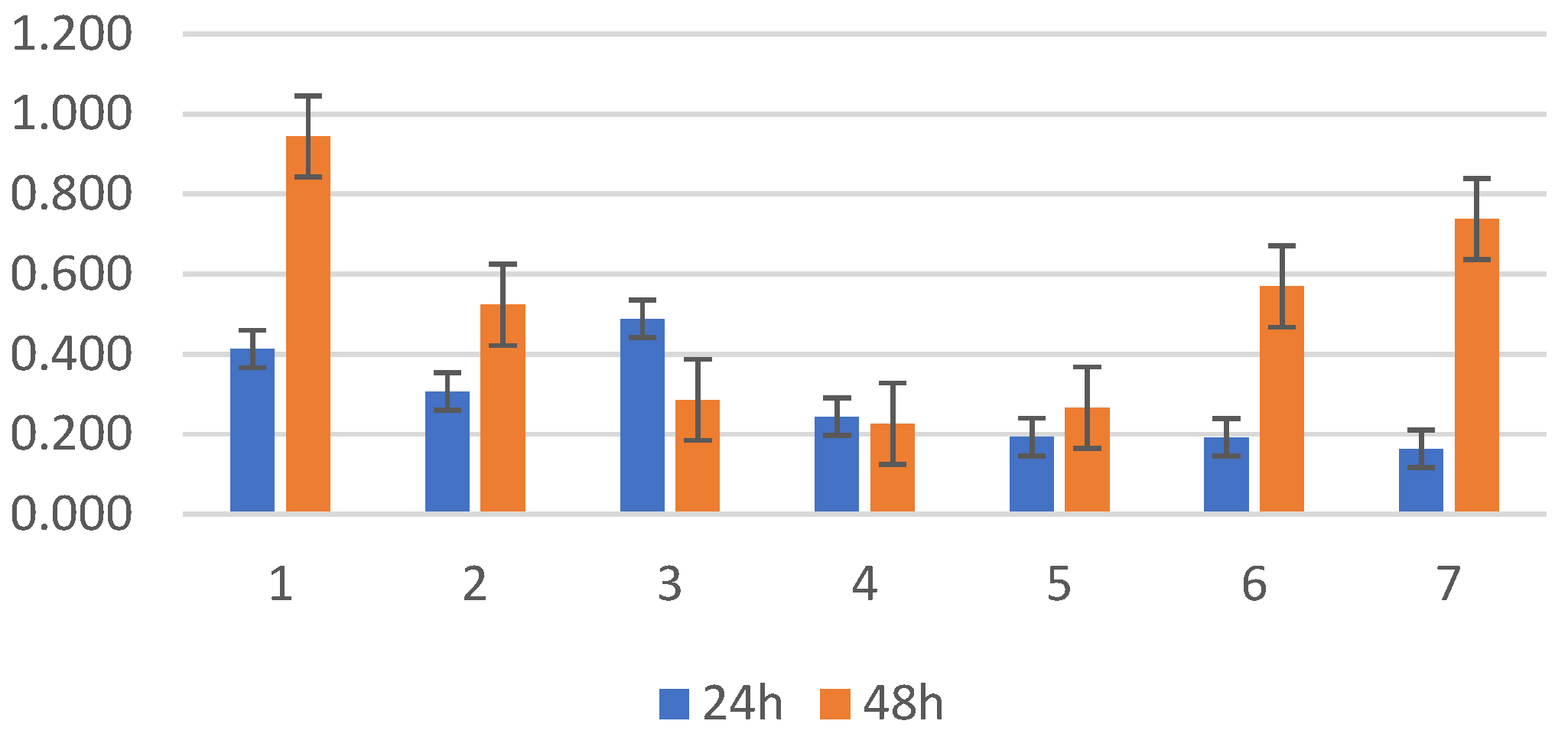
2.1. Biofilm Results
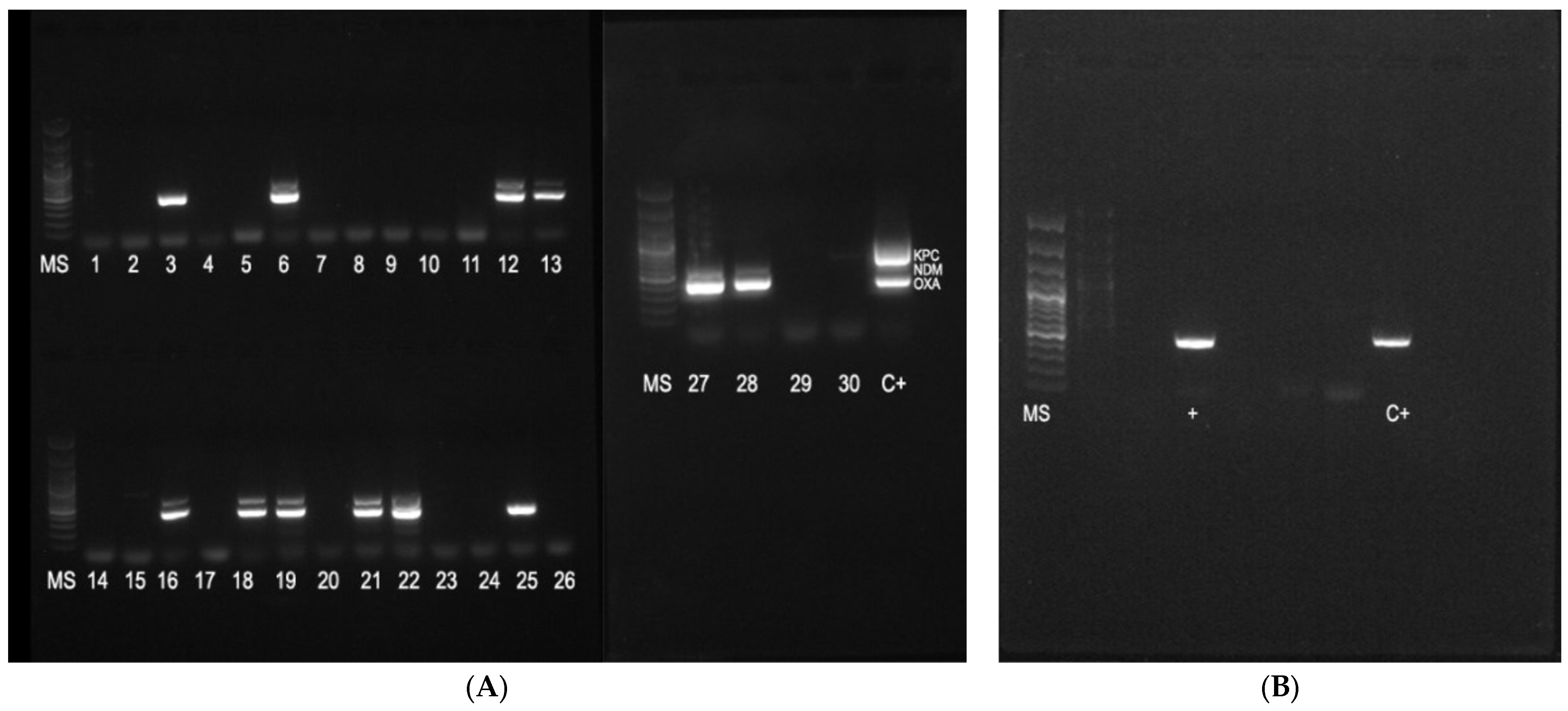
2.2. Genetic Test Results
3. Discussion
4. Materials and Methods
4.1. Sample Selection
- –
- all isolates were grown on CRE Brilliance Agar (Oxoid, Hampshire, UK), from the upper respiratory tract (pharyngeal swab), and confirmed to present a carbapenem-resistance phenotype, identified to the genus and species;
- –
- they were multi-drug-resistant (MDR), extensively resistant (XDR), or pan-resistant (PDR) Gram-negative bacterial isolates from the ICU ward, from the lower respiratory tract.
