Abstract
Rising antimicrobial resistance (AMR) in Salmonella serotypes host-adapted to cattle is of increasing concern to the beef and dairy industry. The bulk of the existing literature focuses on AMR post-slaughter. In comparison, the understanding of AMR in Salmonella among pre-harvest cattle is still limited, particularly in Texas, which ranks top five in beef and dairy exports in the United States; inherently, the health of Texas cattle has nationwide implications for the health of the United States beef and dairy industry. In this study, long-read whole genome sequencing and bioinformatic methods were utilized to analyze antimicrobial resistance genes (ARGs) in 98 isolates from beef and dairy cattle in the Texas Panhandle. Fisher exact tests and elastic net models accounting for population structure were used to infer associations between genomic ARG profiles and antimicrobial phenotypic profiles and metadata. Gene mapping was also performed to assess the role of mobile genetic elements in harboring ARGs. Antimicrobial resistance genes were found to be statistically different between the type of cattle operation and Salmonella serotypes. Beef operations were statistically significantly associated with more ARGs compared to dairy operations. Salmonella Heidelberg, followed by Salmonella Dublin isolates, were associated with the most ARGs. Additionally, specific classes of ARGs were only present within mobile genetic elements.
1. Introduction
Salmonellosis is one of the leading causes of bacterial gastroenteritis in the United States, caused by non-typhoidal Salmonella enterica subspecies enterica. The Centers for Disease Control and Prevention (CDC) estimate that Salmonella accounts for 1.35 million cases and 26,500 hospitalizations in people annually [1]. Non-invasive infections are typically associated with mild cases of fever, diarrhea, vomiting, nausea, or abdominal cramps [2]. However, the symptoms can escalate if an infection becomes systemic, and complications such as meningitis, pancreatitis, or enteric fever that require antimicrobial intervention may ensue [3,4].
Salmonella enterica comprises over 2600 serotypes, but only a fraction of them are relevant in food production systems. Serotype-specific vaccination has been shown to reduce prevalence of dominant Salmonella serotypes in cattle, poultry, and swine, including Typhimurium, Enteritidis, Choleraesuis, and Dublin, among others [5]. However, vaccination may have promoted a disruption of serotype incidence, allowing other serotypes to replace the ecological niches [5].
The increased incidence of antimicrobial-resistant (AMR) bacteria is partially a product of antimicrobial mismanagement on a global scale. The issue is further exacerbated by the lack of new antibiotics to replace compounds with reduced efficacy. For example, one study estimated that resistance to clinically important antimicrobials increased 40% in Salmonella between 2004 and 2008 and 2015 and 2016 for human clinical infections in the United States [6].
Poultry and swine products are the primary sources of Salmonella in humans [7,8]. In the beef industry, economic resources and scientific efforts have traditionally been directed toward evaluating interventions in processing and packaging plants to reduce the risk of human salmonellosis from beef products [5,9]. However, the cattle industry is currently very interested in deepening our understanding of Salmonella ecology in pre-harvest cattle. In recent years, several regional studies have evaluated Salmonella in pre-harvest cattle or their immediate environment [5,10,11,12,13,14,15,16]. Additionally, recent reports have noted higher levels of resistance in Salmonella serotypes that are host-adapted to cattle such as Salmonella Dublin [17,18].
Some studies have evaluated AMR Salmonella and reported strong concordance between antimicrobial resistance genes and phenotypic resistance [10,19,20], prompting subsequent studies to assess AMR solely through genomic data [5,9,11,12,13,14,15,19,21]. Antimicrobial resistance gene prediction has been used to monitor antimicrobial resistance dynamics in Salmonella at the state level [5,11,19]. For instance, Carroll et al. (2020, 2021), carried out two experiments that relied solely on AMR genes to characterize Salmonella resistance for cattle in the state of New York [12,14]. These studies focused on groups of Salmonella serotypes to monitor herd population health, as antimicrobial resistance determinants are often serotype-associated [14]. Source tracking is also regularly applied in outbreak investigations to trace an outbreak back to a source so the corresponding regulations can be enforced to prevent future outbreaks [13,21]. More recently, the use of whole genome sequencing has been expanded to explore the role of mobile genetic elements on the dissemination of antimicrobial resistance genes to draw conclusions on resistance movement [5,15].
Knowledge gaps around antimicrobial resistance in Salmonella from cattle in Texas continue to exist despite the available literature [16,22,23,24,25]. No study has compared antimicrobial resistance in Salmonella from cattle between beef and dairy operations in Texas. Texas ranks the third largest in beef exports at USD 1.57 billion and fourth in dairy exports at USD 704 million [26]. The health of Texas cattle has nationwide implications for the resilience of beef and dairy food supply chains in the United States. To reduce the burden of salmonellosis in humans from food sources, a thorough understanding of the ecology of Salmonella in Texas cattle is necessary. Therefore, this study aimed to evaluate the distribution of antimicrobial resistance genes related to phenotypic resistance in Salmonella from beef and dairy cattle operations and potential associations with cattle age, sex, breed, specimen type, collection year, location, and Salmonella serotypes.
2. Results
2.1. Description of Salmonella Isolates
De novo genome assembly succeeded in 98 out of the 100 Salmonella isolates, which were used for downstream bioinformatics and statistical analyses. The isolates originated from cattle operations in Texas (n = 51), New Mexico (n = 15), or Arizona (n = 1) over a three-year period, i.e., 2021 (n = 35), 2022 (n = 38), and 2023 (n = 25), and were cultured from fecal (n = 77), intestinal (n = 19), or abomasal (n = 2) samples. Isolates originated from dairy operations (n = 71; n= 34 from dairies; n = 37 from calf ranches) and beef operations (n = 16; n = 11 from feedlots; n = 5 from cow-calf operations), and from dairy (n = 56) and beef breeds (n = 8). Isolates from male (n = 10) and female (n = 47) cattle, across different age groups, dichotomized as neonatal (n = 56) and not neonatal (n = 22), were included. Instances of missing data were present, as is usual with data from diagnostic laboratories. There were Salmonella isolates without information for sample origin (n = 31), cattle operation (n = 11), cattle breed (n = 34), sex (n = 41), and age (n = 20). Of the 31 Salmonella isolates without sample origin information, 25 isolates originated from submissions made by Texas-based practitioners, five by New Mexico-based practitioners, and one by a Louisiana-based practitioner. Due to the uncertainty of whether the location of the practitioner is a good proxy for the location of the sampled cattle, we did not use this additional information to replace the missing data for the sample origin.
2.2. Nanopore Sequencing Bioinformatic Pipeline Performance
The long reads maintained an average quality score above Q20 for up to 20,000 bases. The GC content ranged between 51.88% to 52.36% of the genome. A total of 26 isolates assembled into one contig, 47 assembled in two or less, and 85 assembled in five or fewer. The mean NG50 and LG50 was 4,392,366.11 nucleotides and 1.05 contigs, respectively.
Medaka polishing improved the quality of the assemblies as measured by the BUSCO scores. Prior to polishing the average complete, duplicated, fragmented, and missing BUSCO genes were 433.7, 1.46, 1.09, and 3.72, respectively. After polishing with Medaka the total number of complete BUSCO genes increased by 13 genes and missing or fragmented BUSCO genes decreased by 3 and 10 genes, respectively.
