Prevalence of Selected Immune Evasion Genes and Clonal Diversity in Methicillin-Susceptible Staphylococcus aureus Isolated from Nasal Carriers and Outpatients with Cut Wound Infections
Abstract
1. Introduction
2. Results
2.1. PCR Results
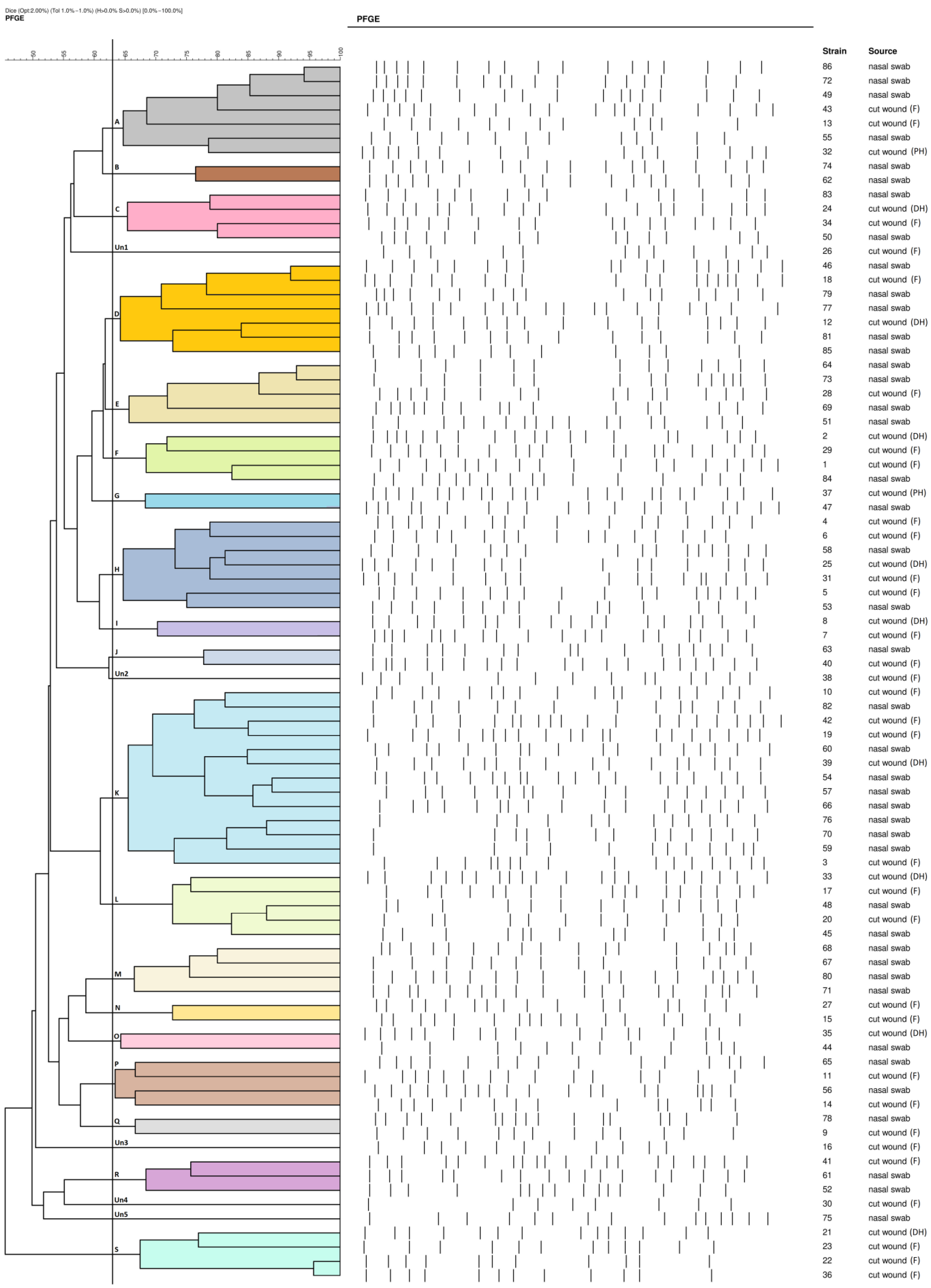
2.2. PFGE Results
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. DNA Isolation and PCR Assay
4.3. PFGE Assay
4.4. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A.; Melles, D.C.; Nouwen, J.; van Leeuwen, W.B.; van Wamel, W.; Vos, M.C.; Wertheim, H.F.L.; Verbrugh, H.A. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect. Genet. Evol. 2009, 9, 32–47. [Google Scholar] [CrossRef]
- Masiuk, H.; Wcisłek, A.; Jursa-Kulesza, J. Determination of nasal carriage and skin colonization, antimicrobial susceptibility and genetic relatedness of Staphylococcus aureus isolated from patients with atopic dermatitis in Szczecin, Poland. BMC Infect. Dis. 2021, 21, 701. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Wang, B.; Zhang, X.; Zhang, J.; Zhang, H.; Liu, X.; Gao, Z.; Yu, Z. The spread of antibiotic resistance to humans and potential protection strategies. Ecotoxicol. Environ. Saf. 2023, 254, 114734. [Google Scholar] [CrossRef]
- Goldmann, O.; Medina, E. Staphylococcus aureus strategies to evade the host acquired immune response. Int. J. Med. Microbiol. 2018, 308, 625–630. [Google Scholar] [CrossRef]
- de Jong, N.W.M.; van Kessel, K.P.M.; van Strijp, J.A.G. Immune evasion by Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, J.; He, Y.; Lv, Z.; Liang, Z.; Chen, J.; Li, P.; Liu, J.; Yang, H.; Tao, A.; et al. Exploring the role of Staphylococcus aureus in inflammatory diseases. Toxins 2022, 14, 464. [Google Scholar] [CrossRef]
- Chai, M.H.; Sukiman, M.Z.; Liew, Y.W.; Shapawi, M.S.; Roslan, F.S.; Hashim, S.N.; Mohamad, N.M.; Ariffin, S.M.Z.; Ghazali, M.F. Detection, molecular characterization, and antibiogram of multi-drug resistant and methicillin-resistant Staphylococcus aureus (MRSA) isolated from pets and pet owners in Malaysia. Iran. J. Vet. Res. 2021, 22, 277–287. [Google Scholar] [CrossRef]