4.2. Evaluation of Ability of Bacteria to Form Biofilms
4.3. Biofilm Culture
4.4. Biofilm Growth Assay
4.5. Acetic Acid Treatment and Spectrophotometry Analysis
4.6. Evaluation of Ability of Bacteria to Produce Carbapenemases
4.7. DNA Extraction
4.8. PCR for Detection of Carbapenemase Genes
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Strain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 1 | s | s | * | * | s | s | * | * | * | s | * | * | s | s | s | s | * | * | * | * | ||
| 2 | s | s | s | * | * | s | s | s | s | s | s | * | * | * | * | s | s | s | ||||
| 3 | s | s | * | * | * | s | * | * | s | s | s | s | * | * | * | * | ||||||
| 4 | s | s | s | s | s | s | s | s | s | s | s | s | s | s | ||||||||
| 5 | * | s | * | * | s | s | s | s | * | * | * | * | ||||||||||
| 6 | * | s | s | s | s | s | * | * | * | * | ||||||||||||
| 7 | s | s | s | s | * | * | * | * | ||||||||||||||
| 8 | * | * | s | s | s | s | ||||||||||||||||
| 9 | s | * | * | * | ||||||||||||||||||
| 10 | * | * | ||||||||||||||||||||
| Strain | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 1 | * | * | * | * | s | * | s | * | * | * | s | * | s | s | s | s | s | * | s | * |
| 2 | s | * | s | * | s | s | * | * | * | s | * | * | s | s | * | * | * | * | ||
| 3 | s | * | s | * | s | * | s | * | s | * | s | s | s | * | s | * | ||||
| 4 | * | * | * | * | * | * | * | s | s | s | * | * | * | * | ||||||
| 5 | s | * | * | * | * | s | s | s | * | * | * | * | ||||||||
| 6 | * | * | * | s | s | s | * | * | * | * | ||||||||||
| 7 | * | s | s | s | * | * | * | * | ||||||||||||
| 8 | s | * | * | * | * | s | ||||||||||||||
| 9 | s | s | s | s | * | |||||||||||||||
| 10 | * | s | ||||||||||||||||||
| Strain | 2 | 3 | 4 | 5 | ||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 1 | s | s | s | s | s | s | s | s |
| 2 | * | * | * | * | s | s | ||
| 3 | * | * | s | s | ||||
| 4 | s | s | ||||||
| Strain | 2 | 3 | 4 | 5 | ||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 1 | s | s | * | * | * | * | s | s |
| 2 | s | s | s | s | s | * | ||
| 3 | * | * | s | s | ||||
| 4 | s | s | ||||||
| Strain | 2 | 3 | 4 | 5 | 6 | 7 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 1 | * | * | * | * | s | s | s | * | s | * | s | * |
| 2 | s | * | * | * | * | * | * | * | s | * | ||
| 3 | s | * | s | * | s | * | s | * | ||||
| 4 | * | * | * | * | * | * | ||||||
| 5 | * | * | * | * | ||||||||
| 6 | * | * | ||||||||||
| Strain | 2 | 3 | 4 | 5 | 6 | 7 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 1 | * | * | * | * | s | s | s | * | s | * | s | * |
| 2 | s | * | * | * | * | * | * | * | s | * | ||
| 3 | s | * | s | * | s | * | s | * | ||||
| 4 | * | * | * | * | * | * | ||||||
| 5 | * | * | * | * | ||||||||
| 6 | * | * | ||||||||||
References