2.3. Overall Distribution of Phenotypic Antimicrobial Resistance among Salmonella Isolates
Guidelines from the Clinical & Laboratory Standards Institute (CLSI) supplement VET01S [27] for Salmonella restricted MIC interpretive standards to the following four antimicrobials (out of the 18 antimicrobials evaluated): ampicillin, gentamicin, tetracycline, and trimethoprim-sulfamethoxazole. Out of the 98 isolates, the highest levels of phenotypic resistance for the antimicrobials with CLSI breakpoints were observed in ampicillin and tetracycline, followed by trimethoprim-sulfamethoxazole and gentamicin (Table 1). Fourteen Salmonella isolates were resistant to three of the antimicrobials with CLSI breakpoints (ampicillin, tetracycline, trimethoprim-sulfamethoxazole), evidence of phenotypic multidrug resistance, since they are from different antimicrobial classes. Tetracycline resistance was observed among all isolates of Salmonella Bredeney, Cerro, Dublin, Heidelberg, and Meleagridis with the same being the case for ampicillin excluding Salmonella Meleagridis (Table 2).

Table 1.
Antimicrobial susceptibility in Salmonella isolates from cattle in the Texas Panhandle for antimicrobials with CLSI clinical cutoffs (n = 98).

Table 2.
Antimicrobial resistance among Salmonella serotypes from cattle in the Texas Panhandle (n = 98 isolates) *.
2.4. Antimicrobial Resistance Genes Detected in Salmonella Isolates
The Salmonella isolates harbored 14 classes of antimicrobial resistance genes (Figure 1). Resistance genes followed serotype patterns. Salmonella Heidelberg carried the largest set of antimicrobial resistance classes. Trimethoprim resistance genes were only found in Salmonella Heidelberg, apart from one Salmonella Typhimurium isolate. Fluoroquinolone, fosfomycin, and tetracycline resistance genes were also most prevalent among Salmonella Heidelberg isolates.
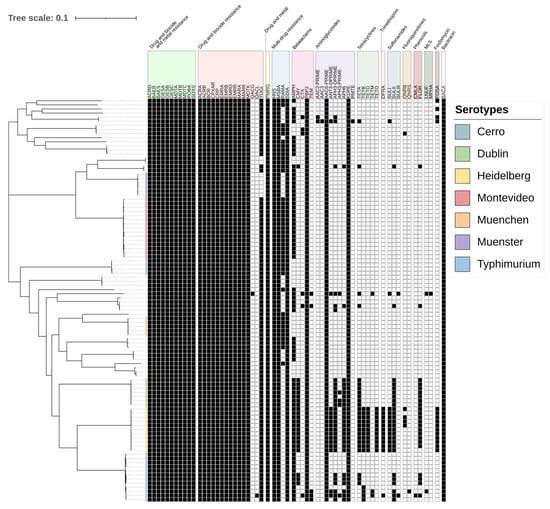
Figure 1.
Maximum likelihood phylogeny of 98 Salmonella isolated from cattle in the Texas Panhandle. The colors in the phylogeny represent the six most common serotypes from SISTR [28]. The table summarizes the presence (black) or absence (white) of resistance for each isolate (row). Antimicrobial resistance genes were grouped by resistance class. MLS: macrolides, lincosamides, and streptogramines. The scale bar represents the average nucleotide substitution per site in the phylogeny tree.
Salmonella Dublin and Salmonella Heidelberg genomes shared many antimicrobial resistance genes. Sulfonamide and phenicol resistance genes were identified in all genomes of those serotypes. Both serotypes along with Typhimurium also carried an array of aminoglycoside resistance genes.
2.5. Mapping Antimicrobial Resistance Genes to Plasmids
Thirty genomes carried plasmids typically associated with antimicrobial resistance genes. Several antimicrobial resistance genes were mapped to the same contigs as the plasmids (Table 3). Of notice, none of the isolates harbored mobile colistin-resistance genes (mcr). Among the 11 Salmonella Heidelberg genomes, all featured an IncA/C2 plasmid, six carried resistance pattern two, two carried resistance pattern four, and the remaining three had similar sets of genes with a few modifications (patterns one, three, and five) (Table 3). Additionally, a col440I plasmid with a qnrB gene was found in addition to the IncA/C2 plasmid in four Salmonella Heidelberg genomes, including two genomes with resistance pattern two, one with resistance pattern three, and one with resistance pattern four. One of three Salmonella Cannstatt genomes was also found to contain a col440I plasmid with the qnrB gene.

Table 3.
Antimicrobial resistance genes constituting resistance patterns for mobile genetic elements in 98 Salmonella isolates from cattle in the Texas Panhandle.
Two incompatibility groups characterized Salmonella Typhimurium isolates. Three genomes contained an IncA/C2 plasmid with resistance pattern seven, while the remaining two isolates carried resistance genes on an IncFIB plasmid, one with resistance pattern six and the other with resistance pattern eight. The former also contained a colRNAI plasmid with the resistance genes aph(3″), aph(3′), and blaCTX-M.
The IncA/C2 plasmid was responsible for all three resistance patterns detected in Salmonella Dublin isolates. Three isolates had resistance pattern 10, two had resistance pattern 9 and two had resistance pattern 7. One of two Salmonella Bredeney genomes also carried an IncA/C2 plasmid with resistance pattern 12.
Both Salmonella Meleagridis genomes carried resistance plasmids. One carried a colRNAI plasmid with the resistance genes aph(3″), aph(3′), and blaCTX-M, while the other had an IncHI2 plasmid with resistance pattern 11. A Salmonella Uganda isolate was also found to contain an IncHI2 plasmid with resistance pattern 13. Two Salmonella Anatum isolates carried resistance plasmids. One genome included an IncR plasmid with resistance pattern 12, and the other carried a colRNAI plasmid with aph(3″), and aph(3′).
2.6. Antimicrobial Resistance Genes Significantly Associated with Serotypes
This study identified a total of 23 serotypes in silico. The results for core genome multilocus sequencing typing (cgMLST) are illustrated in Figure 2. Salmonella isolates clustered mostly by serotype, with the exception of one Salmonella Anatum isolate clustering with the Salmonella Muenchen isolates, and one Salmonella Rostock isolate clustering with the Salmonella Dublin isolates. However, only four Salmonella serotypes were statistically significantly associated with antimicrobial resistance genes. Table 4 highlights the genes statistically significantly associated with serotypes. Salmonella Heidelberg carried the largest set of antimicrobial resistance genes, with 12 out of 14 genes exhibiting complete separability (the gene predicted the phenotype perfectly). Among the statistically significant genes, 70% belonged to genes encoding for resistance against aminoglycosides, sulfonamides, or tetracyclines. Salmonella Dublin was the second most significant reservoir for antimicrobial resistance genes. In addition to sharing six genes with Salmonella Heidelberg, the odds of carrying aph(3′) was 33.4-times higher among Salmonella Dublin isolates compared to all other serotypes (p = 0.01).
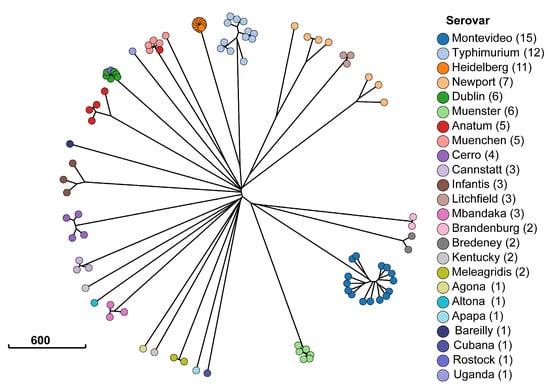
Figure 2.