- Cuny, C.; Abdelbary, M.; Layer, F.; Werner, G.; Witte, W. Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 2015, 177, 219–223. [Google Scholar] [CrossRef]
- Baptista, L.G.; Silva, N.C.C.; Bonsaglia, E.C.R.; Rossi, B.F.; Castilho, I.G.; Junior, A.F.; Rall, V.L.M. Presence of immune evasion cluster and molecular typing of methicillin-susceptible Staphylococcus aureus isolated from food handlers. J. Food Prot. 2016, 79, 682–686. [Google Scholar] [CrossRef]
- Kwiatkowski, P.; Masiuk, H.; Pruss, A.; Łopusiewicz, Ł.; Sienkiewicz, M.; Wojciechowska-Koszko, I.; Roszkowska, P.; Bania, J.; Guenther, S.; Dołęgowska, B. Clonal diversity, antimicrobial susceptibility and presence of genes encoding virulence factors in Staphylococcus aureus strains isolated from cut wound infections. Curr. Microbiol. 2022, 79, 144. [Google Scholar] [CrossRef] [PubMed]
- Holtfreter, S.; Grumann, D.; Schmudde, M.; Nguyen, H.T.T.; Eichler, P.; Strommenger, B.; Kopron, K.; Kolata, J.; Giedrys-Kalemba, S.; Steinmetz, I.; et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 2007, 45, 2669–2680. [Google Scholar] [CrossRef] [PubMed]
- Masiuk, H.; Ostrowski, P.; Masiuk, M.; Pruss, A.; Kwiatkowski, P.; Bilska, I.; Rusińska, K.; Skoryk, A.; Jursa-Kulesza, J. The antimicrobial susceptibility and contribution of Staphylococcus aureus to surgical site infections in patients hospitalized in the West Pomeranian Region (Poland) during the COVID-19 pandemic period—A 3-year follow-up. Pomeranian J. Life Sci. 2023, 69, 6–13. [Google Scholar] [CrossRef]
- Neoh, H.-M.; Tan, X.-E.; Sapri, H.F.; Tan, T.L. Pulsed-Field Gel Electrophoresis (PFGE): A review of the “Gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 74, 103935. [Google Scholar] [CrossRef] [PubMed]
- Żabicka, D.; Hryniewicz, W. Rekomendacje Doboru Testów do Oznaczania Wrażliwości Bakterii na Antybiotyki i Chemioterapeutyki 2010. Oznaczanie Wrażliwości Ziarniaków Gram-Dodatnich z Rodzaju Staphylococcus spp.; Korld: Warsaw, Poland, 2010; pp. 1–20.