- Ilan, M.B.; Kjerulf, A. Who Should Be Screened for Carbapenemase-Producing Enterobacterales and When? A Systematic Review. J. Hosp. Infect. 2023, 142, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Andremont, O.; Armand-Lefevre, L.; Dupuis, C.; de Montmollin, E.; Ruckly, S.; Lucet, J.-C.; Smonig, R.; Magalhaes, E.; Ruppé, E.; Mourvillier, B.; et al. Semi-Quantitative Cultures of Throat and Rectal Swabs Are Efficient Tests to Predict ESBL-Enterobacterales Ventilator-Associated Pneumonia in Mechanically Ventilated ESBL Carriers. Intensive Care Med. 2020, 46, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Pirrone, M.; Pinciroli, R.; Berra, L. Microbiome, Biofilms, and Pneumonia in the ICU. Curr. Opin. Infect. Dis. 2016, 29, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Berlanga, M.; Domènech, Ò.; Guerrero, R. Biofilm Formation on Polystyrene in Detached vs. Planktonic Cells of Polyhydroxyalkanoate-Accumulating Halomonas venusta. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2014, 17, 205–212. [Google Scholar] [CrossRef]
- Marquès, C.; Tasse, J.; Pracros, A.; Collin, V.; Franceschi, C.; Laurent, F.; Chatellier, S.; Forestier, C. Effects of Antibiotics on Biofilm and Unattached Cells of a Clinical Staphylococcus aureus Isolate from Bone and Joint Infection. J. Med. Microbiol. 2015, 64, 1021–1026. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, S.; He, J.; Zhang, Y.; Su, L.; Tong, L.; Sun, Y.; Zhou, M.; Chen, Z. The Correlation Between Biofilm-Forming Ability of Community-Acquired Methicillin-Resistant Staphylococcus aureus Isolated from the Respiratory Tract and Clinical Characteristics in Children. Infect. Drug Resist. 2022, 15, 3657–3668. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M. Fetal Bovine Serum. Mater. Methods 2012, 2, 117. [Google Scholar] [CrossRef]
- Romero, A.; Pérez-Arellano, J.L.; González-Villarón, L.; Brock, J.H.; Bellido, J.L.M.; Castro, S.D. Effect of Transferrin Concentration on Bacterial Growth in Human Ascitic Fluid from Cirrhotic and Neoplastic Patients. Eur. J. Clin. Investig. 1993, 23, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Prasad, J.M.; Negrón, O.; Du, X.; Mullins, E.S.; Palumbo, J.S.; Gilbertie, J.M.; Höök, M.; Grover, S.P.; Pawlinski, R.; Mackman, N.; et al. Host Fibrinogen Drives Antimicrobial Function in Staphylococcus aureus Peritonitis through Bacterial-Mediated Prothrombin Activation. Proc. Natl. Acad. Sci. USA 2021, 118, e2009837118. [Google Scholar] [CrossRef]
- Ghaffarlou, M.; İlk, S.; Rahimi, H.; Danafar, H.; Barsbay, M.; Sharafi, A. Bovine Serum Albumin-Mediated Synthesis and Quorum Sensing Inhibitory Properties of Ag-Ag2S Nanoparticles. Nanomedicine 2022, 17, 2145–2155. [Google Scholar] [CrossRef]
- Fernández-Barat, L.; Ben-Aicha, S.; Motos, A.; Vila, J.; Marco, F.; Rigol, M.; Muñoz, L.; Li Bassi, G.; Ferrer, M.; Torres, A. Assessment of in Vivo versus in Vitro Biofilm Formation of Clinical Methicillin-Resistant Staphylococcus aureus Isolates from Endotracheal Tubes. Sci. Rep. 2018, 8, 11906. [Google Scholar] [CrossRef] [PubMed]