Minimum spanning tree of Enterobase sequence typing [33]. Each dot represents one of the 98 Salmonella isolates from cattle in the Texas Panhandle. The colors indicate the serotype classification from SISTR [28] and the scale bar indicates the number of allele sequence type differences between isolates in the Salmonella cgMLST scheme.

Table 4.
Antimicrobial resistance genes associated with Salmonella serotypes in Texas Panhandle cattle.
2.7. Antimicrobial Resistance Genes Associated with Salmonella Isolates from Beef and Dairy Operations
The findings for the Scoary analysis for genes significantly associated with beef or dairy operations are shown in Figure 3. Among the 12 statistically significant genes, 11 exhibited stronger associations with Salmonella isolates from beef operations. The qnrB gene had the strongest association (p = 0.014). The odds of carrying the gene were 23.3-times higher in Salmonella isolates from beef operations than dairy.
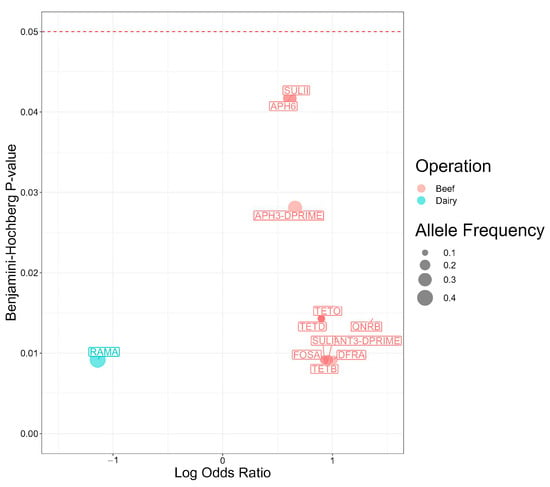
Figure 3.
Antimicrobial resistance genes present in 98 Salmonella isolates from Texas Panhandle cattle associated with beef or dairy operations (Scoary analysis). Log Odds Ratios: natural log-transformed odds ratios were obtained from a main effects model and adjusted for population structure. Benjamini–Hochberg p-value: p value adjusted for false discovery rate using the Benjamini-Hochberg method.
Genes encoding for resistance against aminoglycosides emerged as the most prevalent class of antimicrobial resistance genes found in Salmonella isolates from beef operations. The odds for carrying aminoglycoside resistance genes were 9.1-times higher for ant(3″), 4.6-times higher for aph(3″), and 4.1-times higher with aph(6) in Salmonella isolates from beef operations compared to dairy (p = 0.009, 0.028, and 0.042, respectively).
Two sulfonamide resistance genes exhibited statistical associations with beef operations. The odds ratio of carrying sul1 (OR = 8.4, p = 0.009) was twice the odds ratio of carrying sul2 (4.1, p = 0.042) in beef operations compared with dairy operations. Given trimethoprim is typically administered in conjunction with sulfonamides to treat salmonellosis, the odds of harboring dfrA were 10.3-times higher in Salmonella isolates from cattle on beef operations compared to dairy operations (p = 0.009).
Multiple tetracycline resistance genes were statistically significantly associated with beef operations. The odds of carrying tet(B) were 9.1-times higher in Salmonella isolates from cattle on beef operations (p = 0.009) and the odds of carrying tet(D) or tet(O) were 7.9-times higher (p = 0.014). The gene ramA was the only gene with a statistically significant association with dairy operations. The odds of carrying ramA were 92.7% higher in Salmonella isolates from cattle on dairy operation than beef operations (p = 0.009).
Given the nature of penalized logistic regression, Pyseer analysis yielded a truncated set of genes associated with cattle operations (Figure 4). Among these, two genes were statistically significant. Notably, the odds of originating in a beef operation were 3.2-times higher in Salmonella isolates carrying qnrB compared to isolates without the gene (p = 0.045). By contrast, the odds of originating in a dairy operation were 6% higher in Salmonella isolates carrying tet(A) (p = 0.0007).
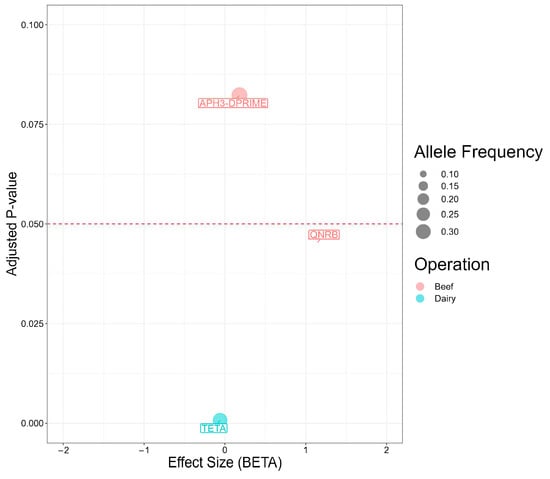
Figure 4.
Antimicrobial resistance genes present in 98 Salmonella isolates from Texas Panhandle cattle associated with beef or dairy operations (Pyseer analysis). Effect size (BETA) represents the natural log-transformed odds ratio (that is the coefficient of the penalized regression) adjusted for overfitting, population structure, and the other antimicrobial resistance genes in the model. Adjusted p-value: p value adjusted for population structure.
2.8. Association between Antimicrobial Resistance Genes and Antimicrobial Susceptibility Testing
Pyseer detected associations between antimicrobial resistance genes and antimicrobial susceptibility results (Figure 5). The phenotypic resistance to 10 out of 18 antimicrobials had significant associations with resistance genes. For five antimicrobials a significant association between the phenotypic resistance and the corresponding antimicrobial resistance gene was observed.
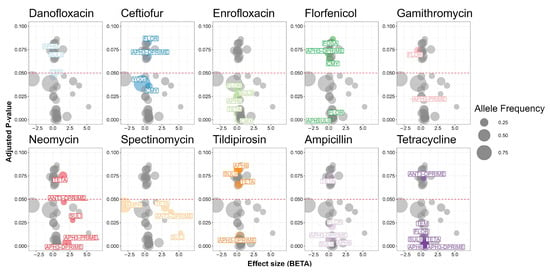
Figure 5.
Antimicrobial resistance genes in 98 Salmonella isolates from Texas Panhandle cattle associated with phenotypic antimicrobial resistance. Effect size (BETA) represents the natural log-transformed odds ratio (that is the coefficient of the penalized regression) adjusted for overfitting, population structure, and the other antimicrobial resistance genes in the model. Adjusted p-value: p value adjusted for population structure.
Phenotypic resistance to neomycin had the most biologically relevant significant associations with antimicrobial resistance genes. Two aminoglycoside resistance mechanisms were identified. One aminoglycoside O-nucleotidyltransferase was statistically significant (p = 0.0469). The MIC of neomycin was 1.6 units higher in Salmonella isolates carrying ant(3″) than isolates not carrying the gene. Two aminoglycoside O-phosphotransferase genes also had associations with neomycin. The MIC of neomycin was 2.1 or 6.8 units higher for Salmonella isolates carrying aph(3″) or aph(3′) than isolates not carrying either gene (p = 0.0036 and 0.0066, respectively).
Ceftiofur was the only antimicrobial with a statistically significant positive and negative association. The MIC for ceftiofur was 0.6 units higher in isolates carrying blaCMY, but 0.4 units lower in isolates carrying yogi compared to isolates not carrying either gene (p = 0.0357 and 0.0388, respectively).