- Sabbagh, P.; Riahi, S.M.; Gamble, H.R.; Rostami, A. The global and regional prevalence, burden, and risk factors for methicillin-resistant Staphylococcus aureus colonization in HIV-infected people: A systematic review and meta-analysis. Am. J. Infect. Control 2019, 47, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.-H.; Moon, S.M. Evaluation of methicillin-resistance rates among community-associated Staphylococcus aureus infections in Korean military personnel. J. Korean Med. Sci. 2018, 33, e250. [Google Scholar] [CrossRef] [PubMed]
- Kluytmans-Vandenbergh, M.F.Q.; Kluytmans, J.A.J.W. Community-acquired methicillin-resistant Staphylococcus aureus: Current perspectives. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2006, 12 (Suppl. 1), 9–15. [Google Scholar] [CrossRef] [PubMed]
- Carleton, H.A.; Diep, B.A.; Charlebois, E.D.; Sensabaugh, G.F.; Perdreau-Remington, F. Community-adapted methicillin-resistant Staphylococcus aureus (MRSA): Population dynamics of an expanding community reservoir of MRSA. J. Infect. Dis. 2004, 190, 1730–1738. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Charlebois, E.D.; Bangsberg, D.R.; Moss, N.J.; Moore, M.R.; Moss, A.R.; Chambers, H.F.; Perdreau-Remington, F. Population-based community prevalence of methicillin-resistant Staphylococcus aureus in the urban poor of San Francisco. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2002, 34, 425–433. [Google Scholar] [CrossRef]
- Verkaik, N.J.; Benard, M.; Boelens, H.A.; de Vogel, C.P.; Nouwen, J.L.; Verbrugh, H.A.; Melles, D.C.; van Belkum, A.; van Wamel, W.J.B. Immune evasion cluster-positive bacteriophages are highly prevalent among human Staphylococcus aureus strains, but they are not essential in the first stages of nasal colonization. Clin. Microbiol. Infect. 2011, 17, 343–348. [Google Scholar] [CrossRef]
- van Wamel, W.J.B.; Rooijakkers, S.H.M.; Ruyken, M.; van Kessel, K.P.M.; van Strijp, J.A.G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef] [PubMed]
- Ahmadrajabi, R.; Layegh-Khavidaki, S.; Kalantar-Neyestanaki, D.; Fasihi, Y. Molecular analysis of immune evasion cluster (IEC) genes and intercellular adhesion gene cluster (ICA) among methicillin-resistant and methicillin-sensitive isolates of Staphylococcus aureus. J. Prev. Med. Hyg. 2017, 58, E308–E314. [Google Scholar] [CrossRef] [PubMed]
- Kmiha, S.; Jouini, A.; Zerriaa, N.; Hamrouni, S.; Thabet, L.; Maaroufi, A. Methicillin-resistant Staphylococcus aureus strains isolated from burned patients in a Tunisian hospital: Molecular typing, virulence genes, and antimicrobial resistance. Antibiotics 2023, 12, 1030. [Google Scholar] [CrossRef] [PubMed]
- Allaw, F.; Zakhour, J.; Kanj, S.S. Community-acquired skin and soft-tissue infections in people who inject drugs. Curr. Opin. Infect. Dis. 2023, 36, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Alizai, Q.; Haseeb, A.; Hamayun, S.; Khan, S.; Ali, F.; Roghani, M.; Khan, M.A.; Ullah, F.; Khan, W.; Ijaz, N. Community-acquired skin and soft tissue infections: Epidemiology and management in patients presenting to the emergency department of a Tertiary Care Hospital. Cureus 2023, 15, e34379. [Google Scholar] [CrossRef] [PubMed]
- Ray, G.T.; Suaya, J.A.; Baxter, R. Incidence, microbiology, and patient characteristics of skin and soft-tissue infections in a U.S. population: A retrospective population-based study. BMC Infect. Dis. 2013, 13, 252. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Sukiman, M.Z.; Kamarun Baharin, A.H.; Ramlan, I.; Lai, L.Z.; Liew, Y.; Malayandy, P.; Mohamad, N.M.; Choong, S.; Ariffin, S.M.Z.; et al. Methicillin-resistant Staphylococcus aureus from peninsular Malaysian animal handlers: Molecular profile, antimicrobial resistance, immune evasion cluster and genotypic categorization. Antibiotics 2022, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Bano, A.; Asghar, F.; Ejaz, H.; Junaid, K.; Bashier Eltayeb, L.; Javed, N. Exploring the virulence potential of immune evasion cluster genes in methicillin-resistant Staphylococcus aureus from cancer patients. Saudi J. Biol. Sci. 2023, 30, 103835. [Google Scholar] [CrossRef]