- Goller, C.C.; Romeo, T. Environmental Influences on Biofilm Development. Curr. Top. Microbiol. Immunol. 2008, 322, 37–66. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in Vitro to in Vivo Models of Bacterial Biofilm-Related Infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Banerjee, G.; Garg, R.; Singh, M. Comparative Study of Biofilm Formation in Pseudomonas aeruginosa Isolates from Patients of Lower Respiratory Tract Infection. J. Clin. Diagn. Res. JCDR 2014, 8, DC09-11. [Google Scholar] [CrossRef] [PubMed]
- Cusumano, J.A.; Caffrey, A.R.; Daffinee, K.E.; Luther, M.K.; Lopes, V.; LaPlante, K.L. Weak Biofilm Formation among Carbapenem-Resistant Klebsiella pneumoniae. Diagn. Microbiol. Infect. Dis. 2019, 95, 114877. [Google Scholar] [CrossRef]
- Nasrollahian, S.; Halaji, M.; Hosseini, A.; Teimourian, M.; Armaki, M.T.; Rajabnia, M.; Gholinia, H.; Pournajaf, A. Genetic Diversity, Carbapenem Resistance Genes, and Biofilm Formation in UPEC Isolated from Patients with Catheter-Associated Urinary Tract Infection in North of Iran. Int. J. Clin. Pract. 2022, 2022, 9520362. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Santos, I.C.; da Conceiçāo Neto, O.C.; da Costa, B.S.; Teixeira, C.B.T.; da Silva Pontes, L.; Silveira, M.C.; Rocha-de-Souza, C.M.; Carvalho-Assef, A.P.D. Evaluation of Phenotypic Detection of Carbapenemase-Producing Pseudomonas Spp. from Clinical Isolates. Braz. J. Microbiol. 2023, 54, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Schoevaerdts, D.; Verroken, A.; Huang, T.-D.; Frennet, M.; Berhin, C.; Jamart, J.; Bogaerts, P.; Swine, C.; Glupczynski, Y. Multidrug-Resistant Bacteria Colonization amongst Patients Newly Admitted to a Geriatric Unit: A Prospective Cohort Study. J. Infect. 2012, 65, 109–118. [Google Scholar] [CrossRef]
- Le, M.N.-T.; Kayama, S.; Yoshikawa, M.; Hara, T.; Kashiyama, S.; Hisatsune, J.; Tsuruda, K.; Onodera, M.; Ohge, H.; Tsuga, K.; et al. Oral Colonisation by Antimicrobial-Resistant Gram-Negative Bacteria among Long-Term Care Facility Residents: Prevalence, Risk Factors, and Molecular Epidemiology. Antimicrob. Resist. Infect. Control 2020, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Hogardt, M.; Proba, P.; Mischler, D.; Cuny, C.; Kempf, V.A.; Heudorf, U. Current Prevalence of Multidrug-Resistant Organisms in Long-Term Care Facilities in the Rhine-Main District, Germany, 2013. Eurosurveillance 2015, 20, 21171. [Google Scholar] [CrossRef]
- Araoka, H.; Kimura, M.; Abe, M.; Takahashi, N.; Yoneyama, A. Appropriate Sampling Sites for the Surveillance of Multidrug-Resistant Pseudomonas aeruginosa Colonization. Jpn. J. Infect. Dis. 2014, 67, 118–119. [Google Scholar] [CrossRef][Green Version]
- Marchaim, D.; Navon-Venezia, S.; Schwartz, D.; Tarabeia, J.; Fefer, I.; Schwaber, M.J.; Carmeli, Y. Surveillance Cultures and Duration of Carriage of Multidrug-Resistant Acinetobacter baumannii. J. Clin. Microbiol. 2007, 45, 1551. [Google Scholar] [CrossRef]