Single antimicrobial resistance genes drove resistance against three antimicrobials. The MIC for florfenicol was 0.6 units higher in Salmonella isolates carrying floR than isolates not carrying the gene (p = 0.00327). The MIC for spectinomycin was 3.8 units higher in Salmonella isolates carrying ant(3″) than isolates not carrying the gene (p = 0.0365). The odds of being resistant to tetracycline were 95% higher among Salmonella isolates carrying tet(A) compared to isolates not carrying tet(A) (p = 0.002910).
Scoary detected biologically relevant associations with phenotypic resistance in three of the four antimicrobials with MIC interpretation. Tetracycline had the largest number of genes. The tet(A), tet(B), tet(D), and tet(O) genes all showed complete separability for isolates classified as resistant. The blaCMY gene was also found to have complete separability with the ampicillin resistance phenotype.
Three genes were statistically significantly associated with resistance to trimethoprim-sulfamethoxazole. One gene confers resistance to trimethoprim. The odds of carrying dfrA was 33.9-times higher among Salmonella isolates resistant to trimethoprim-sulfamethoxazole (p < 0.0001). Two genes that confer resistance to sulfamethoxazole were also statistically significant. The odds of carrying sul1 or sul2 were 25.1- and 9.8-times higher among Salmonella isolates resistant to trimethoprim-sulfamethoxazole (p < 0.0001 and 0.0004, respectively).
2.9. Other Associations
Pyseer also found associations between antimicrobial resistance genes and the remaining phenotypes. The gene tet(B) was the only one with geographical associations. The odds of originating from New Mexico (versus Texas or Arizona) were 1.9-times higher in Salmonella isolates carrying tet(B) compared to those without the gene (p = 0.0499); additionally, the odds of originating in Texas (versus New Mexico or Arizona) were 4.5-times lower in Salmonella isolates carrying tet(B) compared to the isolates without the gene (p = 0.0466). Cattle sex was also linked to antimicrobial resistance genes. The odds of a cattle being male were 28.2-times higher in Salmonella isolates carrying fosA compared to isolates without the gene (p = 0.0482), while the odds of cattle being female were 5.3-times higher in Salmonella isolates carrying ampH compared to isolates not carrying the gene (p = 0.00912). Three genes had negative associations with Salmonella isolates collected in 2022. The odds of isolates being collected in 2022 (versus in 2021 or 2023) were 74%, 12%, and 75% lower in Salmonella isolates carrying aph(6), floR, and sul2, respectively, than in isolates without those genes (p = 0.0081, 0.0307, and 0.0081, respectively).
3. Discussion
This study aimed to identify antimicrobial resistance genes in Salmonella from cattle residing in the Texas Panhandle region. The results of this study highlighted the differences in the prevalence of antimicrobial resistance genes in Salmonella between beef and dairy operations and serotypes. Salmonella isolated from cattle in beef operations had more antimicrobial resistance genes. Salmonella Heidelberg, followed by Salmonella Dublin, harbored the most antimicrobial resistant genes. Additionally, specific classes of antimicrobial resistance genes were only present within mobile genetic elements.
Several studies have reported strong concordance between antimicrobial resistance genes and resistance phenotypes in the past [10,19,20,34]. For example, Carroll et al. (2017) reported a prediction sensitivity of 97.2% and a specificity of 85.2% [10]. However, the statistical methods employed by these studies do not adjust for common confounders in bacterial genome-wide association studies such as population structure [35,36]. In our study, only five antimicrobials resistance determinants (against ceftiofur, florfenicol, neomycin, spectinomycin, and tetracycline) were significantly associated with higher MICs for the corresponding antimicrobials. We attribute these discrepancies, in part, to the differences in the bioinformatics processing and statistical analysis. The presence or absence of a resistance gene is not the sole indicator of resistance. Enzyme activation, target modification, gene expression regulation, or cell wall configuration changes also influence phenotypic resistance [20,37]. Antimicrobial resistance is a multifactorial problem involving management practices and mineral deficiency, in addition to transmission of antimicrobial resistant bacteria [38,39,40,41,42].
Salmonella isolates carrying IncA/C2 plasmids often originate from cattle sources [9,43]. The rise of multidrug-resistant Salmonella in 2010 coincided with the emergence of IncA/C2 plasmids [44,45]. In this study, IncA/C2 plasmids were responsible for conferring resistance in most (22 of 30) isolates for all antimicrobial resistance genes that predicted the correct corresponding antimicrobial resistance phenotype. This is not surprising as antimicrobial resistance stemming from IncA/C2 has been often reported in Salmonella [19,46,47,48].
A quinolone gene was only present on col440I plasmids. Plasmid-mediated quinolone resistance (PMQR) is well characterized in Salmonella. Col plasmids often carry PMQR genes that mediate reduced susceptibility to quinolones in Enterobacteriaceae, but they do not typically manifest into resistance [49,50,51,52]. While identifying point mutations was beyond the scope of this study, resistance is not achieved without point mutations in the quinolone resistance determining region of parC or gyrA [9,10,16,19,53,54,55]. Additional point mutations lead to higher levels of phenotypic resistance [45,56,57].
Salmonella serotypes often follow particular antimicrobial resistance patterns, and many factors can be associated with their prevalence and distribution. A study by Levent et al. (2019) found that the pen was the most important factor contributing to the prevalence of specific serotypes in cattle herds [22]. Given our study acquired isolates through convenience sampling, they did not originate from the same operation and therefore we cannot evaluate the pen effect. However, we similarly observed that antimicrobial resistance patterns were associated with specific serotypes. Salmonella Dublin and Salmonella Heidelberg harbored the most antimicrobial resistance genes. Both serotypes are of high concern because they are responsible for the third (0.31) and fourth (0.27) highest hospitalization to illness ratios among non-typhoidal Salmonella infections [58]. The rise of multidrug-resistant Salmonella Dublin, the most prominent serotype found in clinical cases from cattle, has been documented to have arisen from the emergence of IncA/C2 plasmids [17,37,45,59,60]. Our study only detected Salmonella Dublin from dairy operations and Salmonella Dublin has been reported to have increasing prevalence in dairy facilities [17]. Interestingly, we did not find genotypic multidrug resistance in either Salmonella Cerro or Salmonella Montevideo, which represent over 27% of Salmonella isolates in bovine cases [45]. Pan-susceptibility from Salmonella Cerro and Salmonella Montevideo is consistent with the existing literature on cattle [61,62,63].
In recent decades, Salmonella Heidelberg has broadened its resistance profile, leading to increased hospitalizations in people [13]. The relationship between elevated virulence and co-selection with antimicrobial resistance may be responsible for the virulence factors and antimicrobial resistance genes consistently expressed by this serotype [13,45,64,65]. In our study, Salmonella Heidelberg was identified in cattle on both beef and dairy cattle operations. The multidrug resistant phenotype in this serotype is commonly associated with dairy beef calves [66].
Salmonella Infantis is well known for exhibiting multidrug resistance in poultry [67]. In our study, all three isolates were phenotypically susceptible to the four antimicrobials with CLSI breakpoints but harbored an important set of antimicrobial resistance genes, suggesting they may be multidrug-resistant isolates, findings consistent with the existing literature [68,69].