- Hau, S.J.; Sun, J.; Davies, P.R.; Frana, T.S.; Nicholson, T.L. Comparative prevalence of immune evasion complex genes associated with β-hemolysin converting bacteriophages in MRSA ST5 isolates from swine, swine facilities, humans with swine contact, and humans with no swine contact. PLoS ONE 2015, 10, e0142832. [Google Scholar] [CrossRef]
- Rooijakkers, S.H.M.; van Wamel, W.J.B.; Ruyken, M.; van Kessel, K.P.M.; van Strijp, J.A.G. Anti-opsonic properties of staphylokinase. Microbes Infect. 2005, 7, 476–484. [Google Scholar] [CrossRef]
- Ellis, S.; Lin, E.J.; Tartar, D. Immunology of wound healing. Curr. Dermatol. Rep. 2018, 7, 350–358. [Google Scholar] [CrossRef]
- MacLeod, A.S.; Mansbridge, J.N. The innate immune system in acute and chronic wounds. Adv. Wound Care 2016, 5, 65–78. [Google Scholar] [CrossRef]
- Holtfreter, S.; Nguyen, T.T.H.; Wertheim, H.; Steil, L.; Kusch, H.; Truong, Q.P.; Engelmann, S.; Hecker, M.; Völker, U.; van Belkum, A.; et al. Human immune proteome in experimental colonization with Staphylococcus aureus. Clin. Vaccine Immunol. 2009, 16, 1607–1614. [Google Scholar] [CrossRef]
- Rozgonyi, F.; Kocsis, E.; Kristóf, K.; Nagy, K. Is MRSA more virulent than MSSA? Clin. Microbiol. Infect. 2007, 13, 843–845. [Google Scholar] [CrossRef]
- Correa-Jiménez, O.; Pinzón-Redondo, H.; Reyes, N. High frequency of Panton-Valentine leukocidin in Staphylococcus aureus causing pediatric infections in the city of Cartagena-Colombia. J. Infect. Public Health 2016, 9, 415–420. [Google Scholar] [CrossRef]
- Holmgren, A.M.; McConkey, C.A.; Shin, S. Outrunning the Red Queen: Bystander activation as a means of outpacing innate immune subversion by intracellular pathogens. Cell. Mol. Immunol. 2017, 14, 14–21. [Google Scholar] [CrossRef]
- Fijałkowski, K.; Czernomysy-Furowicz, D.; Ferlas, M. Staphylococcus aureus kontra układ immunologiczny. Postep. Mikrobiol. 2008, 47, 497–501. [Google Scholar]
- de Jong, N.W.M.; Ramyar, K.X.; Guerra, F.E.; Nijland, R.; Fevre, C.; Voyich, J.M.; McCarthy, A.J.; Garcia, B.L.; van Kessel, K.P.M.; van Strijp, J.A.G.; et al. Immune evasion by a staphylococcal inhibitor of myeloperoxidase. Proc. Natl. Acad. Sci. USA 2017, 114, 9439–9444. [Google Scholar] [CrossRef]
- Amdahl, H.; Jongerius, I.; Meri, T.; Pasanen, T.; Hyvärinen, S.; Haapasalo, K.; van Strijp, J.A.; Rooijakkers, S.H.; Jokiranta, T.S. Staphylococcal ECB protein and host complement regulator factor h enhance functions of each other in bacterial immune evasion. J. Immunol. 2013, 191, 1775–1784. [Google Scholar] [CrossRef]
- de Haas, C.J.C.; Veldkamp, K.E.; Peschel, A.; Weerkamp, F.; Van Wamel, W.J.B.; Heezius, E.C.J.M.; Poppelier, M.J.J.G.; Van Kessel, K.P.M.; van Strijp, J.A.G. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 2004, 199, 687–695. [Google Scholar] [CrossRef]
- Smith, E.J.; Visai, L.; Kerrigan, S.W.; Speziale, P.; Foster, T.J. The SBI protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect. Immun. 2011, 79, 3801–3809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jacobson, K.; Vasi, J.; Lindberg, M.; Frykberg, L. Second IgG-binding protein in Staphylococcus aureus. Microbiology 1998, 408, 985–991. [Google Scholar] [CrossRef]
- Burman, J.D.; Leung, E.; Atkins, K.L.; O’Seaghdha, M.N.; Lango, L.; Bernadó, P.; Bagby, S.; Svergun, D.I.; Foster, T.J.; Isenman, D.E.; et al. Interaction of human complement with sbi, a staphylococcal immunoglobulin-binding protein. J. Biol. Chem. 2008, 283, 17579–17593. [Google Scholar] [CrossRef]
- Thomas, D.; Chou, S.; Dauwalder, O.; Lina, G. Diversity in Staphylococcus aureus enterotoxins. Chem. Immunol. Allergy 2007, 93, 24–41. [Google Scholar] [CrossRef]
- Paszko, K.; Michnowska, E.; Kurlenda, J.; Grinholc, M.; Nakonieczna, J.; Bielawski, K.P. MRSA distribution and epidemiological procedures evaluation at two hospitals in northern Poland. GMS Krankenhhyg. Interdiszip. 2011, 6, Doc19. [Google Scholar] [CrossRef]
- Naimi, T.S.; LeDell, K.H.; Como-Sabetti, K.; Borchardt, S.M.; Boxrud, D.J.; Etienne, J.; Johnson, S.K.; Vandenesch, F.; Fridkin, S.; O’Boyle, C.; et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 2003, 290, 2976–2984. [Google Scholar] [CrossRef]
- Public Health Agency. S. aureus Bacteraemia Surveillance, Quarterly Report; Public Health Agency: Belfast, UK, 2023.