- Mahapatra, A.; Nikitha, K.; Rath, S.; Behera, B.; Gupta, K. Evaluation of HiCrome KPC Agar for the Screening of Carbapenem-Resistant Enterobacterales Colonization in the ICU Setting of a Tertiary Care Hospital. J. Lab. Phys. 2021, 13, 358–361. [Google Scholar] [CrossRef]
- Pericolini, E.; Colombari, B.; Ferretti, G.; Iseppi, R.; Ardizzoni, A.; Girardis, M.; Sala, A.; Peppoloni, S.; Blasi, E. Real-Time Monitoring of Pseudomonas aeruginosa Biofilm Formation on Endotracheal Tubes in Vitro. BMC Microbiol. 2018, 18, 84. [Google Scholar] [CrossRef]
- Nirwati, H.; Sinanjung, K.; Fahrunissa, F.; Wijaya, F.; Napitupulu, S.; Hati, V.P.; Hakim, M.S.; Meliala, A.; Aman, A.T.; Nuryastuti, T. Biofilm Formation and Antibiotic Resistance of Klebsiella pneumoniae Isolated from Clinical Samples in a Tertiary Care Hospital, Klaten, Indonesia. BMC Proc. 2019, 13, 1–8. [Google Scholar] [CrossRef]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Krzyściak, P.; Chmielarczyk, A.; Pobiega, M.; Romaniszyn, D.; Wójkowska-Mach, J. Acinetobacter baumannii Isolated from Hospital-Acquired Infection: Biofilm Production and Drug Susceptibility. APMIS 2017, 125, 1017–1026. [Google Scholar] [CrossRef]
- Gedefie, A.; Demsis, W.; Ashagrie, M.; Kassa, Y.; Tesfaye, M.; Tilahun, M.; Bisetegn, H.; Sahle, Z. Acinetobacter baumannii Biofilm Formation and Its Role in Disease Pathogenesis: A Review. Infect. Drug Resist. 2021, 14, 3711. [Google Scholar] [CrossRef]
- Runci, F.; Bonchi, C.; Frangipani, E.; Visaggio, D.; Visca, P. Acinetobacter baumannii Biofilm Formation in Human Serum and Disruption by Gallium. Antimicrob. Agents Chemother. 2016, 61, e01563-16. [Google Scholar] [CrossRef]
- Yang, C.-H.; Su, P.-W.; Moi, S.-H.; Chuang, L.-Y. Biofilm Formation in Acinetobacter baumannii: Genotype-Phenotype Correlation. Molecules 2019, 24, 1849. [Google Scholar] [CrossRef] [PubMed]
- Dawan, J.; Ahn, J. Bacterial Stress Responses as Potential Targets in Overcoming Antibiotic Resistance. Microorganisms 2022, 10, 1385. [Google Scholar] [CrossRef] [PubMed]
- Didelot, X.; Walker, A.S.; Peto, T.E.; Crook, D.W.; Wilson, D.J. Within-Host Evolution of Bacterial Pathogens. Nat. Rev. Microbiol. 2016, 14, 150–162. [Google Scholar] [CrossRef]
- Hammond, A.; Dertien, J.; Colmer-Hamood, J.A.; Griswold, J.A.; Hamood, A.N. Serum Inhibits P. aeruginosa Biofilm Formation on Plastic Surfaces and Intravenous Catheters. J. Surg. Res. 2010, 159, 735–746. [Google Scholar] [CrossRef]
- Zarzosa-Moreno, D.; Avalos-Gómez, C.; Ramírez-Texcalco, L.S.; Torres-López, E.; Ramírez-Mondragón, R.; Hernández-Ramírez, J.O.; Serrano-Luna, J.; de la Garza, M. Lactoferrin and Its Derived Peptides: An Alternative for Combating Virulence Mechanisms Developed by Pathogens. Molecules 2020, 25, 5763. [Google Scholar] [CrossRef] [PubMed]
- Arenas, J.; Szabo, Z.; van der Wal, J.; Maas, C.; Riaz, T.; Tønjum, T.; Tommassen, J. Serum Proteases Prevent Bacterial Biofilm Formation: Role of Kallikrein and Plasmin. Virulence 2021, 12, 2902–2917. [Google Scholar] [CrossRef] [PubMed]