Host age is a major contributing factor to the prevalence of antimicrobial resistance in Salmonella. Bacteria recovered from dairy calves often exhibit higher levels of antimicrobial resistance than bacteria from adults in part due to early exposure to antimicrobials as prevention measures against diseases [38,39,40,70,71,72,73]. Administration of antimicrobials is limited during lactation to prevent antimicrobial residues from contaminating milk, leading to lower level of antimicrobial resistance genes in Salmonella from adult dairy cows [74,75]. However, cows in dry off are often administered antimicrobials to manage mastitis and protect the performance of future lactation cycles [76,77].
Hille et al. (2017) found that less rigorous practices led to lower prevalence of antimicrobial resistant bacteria in beef operations, which Tello et al. (2020) claims is due to lower stress and infection pressure [74,78]. Similar to dairy practices, antimicrobials are also used sparingly in beef cattle approaching slaughter because if antimicrobials are administered close to slaughter, additional resources are required to support an extended withdrawal period to prevent antimicrobial residues in the meat [79]. We did not find any significant associations between antimicrobial resistance genes and the age of cattle. We attribute this lack of association to sparsity of the metadata. Cattle could only be dichotomized based on neonatal status (age < 1 month) and even then 20% of samples were excluded due to missing data.
Recent studies have reported differentially abundant antimicrobial resistance genes in the resistomes of beef compared to dairy cattle. Rovira et al. (2019) reported that beef cattle feces had more relatively abundant antimicrobial resistance genes than dairy cattle feces, but relative abundance differed by resistance class. Beef cattle resistomes featured a higher relative abundance of genes conferring resistance to tetracycline and macrolides, but dairy cattle resistomes had higher relative abundance of beta-lactamases [80]. Wang et al. (2021) also found that beef cattle gut samples carried more antimicrobial resistance genes than dairy. Beef cattle resistomes had a higher relative abundance of macrolides, beta-lactamases, and multidrug resistance genes and dairy cattle resistomes had more quinolone and aminoglycoside resistance genes [81]. While the resistome of cattle feces was not the focus of this study, we also observed that Salmonella isolates from beef operations showed disproportionally more associations with antimicrobial resistance genes than those from dairy operations with both statistical approaches. However, associations with Salmonella from beef and dairy operations could have been confounded by age, considering most isolates from beef operations did not have a reported age (9/16), and three out of the seven isolates with age information were from calves under one month old.
This study contributes to federal food safety initiatives to reduce Salmonella in the beef industry continuum, particularly in the pre-harvest stage, which is underrepresented relative to poultry or swine in research. However, this study does have limitations. The isolates from this study originated from sample submissions positive for Salmonella. Consequently, there may be selection bias because no isolates from healthy hosts were included. Since it was a convenience sampling, it is difficult to generalize findings to the cattle populations of the Texas Panhandle cattle. Additionally, the sample size was small, and the diversity of the isolates was constrained to what clients submitted to the Texas A&M Veterinary Medical Diagnostic Laboratory, leading to a disproportionate number of isolates from dairy operations. Many samples also were not submitted with complete descriptions of the sample’s origin because the level of detail for the metadata was at the will of the client, creating instances of missing data. We could not determine whether the submissions were related to clinical salmonellosis cases. The search for antimicrobial resistance genes was conducted with a minimum percent identity and coverage of 80%, which may have influenced the detection of some antimicrobial resistance genes not commonly reported in Salmonella. However, 42% of genes had a coverage of 100% and an identity of 90%. Future studies should adopt probabilistic sampling approaches that include a representative sampling of all stages of pre-harvested beef and dairy production to gain a deeper understanding of the ecology of Salmonella from cattle in Texas.
4. Materials and Methods
4.1. Isolate Selection and Antimicrobial Susceptibility Testing
A total of 100 Salmonella isolates that were recovered from different bovine enteric samples (intestinal tissues, feces or fecal swabs) were used in this study. All specimens were obtained from clinically ill cattle and submitted to the American Association of Veterinary Laboratory Diagnosticians (AAVLD)-accredited Texas A&M Veterinary Medical Diagnostic Laboratory at Canyon, TX, USA (TVMDL-Canyon) for culture. Submissions ranged between the years 2021 (n = 36), 2022 (n = 38), and 2023 (n = 26). Isolation procedures, biochemical characterization, and detection of different Salmonella isolates were performed utilizing well-established morphological and biochemical identification methods for Salmonella at the TVMDL-Canyon [82,83,84]. Antimicrobial susceptibility testing for all 100 isolates was performed using broth microdilution MIC plate method (Sensititre™ Vet Bovine BOPO7F Plate; ThermoFisher, Waltham, MS, USA), following the manufacturer’s instructions [85,86] at the TVMDL-Canyon. Antimicrobial resistance patterns (phenotypes) for Salmonella isolates were determined by the BIOMIC V3 (Giles Scientific, Santa Barbara, CA, USA) instrument employing the most recently breakpoints published by CLSI in the VET01S supplement for accurate analysis [27,87,88]. Frozen Salmonella isolates were transferred to the Texas A&M Veterinary Education, Research, & Outreach (VERO) Research Laboratory (Canyon, TX, USA ) for bacterial DNA extraction and whole genome sequencing.
4.2. Bacteriological Culture
Salmonella isolates underwent a series of microbiological culture steps to confirm the identity of Salmonella and assess the purity of the isolates. Isolates were inoculated in Brain Heart Infusion (BHI) broth (Becton Dickinson, Franklin Lakes, NJ, USA) and incubated for 24 h at 37 °C under aerobic conditions. If bacterial growth was evidenced via turbidity, an aliquot was subcultured onto Xylose-Lysine-Tergitol 4 (XLT4) (Hardy Diagnostics, Santa Maria, CA, USA) and MacConkey (Hardy Diagnostics) agar plates, and incubated for 24 h at 37 °C under aerobic conditions to confirm the presence of Salmonella. Isolates with Salmonella-like colonies (black colonies on XLT4 and pale colonies on MacConkey agar plates) were subcultured to assess their purity, picking one colony from the XLT4 plate and streaking it onto a Columbia blood agar plate (Hardy Diagnostics) with 24-hour incubation at 37 °C under aerobic conditions. One colony per isolate was inoculated from Columbia blood agar plate into BHI broth and incubated for 24 h at 37 °C under aerobiosis before DNA extraction.
4.3. DNA Extraction
Each pure Salmonella isolate in BHI broth was centrifuged in 1.5 mL vials at 10,000× g for 1 min at 4 °C in two consecutive rounds (using 1 mL and 0.5 mL of BHI broth, respectively) to pellet the bacterial cells before extracting the DNA. DNA was extracted using the DNeasy Ultraclean Microbial Kit on a QIAcube Connect RNA/DNA extraction instrument (Qiagen, Hilden, Germany) using the manufacturer’s instructions. The heat treatment (65 °C for 10 min) was chosen instead of the bead beating step as recommended by the manufacturer to reduce DNA shearing.
4.4. Whole Genome Sequencing
The extracted DNA (800–1000 ng per isolate) was sheared using g-TUBES (Covaris, Woborn, MA, USA) to obtain an expected fragment length of 8 kbp, following manufacturer’s instructions. The sheared DNA (in 150 μL of low EDTA TE buffer) was cleaned using AMPure XP beads (Beckman Coulter, Brea, CA, USA) at a ratio of 0.4× and eluted in 14 μL of molecular grade water (Corning, Corning, NY, USA). The sheared DNA was assessed via visualization of the electropherogram in a genomic ScreenTape (Agilent, Santa Clara, CA, USA) and its concentration and quality were evaluated using a NanoDrop Eight (ThermoFisher, Waltham, MA, USA). DNA libraries were prepared using ONT Native Barcoding Kit V14 (SQK-NBD114-24; ONT, Oxford, UK), multiplexing 24 Salmonella isolates per DNA library. Three extraction blanks were also processed for sequencing to assess the presence of background DNA. Sequencing was performed on a MinION Mk1C (ONT) device with a flow cell version 10.4.1 at a speed of 260 bases per second for 72 h, using the Fast model (MinKNOW version 22.10.7, Guppy version 6.3.9) due to the constraints in the computational resources of the device. After the sequencing run, the corresponding FAST5 files were base called again with the Super Accurate Model (standalone Guppy version 6.5.7) on a dedicated workstation for downstream analysis.