- Kaasch, A.J.; Barlow, G.; Edgeworth, J.D.; Fowler, V.G.J.; Hellmich, M.; Hopkins, S.; Kern, W.V.; Llewelyn, M.J.; Rieg, S.; Rodriguez-Baño, J.; et al. Staphylococcus aureus bloodstream infection: A pooled analysis of five prospective, observational studies. J. Infect. 2014, 68, 242–251. [Google Scholar] [CrossRef]
- Asgeirsson, H.; Thalme, A.; Weiland, O. Staphylococcus aureus bacteraemia and endocarditis—Epidemiology and outcome: A review. Infect. Dis. 2018, 50, 175–192. [Google Scholar] [CrossRef]
- Fowler, V.G.J.; Olsen, M.K.; Corey, G.R.; Woods, C.W.; Cabell, C.H.; Reller, L.B.; Cheng, A.C.; Dudley, T.; Oddone, E.Z. Clinical identifiers of complicated Staphylococcus aureus bacteremia. Arch. Intern. Med. 2003, 163, 2066–2072. [Google Scholar] [CrossRef]
- Paulsen, J.; Mehl, A.; Askim, Å.; Solligård, E.; Åsvold, B.O.; Damås, J.K. Epidemiology and outcome of Staphylococcus aureus bloodstream infection and sepsis in a Norwegian country 1996-2011: An Observational Study. BMC Infect. Dis. 2015, 15, 116. [Google Scholar] [CrossRef]
- Cosgrove, S.E.; Sakoulas, G.; Perencevich, E.N.; Schwaber, M.J.; Karchmer, A.W.; Carmeli, Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2003, 36, 53–59. [Google Scholar] [CrossRef]
- Goto, M.; Schweizer, M.L.; Vaughan-Sarrazin, M.S.; Perencevich, E.N.; Livorsi, D.J.; Diekema, D.J.; Richardson, K.K.; Beck, B.F.; Alexander, B.; Ohl, M.E. Association of evidence-based care processes with mortality in Staphylococcus aureus bacteremia at Veterans Health Administration Hospitals, 2003–2014. JAMA Intern. Med. 2017, 177, 1489–1497. [Google Scholar] [CrossRef]
- Vogel, M.; Schmitz, R.P.H.; Hagel, S.; Pletz, M.W.; Gagelmann, N.; Scherag, A.; Schlattmann, P.; Brunkhorst, F.M. Infectious disease consultation for Staphylococcus aureus bacteremia—A systematic review and meta-analysis. J. Infect. 2016, 72, 19–28. [Google Scholar] [CrossRef]
- Saunderson, R.B.; Gouliouris, T.; Nickerson, E.K.; Cartwright, E.J.P.; Kidney, A.; Aliyu, S.H.; Brown, N.M.; Limmathurotsakul, D.; Peacock, S.J.; Török, M.E. Impact of routine bedside infectious disease consultation on clinical management and outcome of Staphylococcus aureus bacteraemia in adults. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2015, 21, 779–785. [Google Scholar] [CrossRef]
- Gómez, P.; Ruiz-Ripa, L.; Fernández-Fernández, R.; Gharsa, H.; Ben Slama, K.; Höfle, U.; Zarazaga, M.; Holmes, M.A.; Torres, C. Genomic analysis of Staphylococcus aureus of the lineage CC130, including MecC-Carrying MRSA and MSSA isolates recovered of animal, human, and environmental origins. Front. Microbiol. 2021, 12, 655994. [Google Scholar] [CrossRef]