- Post, S.J.; Shapiro, J.A.; Wuest, W.M. Connecting Iron Acquisition and Biofilm Formation in the ESKAPE Pathogens as a Strategy for Combatting Antibiotic Resistance. MedChemComm 2019, 10, 505–512. [Google Scholar] [CrossRef]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. Iron and Pseudomonas Aeruginosa Biofilm Formation. Proc. Natl. Acad. Sci. USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Liu, H.; Zhang, X.; Dong, G.; Li, J.; Tian, X.; Wu, Z.; Zhou, J.; Cao, J.; Zhou, T. Difference in Biofilm Formation between Carbapenem-Resistant and Carbapenem-Sensitive Klebsiella pneumoniae Based on Analysis of mrkH Distribution. Microb. Pathog. 2021, 152, 104743. [Google Scholar] [CrossRef]
- Venkatesan, N.; Perumal, G.; Doble, M. Bacterial Resistance in Biofilm-Associated Bacteria. Future Microbiol. 2015, 10, 1743–1750. [Google Scholar] [CrossRef]
- Candan, E.D.; Aksöz, N. Klebsiella pneumoniae: Characteristics of Carbapenem Resistance and Virulence Factors. Acta Biochim. Pol. 2015, 62, 867–874. [Google Scholar] [CrossRef]
- Crabbé, A.; Liu, Y.; Matthijs, N.; Rigole, P.; De La Fuente-Nùñez, C.; Davis, R.; Ledesma, M.A.; Sarker, S.; Van Houdt, R.; Hancock, R.E.W.; et al. Antimicrobial Efficacy against Pseudomonas aeruginosa Biofilm Formation in a Three-Dimensional Lung Epithelial Model and the Influence of Fetal Bovine Serum. Sci. Rep. 2017, 7, 43321. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.D.; Choudhuri, M.; Basu, T.; Gupta, D.; Datta, A. Decisive Role of Polymer-Bovine Serum Albumin Interactions in Biofilm Substrates on “Philicity” and Extracellular Polymeric Substances Composition. Langmuir ACS J. Surf. Colloids 2022, 38, 1966–1976. [Google Scholar] [CrossRef] [PubMed]
- Coșeriu, R.L.; Mare, A.D.; Toma, F.; Vintilă, C.; Ciurea, C.N.; Togănel, R.O.; Cighir, A.; Simion, A.; Man, A. Uncovering the Resistance Mechanisms in Extended-Drug-Resistant Pseudomonas aeruginosa Clinical Isolates: Insights from Gene Expression and Phenotypic Tests. Microorganisms 2023, 11, 2211. [Google Scholar] [CrossRef] [PubMed]
| % Resistance | IMP | MEM | CN | AK | CIP | LEV | SXT | TZP* | CZA* | IMR* | CS | Other Beta-Lactams |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Screening Non-fermentative | 100% | 100% | 90% | 93% | 100% | 100% | 100% | 100% | 100% | - | - | - |
| Infection Non-fermentative | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 9% | - |
| Screening K. pneumoniae | 100% | 100% | 100% | 75% | 100% | 100% | 100% | - | - | - | - | 100% |
| Infection K. pneumoniae | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 62% | 100% |
| Strain | Sample Type | Number of Strains | blaOXA-48-like | blaKPC | blaNDM | blaIMP | blaVIM | blaSPM |
|---|---|---|---|---|---|---|---|---|
| K. pneumoniae | Screening | n = 5 | 0 | 0 | 0 | 0 | 1 | 0 |
| Infection | n = 13 | 10 | 0 | 4 | 0 | 0 | 0 | |
| A. baumannii | Screening | n = 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| Infection | n = 13 | 1 | 0 | 0 | 0 | 0 | 0 | |