4.5. Bioinformatics and Statistical Analysis
The read quality was assessed with LongQC (version 1.2.0) [89], and long reads were assembled using Flye (version 2.9.1) [90]. In those cases where Flye failed to assemble the Salmonella genome, Canu (version 2.2) [91] was used. Genomes were first polished with Medaka (version 1.7.2) [92], followed by a second round of polishing with Homopolish (version 0.4.1) [93]. Afterward, QUAST (version 5.0.2) [94] and BUSCO (version 5.1.2) [95] were employed to evaluate the structure and biological relevancy of the assemblies using serotype-specific references from NCBI, matching the serotype classification of the isolates obtained in silico with the Salmonella In Silico Typing Resource (SISTR, version 1.1.2) [28]. Salmonella isolates having assemblies with coverages below 60× were re-extracted and re-sequenced to obtain higher coverage.
The phylogenetic relationship between isolates was assessed with two methods. Samples were sequenced typed using the Enterobase Salmonella cgMLST scheme (version 1.2.0) [33] according to allele variant matching with 100% identity to the housekeeping sequences. Additionally, a SNP-based maximum likelihood phylogeny was generated using Snippy (version 4.6.0) [96] and Molecular Evolutionary Genetics Analysis (MEGA) 11 with the Tamura-Nei substitution model and the Nearest-Neighbor-Interchange maximum likelihood heuristic method [97]. ABRicate (release FOFN MOTD 80 80) [30] was employed to search for antimicrobial resistance genes with the MEGARes database (version 2.00) [31]. The genes detected by ABRicate were evaluated against the available metadata (sample type, cattle operation, cattle breed, age, sex, sample origin, and collection year) and antimicrobial susceptibility profiles (ampicillin, ceftiofur, clindamycin, danofloxacin, enrofloxacin, florfenicol, gamithromycin, gentamicin, neomycin, penicillin G, spectinomycin, sulfadimethoxime, tetracycline, tildipirosin, tilmicosin, trimethoprim-sulfamethoxazole, tulathromycin, and tylosin).
The associations between the antimicrobial resistance genes and the metadata were evaluated with two statistical approaches. Scoary (version 1.6.16) [98] was utilized to detect associations between individual genes with each variable in the metadata using a Fisher’s exact test, while controlling the false discovery rate with the Benjamini–Hochberg method according to a p-value cutoff of 0.05 and adjusting for the population structure from the maximum likelihood phylogeny described above. An elastic net logistic regression in Pyseer (version 1.3.11) [99] was also applied to assess the independent associations between the genetic determinants and each outcome, adjusting for the population structure using the maximum likelihood phylogeny described above. Isolates with intermediate resistance were reassigned as susceptible for the analysis. The MICs of the antimicrobials without CLSI breakpoints were assessed with elastic net linear regression, also in Pyseer.
Bandage was used to annotate and map antimicrobial resistance genes and plasmid sequences (from ABRicate with the MEGARes and PlasmidFinder [32] databases, respectively) to Flye and Canu assemblies. In Bandage, BLAST hits were restricted to hits with a query identity and coverage above 80%. In cases where more than one plasmid mapped to a contig, the plasmid sequence with the highest identity and coverage was selected as the consensus [29].
5. Conclusions
This study summarizes antimicrobial resistance genes found in 98 Salmonella isolates from cattle residing in the Texas Panhandle. Antimicrobial resistance genes did not explain all antimicrobial resistance phenotypes. Some antimicrobial resistance genes were present in mobile genetic elements. A few serotypes displayed multidrug resistance and antimicrobial resistance gene profiles differed in beef operations compared to dairy operations. Based on this study, mitigation strategies for multidrug resistant Salmonella should continue to be developed for the beef industry to help curb the rising antimicrobial resistance trends.
Author Contributions
H.K. and R.V.-C.: conceptualization and investigation. R.V.-C.: funding acquisition. M.C., E.D., H.K., A.C.T., M.S., K.N. and R.V.-C.: Methodology. M.C. and R.V.-C.: formal analysis. M.C. and R.V.-C.: visualization. H.K., M.S., K.N. and R.V.-C.: supervision. M.C., K.N. and R.V.-C.: writing—original draft preparation. M.C., E.D., H.K., A.C.T., M.S., K.N. and R.V.-C.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was internally supported by Texas A&M University and did not receive external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
De-identified data sets from this study are available in the SRA archives at NCBI (https://www.ncbi.nlm.nih.gov/sra/PRJNA1135485, accessed on 16 July 2024).
Acknowledgments
We thank the personnel at the Texas A&M University Veterinary Education, Research, and Outreach (VERO) research lab for their support.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- CDC. Salmonella. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 5 May 2024).