- Rooijakkers, S.H.M.; Ruyken, M.; Roos, A.; Daha, M.R.; Presanis, J.S.; Sim, R.B.; van Wamel, W.J.B.; van Kessel, K.P.M.; van Strijp, J.A.G. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005, 6, 920–927. [Google Scholar] [CrossRef]
- Mama, O.M.; Aspiroz, C.; Ruiz-Ripa, L.; Ceballos, S.; Iñiguez-Barrio, M.; Cercenado, E.; Azcona, J.M.; López-Cerero, L.; Seral, C.; López-Calleja, A.I.; et al. Prevalence and genetic characteristics of Staphylococcus aureus CC398 isolates from invasive infections in Spanish hospitals, focusing on the livestock-independent CC398-MSSA clade. Front. Microbiol. 2021, 12, 623108. [Google Scholar] [CrossRef]
- Baumgartner, A.; Niederhauser, I.; Johler, S. Virulence and resistance gene profiles of Staphylococcus aureus strains isolated from ready-to-eat foods. J. Food Prot. 2014, 77, 1232–1236. [Google Scholar] [CrossRef]
- Rinsky, J.L.; Nadimpalli, M.; Wing, S.; Hall, D.; Baron, D.; Price, L.B.; Larsen, J.; Stegger, M.; Stewart, J.; Heaney, C.D. livestock-associated methicillin and multidrug resistant Staphylococcus aureus is present among industrial, not antibiotic-free livestock operation workers in north Carolina. PLoS ONE 2013, 8, e67641. [Google Scholar] [CrossRef]
- Sanfilippo, C.M.; Hesje, C.K.; Haas, W.; Morris, T.W. Topoisomerase mutations that are associated with high-level resistance to earlier fluoroquinolones in Staphylococcus aureus have less effect on the antibacterial activity of besifloxacin. Chemotherapy 2011, 57, 363–371. [Google Scholar] [CrossRef]
- Baba, T.; Bae, T.; Schneewind, O.; Takeuchi, F.; Hiramatsu, K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: Polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008, 190, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, P.W.; Allison, M.E.; Akkaraju, S.; Goodnow, C.C.; Fearon, D.T. C3d of complement as a molecular adjuvant: Bridging innate and acquired immunity. Science 1996, 271, 348–350. [Google Scholar] [CrossRef] [PubMed]
- Fearon, D.T.; Locksley, R.M. The instructive role of innate immunity in the acquired immune response. Science 1996, 272, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Nuutila, J.; Jalava-Karvinen, P.; Hohenthal, U.; Kotilainen, P.; Pelliniemi, T.-T.; Nikoskelainen, J.; Lilius, E.-M. Use of complement regulators, CD35, CD46, CD55, and CD59, on leukocytes as markers for diagnosis of viral and bacterial infections. Hum. Immunol. 2013, 74, 522–530. [Google Scholar] [CrossRef]