| P. aeruginosa | Screening | n = 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Infection | n = 4 | 1 | 0 | 0 | 0 | 0 | 0 | |
| S. maltophilia | Screening | n = 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| Infection | n = 0 | - | - | - | - | - | - |
| Gene Type | Primer Sequence (5′ > 3′) | Amplicon Length (bp) | |
|---|---|---|---|
| Triplex PCR | blaKPC Forward blaKPC Reverse | ATGTCACTGTATCGCCGTCT TTTTCAGAGCCTTACTGCCC | 893 |
| blaOXA48-like Forward blaOXA48-like Reverse | GCGTGGTTAAGGATGAACAC CATCAAGTTCAACCCAACCG | 438 | |
| blaNDM Forward blaNDM Reverse | GGTTTGGCGATCTGGTTTTC CGGAATGGCTCATCACGATC | 621 | |
| Simplex PCR | blaIMP Forward blaIMP Reverse | GGAATAGAGTGGCTTAAYTCTC GGTTTAAYAAAACAACCACC | 232 |
| blaVIM Forward blaVIM Reverse | GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG | 390 | |
| blaSPM Forward blaSPM Reverse | AAAATCTGGGTACGCAAACG ACATTATCCGCTGGAACAGG | 271 | |
| mcr-1 Forward mcr-1 Reverse | CGGTCAGTCCGTTTGTTC CTTGGTCGGTCTGTAGGG | 309 |
| Initial Denaturation | Denaturation | Annealing | Elongation | Final Elongation | |
|---|---|---|---|---|---|
| 1 Cycle | 36 Cycles | 1 Cycle | |||
| SPM, IMP | 95 °C | 95 °C | 52 °C | 72 °C | 72 °C |
| 5 min | 30 s | 60 s | 2 min | 8 min | |
| VIM | 95 °C | 95 °C | 59 °C | 72 °C | 72 °C |
| 15 min | 30 s | 90 s | 90 s | 10 min | |
| KPC, OXA-48-LIKE, NDM | 95 °C | 95 °C | 57.3 °C | 72 °C | 72 °C |
| 10 min | 30 s | 40 s | 50 s | 5 min | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vintilă, C.; Coșeriu, R.L.; Mare, A.D.; Ciurea, C.N.; Togănel, R.O.; Simion, A.; Cighir, A.; Man, A. Biofilm Formation and Antibiotic Resistance Profiles in Carbapenemase-Producing Gram-Negative Rods—A Comparative Analysis between Screening and Pathological Isolates. Antibiotics 2024, 13, 687. https://doi.org/10.3390/antibiotics13080687
Vintilă C, Coșeriu RL, Mare AD, Ciurea CN, Togănel RO, Simion A, Cighir A, Man A. Biofilm Formation and Antibiotic Resistance Profiles in Carbapenemase-Producing Gram-Negative Rods—A Comparative Analysis between Screening and Pathological Isolates. Antibiotics. 2024; 13(8):687. https://doi.org/10.3390/antibiotics13080687
Chicago/Turabian StyleVintilă, Camelia, Răzvan Lucian Coșeriu, Anca Delia Mare, Cristina Nicoleta Ciurea, Radu Ovidiu Togănel, Anastasia Simion, Anca Cighir, and Adrian Man. 2024. "Biofilm Formation and Antibiotic Resistance Profiles in Carbapenemase-Producing Gram-Negative Rods—A Comparative Analysis between Screening and Pathological Isolates" Antibiotics 13, no. 8: 687. https://doi.org/10.3390/antibiotics13080687
APA StyleVintilă, C., Coșeriu, R. L., Mare, A. D., Ciurea, C. N., Togănel, R. O., Simion, A., Cighir, A., & Man, A. (2024). Biofilm Formation and Antibiotic Resistance Profiles in Carbapenemase-Producing Gram-Negative Rods—A Comparative Analysis between Screening and Pathological Isolates. Antibiotics, 13(8), 687. https://doi.org/10.3390/antibiotics13080687