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Razafindrazoto, C.I.; Rakotomalala, J.A.; Randriamifidy, N.H.; Ralaizanaka, B.M.; Maherison, S.; Hasina Laingonirina, D.H.; Rakotomaharo, M.; Rasolonjatovo, A.S.; Rakotozafindrabe, A.L.R.; Rabenjanahary, T.H.; et al. Acute infectious pancreatitis due to Salmonella typhi: Case report and literature review. JGH Open 2021, 5, 1106–1107. [Google Scholar] [CrossRef]
- Elouali, A.; Ouerradi, N.; Ayad, G.; Babakhouya, A.; Rkain, M. Salmonella Meningitis in a Young Infant: A Case Report. Cureus 2023, 15, e44147. [Google Scholar] [CrossRef]
- Hull, D.M.; Harrell, E.; Harden, L.; Thakur, S. Multidrug resistance and virulence genes carried by mobile genomic elements in Salmonella enterica isolated from live food animals, processed, and retail meat in North Carolina, 2018–2019. Int. J. Food Microbiol. 2022, 378, 109821. [Google Scholar] [CrossRef]
- Medalla, F.; Gu, W.; Friedman, C.R.; Judd, M.; Folster, J.; Griffin, P.M.; Hoekstra, R.M. Increased Incidence of Antimicrobial-Resistant Nontyphoidal Salmonella Infections, United States, 2004–2016. Emerg. Infect. Dis. 2021, 27, 1662–1672. [Google Scholar] [CrossRef] [PubMed]
- Scharff, R.L. Food Attribution and Economic Cost Estimates for Meat- and Poultry-Related Illnesses. J. Food Prot. 2020, 83, 959–967. [Google Scholar] [CrossRef] [PubMed]
- NACMCF. Response to Questions Posed by the Food Safety and Inspection Service: Enhancing Salmonella Control in Poultry Products. J. Food Prot. 2024, 87, 100168. [Google Scholar] [CrossRef]
- Tate, H.; Li, C.; Nyirabahizi, E.; Tyson, G.H.; Zhao, S.; Rice-Trujillo, C.; Jones, S.B.; Ayers, S.; M’Ikanatha, N.M.; Hanna, S.; et al. A National Antimicrobial Resistance Monitoring System Survey of Antimicrobial-Resistant Foodborne Bacteria Isolated from Retail Veal in the United States. J. Food Prot. 2021, 84, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Carroll, L.M.; Wiedmann, M.; den Bakker, H.; Siler, J.; Warchocki, S.; Kent, D.; Lyalina, S.; Davis, M.; Sischo, W.; Besser, T.; et al. Whole-Genome Sequencing of Drug-Resistant Salmonella enterica Isolates from Dairy Cattle and Humans in New York and Washington States Reveals Source and Geographic Associations. Appl. Environ. Microbiol. 2017, 83, e00140-17. [Google Scholar] [CrossRef]
- Pornsukarom, S.; van Vliet, A.H.M.; Thakur, S. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genom. 2018, 19, 801. [Google Scholar] [CrossRef]
- Carroll, L.M.; Huisman, J.S.; Wiedmann, M. Twentieth-century emergence of antimicrobial resistant human- and bovine-associated Salmonella enterica serotype Typhimurium lineages in New York State. Sci. Rep. 2020, 10, 14428. [Google Scholar] [CrossRef] [PubMed]
- Nichols, M.; Gollarza, L.; Sockett, D.; Aulik, N.; Patton, E.; Francois Watkins, L.K.; Gambino-Shirley, K.J.; Folster, J.P.; Chen, J.C.; Tagg, K.A.; et al. Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Dairy Calf Exposure, United States, 2015–2018. Foodborne Pathog. Dis. 2022, 19, 199–208. [Google Scholar] [CrossRef]
- Carroll, L.M.; Buehler, A.J.; Gaballa, A.; Siler, J.D.; Cummings, K.J.; Cheng, R.A.; Wiedmann, M. Monitoring the Microevolution of Salmonella enterica in Healthy Dairy Cattle Populations at the Individual Farm Level Using Whole-Genome Sequencing. Front. Microbiol. 2021, 12, 763669. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.L.; Fenton, R.J.; Moriyama, E.N.; Loy, J.D.; Moxley, R.A. Association of ISVsa3 with Multidrug Resistance in Salmonella enterica Isolates from Cattle (Bos taurus). Microorganisms 2023, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Nickodem, C.; Arnold, A.; Gehring, K.B.; Gill, J.J.; Richeson, J.T.; Samuelson, K.L.; Scott, H.; Smith, J.; Taylor, T.; Vinasco, J.; et al. A longitudinal study on the dynamics of Salmonella enterica prevalence and serovar composition in beef cattle feces and lymph nodes and potential contributing sources from the feedlot environment. Appl. Environ. Microbiol. 2023, 89, e0003323. [Google Scholar] [CrossRef] [PubMed]
- Velasquez-Munoz, A.; Castro-Vargas, R.; Cullens-Nobis, F.M.; Mani, R.; Abuelo, A. Review: Salmonella Dublin in dairy cattle. Front. Vet. Sci. 2023, 10, 1331767. [Google Scholar] [CrossRef]
- Cornell University CVM. Salmonellosis: Background, Management and Control. Available online: https://www.vet.cornell.edu/animal-health-diagnostic-center/programs/nyschap/modules-documents/salmonellosis-background-management-and-control#:~:text=Overall%2C%2028%25%20of%20dairy%20farms,a%20short%20period%20of%20time (accessed on 5 May 2024).
- Eyler, A.B.; M’Ikanatha, N.M.; Xiaoli, L.; Dudley, E.G. Whole-genome sequencing reveals resistome of highly drug-resistant retail meat and human Salmonella Dublin. Zoonoses Public Health 2020, 67, 251–262. [Google Scholar] [CrossRef]
- Srednik, M.E.; Morningstar-Shaw, B.R.; Hicks, J.A.; Mackie, T.A.; Schlater, L.K. Antimicrobial resistance and genomic characterization of Salmonella enterica serovar Senftenberg isolates in production animals from the United States. Front. Microbiol. 2022, 13, 979790. [Google Scholar] [CrossRef]
- Marshall, K.E.H.; Tewell, M.; Tecle, S.; Leeper, M.; Sinatra, J.; Kissler, B.; Fung, A.; Brown, K.; Wagner, D.; Trees, E.; et al. Protracted Outbreak of Salmonella Newport Infections Linked to Ground Beef: Possible Role of Dairy Cows—21 States, 2016–2017. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 443–446. [Google Scholar] [CrossRef]
- Levent, G.; Schlochtermeier, A.; Ives, S.E.; Norman, K.N.; Lawhon, S.D.; Loneragan, G.H.; Anderson, R.C.; Vinasco, J.; Scott, H.M. Population Dynamics of Salmonella enterica within Beef Cattle Cohorts Followed from Single-Dose Metaphylactic Antibiotic Treatment until Slaughter. Appl. Environ. Microbiol. 2019, 85, e01386-19. [Google Scholar] [CrossRef]
- Manishimwe, R.; Moncada, P.M.; Bugarel, M.; Scott, H.M.; Loneragan, G.H. Antibiotic resistance among Escherichia coli and Salmonella isolated from dairy cattle feces in Texas. PLoS ONE 2021, 16, e0242390. [Google Scholar] [CrossRef] [PubMed]
- Levent, G.; Schlochtermeier, A.; Ives, S.E.; Norman, K.N.; Lawhon, S.D.; Loneragan, G.H.; Anderson, R.C.; Vinasco, J.; den Bakker, H.C.; Scott, H.M. High-Resolution Genomic Comparisons within Salmonella enterica Serotypes Derived from Beef Feedlot Cattle: Parsing the Roles of Cattle Source, Pen, Animal, Sample Type, and Production Period. Appl. Environ. Microbiol. 2021, 87, e0048521. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.A.; Ossa-Trujillo, C.; Vinasco, J.; Jordan, E.R.; García Buitrago, J.A.; Hagevoort, R.; Norman, K.N.; Lawhon, S.D.; Piñeiro, J.M.; Levent, G.; et al. Use of critically important antimicrobial classes early in life may adversely impact bacterial resistance profiles during adult years: Potential co-selection for plasmid-borne fluoroquinolone and macrolide resistance via extended-spectrum beta-lactam use in dairy cattle. Lett. Appl. Microbiol. 2021, 72, 220–224. [Google Scholar] [CrossRef] [PubMed]
- USDA. Annual State Agricultural Exports Interactive Chart. Available online: https://www.ers.usda.gov/data-products/state-agricultural-trade-data/annual-state-agricultural-exports/ (accessed on 5 May 2024).
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 6th ed.; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2023; Volume CLSI Supplement VET01S. [Google Scholar]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Seemann, T. Abricate. Available online: https://github.com/tseemann/abricate (accessed on 7 May 2024).