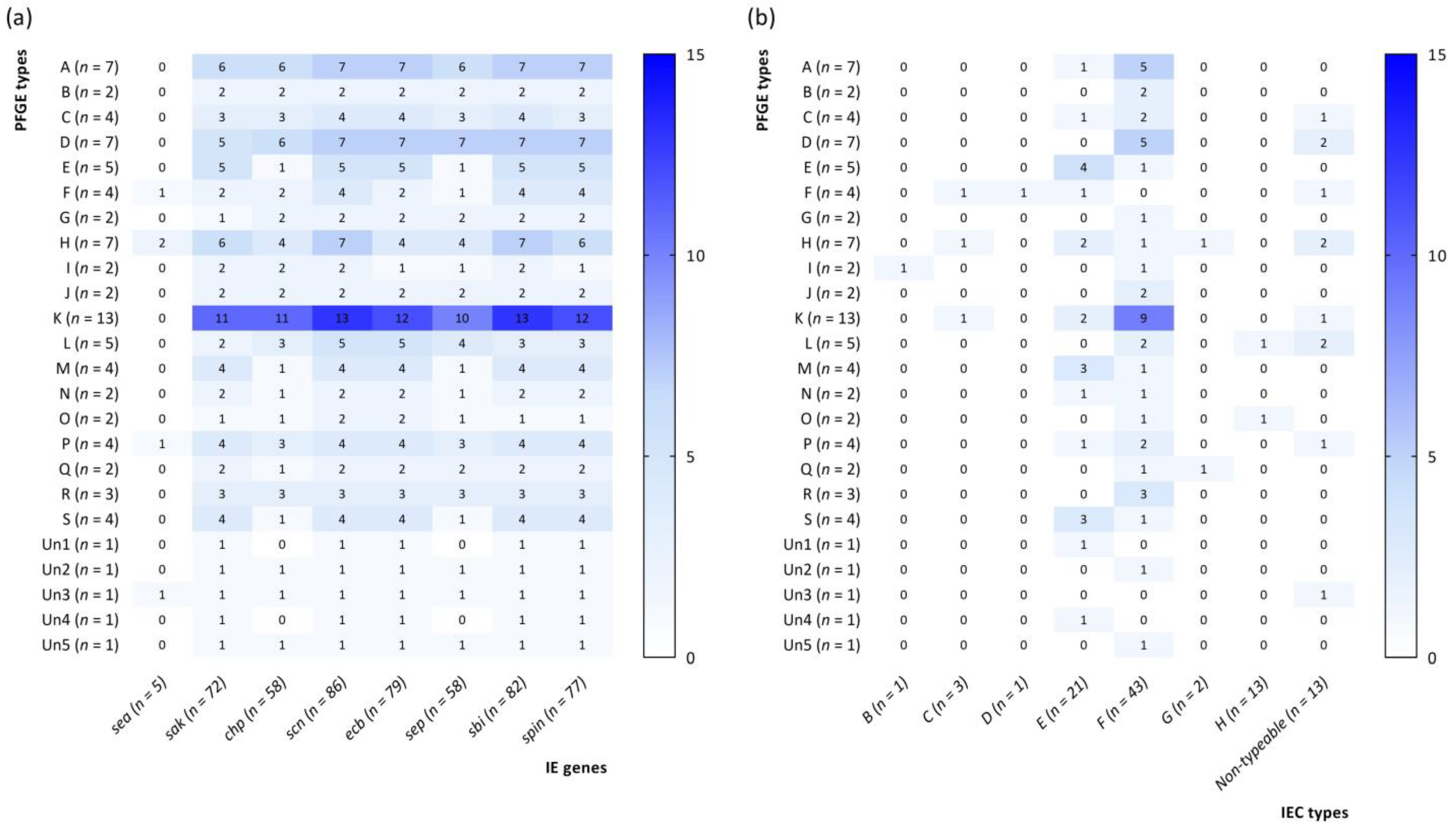
| IE Gene | Result | Total (n = 86) n (%) | Cut Wounds (n = 43) n (%) | Nasal Carriers (n = 43) n (%) | p-Value |
|---|---|---|---|---|---|
| sea | Positive | 5 (5.81) | 3 (6.98) | 2 (4.65) | 0.6449 |
| Negative | 81 (94.19) | 40 (93.02) | 41 (95.35) | ||
| sak | Positive | 72 (83.72) | 39 (90.7) | 33 (76.74) | 0.0797 |
| Negative | 14 (6.28) | 4 (9.3) | 10 (23.26) | ||
| chp | Positive | 58 (67.44) | 29 (67.44) | 29 (67.44) | 1.0 |
| Negative | 28 (32.56) | 14 (32.56) | 14 (32.56) | ||
| scn | Positive | 86 (100.0) | 43 (100.0) | 43 (100.0) | 1.0 |
| Negative | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| sep | Positive | 58 (67.44) | 26 (60.47) | 32 (74.42) | 0.1674 |
| Negative | 28 (32.56) | 17 (39.53) | 11 (25.58) | ||
| ecb | Positive | 79 (91.86) | 36 (83.72) | 43 (100.0) | 0.0058 |
| Negative | 7 (8.14) | 7 (16.28) | 0 (0.0) | ||
| sbi | Positive | 82 (95.35) | 42 (97.67) | 40 (93.02) | 0.3058 |
| Negative | 4 (4.65) | 1 (2.33) | 3 (6.98) | ||
| spin | Positive | 77 (89.53) | 38 (88.37) | 39 (90.7) | 0.7246 |
| Negative | 9 (10.47) | 5 (11.63) | 4 (9.3) |
| IEC Type | Result | Total (n = 86) n (%) | Cut Wounds (n = 43) n (%) | Nasal Carriers (n = 43) n (%) | p-Value |
|---|---|---|---|---|---|
| A (sea-sak-chp-scn) | Positive | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.0 |
| Negative | 86 (100.0) | 43 (100.0) | 43 (100.0) | ||
| B (sak-chp-scn) | Positive | 1 (1.16) | 1 (2.33) | 0 (0.00) | 0.3145 |
| Negative | 85 (98.84) | 42 (97.67) | 43 (100.0) | ||
| C (chp-scn) | Positive | 3 (3.49) | 3 (6.98) | 0 (0.00) | 0.0779 |
| Negative | 83 (96.51) | 40 (93.02) | 43 (100.0) | ||
| D (sea-sak-scn) | Positive | 1 (1.16) | 1 (2.33) | 0 (0.00) | 0.3145 |
| Negative | 85 (98.84) | 42 (97.67) | 43 (100.0) | ||
| E (sak-scn) | Positive | 21 (24.42) | 11 (25.58) | 10 (23.26) | 0.8018 |
| Negative | 65 (75.58) | 32 (74.42) | 33 (76.74) | ||
| F (sep-sak-chp-scn) | Positive | 43 (50.0) | 23 (53.49) | 20 (46.51) | 0.5176 |
| Negative | 43 (50.0) | 20 (46.51) | 23 (53.49) | ||
| G (sep-sak-scn) | Positive | 2 (2.33) | 1 (2.33) | 1 (2.33) | 1.0 |
| Negative | 83 (96.51) | 42 (97.67) | 42 (97.67) | ||
| H (scn) | Positive | 2 (2.33) | 1 (2.33) | 1 (2.33) | 1.0 |
| Negative | 83 (96.51) | 42 (97.67) | 42 (97.67) | ||
| Non-typeable (all IEC genes detected) | Positive | 13 (15.12) | 2 (4.65) | 11 (25.58) | 0.0067 |
| Negative | 73 (84.88) | 41 (95.35) | 32 (74.42) |
| Gene | Sequence (5′→3′) | Size (bp) | Reference |
|---|---|---|---|
| sak | F: TGC GAC AGC ATA TAA AGA GTT TAG AGT A R: TCT GGG ACA ACA AAA CCT TTT TC | 137 | This study |
| sbi | F: GAA GAA CAA CGT AAC CAA TAC ATC AAA R: GGG TTC TTG CTG TCT TTA AGT GAT T | 94 | This study |
| sea | F: CAA ATA AAT CGT AAT TAA CCG AAG GTT C R: GAA AAA AGT CTG AAT TGC AGG GAA CA | 560 | [62] |
| spin | F: CAT GTA GTA CGA GTC CAT TTT GAG AAT AA R: GCT ACG GCA ATG GTA GGT GTT T | 96 | This study |
| chp | F: GCG AAA GCT TTT ACT TTT GAA CC R: CCT AGC GTT GTA GGA AGA CCA | 125 | This study |
| sep | F: CCG CCA TAC ATA CAA GCT GTT R: GTT CAA AAG ACA CCG CCA AT | 115 | This study |
| scn | F: ATT CAT TCG ATG TTG GCA AG R: ACT TGC GGG AAC TTT AGC AA | 101 | This study |
| ecb | F: ACT AGA TCG ATT TGT CTT TGT AAT TTT R: GTT GCA ACA CAC CGT AAA GC | 120 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jura, G.; Masiuk, H.; Pruss, A.; Kurzawski, M.; Sienkiewicz, M.; Wojciechowska-Koszko, I.; Kwiatkowski, P. Prevalence of Selected Immune Evasion Genes and Clonal Diversity in Methicillin-Susceptible Staphylococcus aureus Isolated from Nasal Carriers and Outpatients with Cut Wound Infections. Antibiotics 2024, 13, 730. https://doi.org/10.3390/antibiotics13080730
Jura G, Masiuk H, Pruss A, Kurzawski M, Sienkiewicz M, Wojciechowska-Koszko I, Kwiatkowski P. Prevalence of Selected Immune Evasion Genes and Clonal Diversity in Methicillin-Susceptible Staphylococcus aureus Isolated from Nasal Carriers and Outpatients with Cut Wound Infections. Antibiotics. 2024; 13(8):730. https://doi.org/10.3390/antibiotics13080730
Chicago/Turabian StyleJura, Gabriela, Helena Masiuk, Agata Pruss, Mateusz Kurzawski, Monika Sienkiewicz, Iwona Wojciechowska-Koszko, and Paweł Kwiatkowski. 2024. "Prevalence of Selected Immune Evasion Genes and Clonal Diversity in Methicillin-Susceptible Staphylococcus aureus Isolated from Nasal Carriers and Outpatients with Cut Wound Infections" Antibiotics 13, no. 8: 730. https://doi.org/10.3390/antibiotics13080730
APA StyleJura, G., Masiuk, H., Pruss, A., Kurzawski, M., Sienkiewicz, M., Wojciechowska-Koszko, I., & Kwiatkowski, P. (2024). Prevalence of Selected Immune Evasion Genes and Clonal Diversity in Methicillin-Susceptible Staphylococcus aureus Isolated from Nasal Carriers and Outpatients with Cut Wound Infections. Antibiotics, 13(8), 730. https://doi.org/10.3390/antibiotics13080730