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Veeraraghavan, B.; Jacob, J.J.; Prakash, J.A.J.; Pragasam, A.K.; Neeravi, A.; Narasimman, V.; Anandan, S. Extensive drug resistant Salmonella enterica serovar Senftenberg carrying bla(NDM) encoding plasmid p5558 (IncA/C) from India. Pathog. Glob. Health 2019, 113, 20–26. [Google Scholar] [CrossRef]
- Power, R.A.; Parkhill, J.; de Oliveira, T. Microbial genome-wide association studies: Lessons from human GWAS. Nat. Rev. Genet. 2017, 18, 41–50. [Google Scholar] [CrossRef]
- Sul, J.H.; Martin, L.S.; Eskin, E. Population structure in genetic studies: Confounding factors and mixed models. PLoS Genet. 2018, 14, e1007309. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, N.; Pan, H.; Elbediwi, M.; Zhou, X.; Peng, X.; Li, X.; Fang, W.; Yue, M. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. 2019, 19, 226. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Wickramasingha, D.; Abdelfattah, E.M.; Pereira, R.V.; Okello, E.; Maier, G. Prevalence of antimicrobial resistance in fecal Escherichia coli and Enterococcus spp. isolates from beef cow-calf operations in northern California and associations with farm practices. Front. Microbiol. 2023, 14, 1086203. [Google Scholar] [CrossRef]
- Berge, A.C.; Hancock, D.D.; Sischo, W.M.; Besser, T.E. Geographic, farm, and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the western United States. J. Am. Vet. Med. Assoc. 2010, 236, 1338–1344. [Google Scholar] [CrossRef]
- Yamamoto, S.; Iwabuchi, E.; Hasegawa, M.; Esaki, H.; Muramatsu, M.; Hirayama, N.; Hirai, K. Prevalence and molecular epidemiological characterization of antimicrobial-resistant Escherichia coli isolates from Japanese black beef cattle. J. Food Prot. 2013, 76, 394–404. [Google Scholar] [CrossRef]
- Markland, S.; Weppelmann, T.A.; Ma, Z.; Lee, S.; Mir, R.A.; Teng, L.; Ginn, A.; Lee, C.; Ukhanova, M.; Galindo, S.; et al. High Prevalence of Cefotaxime Resistant Bacteria in Grazing Beef Cattle: A Cross Sectional Study. Front. Microbiol. 2019, 10, 176. [Google Scholar] [CrossRef]
- University of Georgia Extension. Mineral Supplements for Beef Cattle. Available online: https://extension.uga.edu/publications/detail.html?number=B895&title=mineral-supplements-for-beef-cattle (accessed on 5 May 2024).
- Folster, J.P.; Pecic, G.; Bolcen, S.; Theobald, L.; Hise, K.; Carattoli, A.; Zhao, S.; McDermott, P.F.; Whichard, J.M. Characterization of extended-spectrum cephalosporin-resistant Salmonella enterica serovar Heidelberg isolated from humans in the United States. Foodborne Pathog. Dis. 2010, 7, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Mangat, C.S.; Bekal, S.; Avery, B.P.; Côté, G.; Daignault, D.; Doualla-Bell, F.; Finley, R.; Lefebvre, B.; Bharat, A.; Parmley, E.J.; et al. Genomic Investigation of the Emergence of Invasive Multidrug-Resistant Salmonella enterica Serovar Dublin in Humans and Animals in Canada. Antimicrob. Agents Chemother. 2019, 63, e00108-19. [Google Scholar] [CrossRef] [PubMed]
- Srednik, M.E.; Lantz, K.; Hicks, J.A.; Morningstar-Shaw, B.R.; Mackie, T.A.; Schlater, L.K. Antimicrobial resistance and genomic characterization of Salmonella Dublin isolates in cattle from the United States. PLoS ONE 2021, 16, e0249617. [Google Scholar] [CrossRef]
- Fricke, W.F.; Welch, T.J.; McDermott, P.F.; Mammel, M.K.; LeClerc, J.E.; White, D.G.; Cebula, T.A.; Ravel, J. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 2009, 191, 4750–4757. [Google Scholar] [CrossRef]
- Lindsey, R.L.; Fedorka-Cray, P.J.; Frye, J.G.; Meinersmann, R.J. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 2009, 75, 1908–1915. [Google Scholar] [CrossRef] [PubMed]
- Welch, T.J.; Fricke, W.F.; McDermott, P.F.; White, D.G.; Rosso, M.L.; Rasko, D.A.; Mammel, M.K.; Eppinger, M.; Rosovitz, M.J.; Wagner, D.; et al. Multiple antimicrobial resistance in plague: An emerging public health risk. PLoS ONE 2007, 2, e309. [Google Scholar] [CrossRef] [PubMed]
- Akinyemi, K.O.; Fakorede, C.O.; Linde, J.; Methner, U.; Wareth, G.; Tomaso, H.; Neubauer, H. Whole genome sequencing of Salmonella enterica serovars isolated from humans, animals, and the environment in Lagos, Nigeria. BMC Microbiol. 2023, 23, 164. [Google Scholar] [CrossRef] [PubMed]
- Raufu, I.A.; Ahmed, O.A.; Aremu, A.; Ameh, J.A.; Timme, R.E.; Hendriksen, R.S.; Ambali, A.G. Occurrence, antimicrobial resistance and whole genome sequence analysis of Salmonella serovars from pig farms in Ilorin, North-central Nigeria. Int. J. Food Microbiol. 2021, 350, 109245. [Google Scholar] [CrossRef]
- Aworh, M.K.; Nilsson, P.; Egyir, B.; Owusu, F.A.; Hendriksen, R.S. Rare serovars of non-typhoidal Salmonella enterica isolated from humans, beef cattle and abattoir environments in Nigeria. PLoS ONE 2024, 19, e0296971. [Google Scholar] [CrossRef]
- Cummings, K.J.; Rodriguez-Rivera, L.D.; Norman, K.N.; Ohta, N.; Scott, H.M. Identification of a Plasmid-Mediated Quinolone Resistance Gene in Salmonella Isolates from Texas Dairy Farm Environmental Samples. Zoonoses Public Health 2017, 64, 305–307. [Google Scholar] [CrossRef]
- Slowey, R.; Kim, S.W.; Prendergast, D.; Madigan, G.; Van Kessel, J.A.S.; Haley, B.J. Genomic diversity and resistome profiles of Salmonella enterica subsp. enterica serovar Kentucky isolated from food and animal sources in Ireland. Zoonoses Public Health 2022, 69, 1–12. [Google Scholar] [CrossRef]
- Cloeckaert, A.; Chaslus-Dancla, E. Mechanisms of quinolone resistance in Salmonella. Vet. Res. 2001, 32, 291–300. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- McDermott, P.F.; Tyson, G.H.; Kabera, C.; Chen, Y.; Li, C.; Folster, J.P.; Ayers, S.L.; Lam, C.; Tate, H.P.; Zhao, S. Whole-Genome Sequencing for Detecting Antimicrobial Resistance in Nontyphoidal Salmonella. Antimicrob. Agents Chemother. 2016, 60, 5515–5520. [Google Scholar] [CrossRef]
- Day, M.R.; Doumith, M.; Do Nascimento, V.; Nair, S.; Ashton, P.M.; Jenkins, C.; Dallman, T.J.; Stevens, F.J.; Freedman, J.; Hopkins, K.L.; et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Salmonella enterica serovars Typhi and Paratyphi. J. Antimicrob. Chemother. 2018, 73, 365–372. [Google Scholar] [CrossRef]
- Katz, T.S.; Harhay, D.M.; Schmidt, J.W.; Wheeler, T.L. Identifying a list of Salmonella serotypes of concern to target for reducing risk of salmonellosis. Front. Microbiol. 2024, 15, 1307563. [Google Scholar] [CrossRef]
- Fritz, H.M.; Pereira, R.V.; Toohey-Kurth, K.; Marshall, E.; Tucker, J.; Clothier, K.A. Salmonella enterica Serovar Dublin from Cattle in California from 1993-2019: Antimicrobial Resistance Trends of Clinical Relevance. Antibiotics 2022, 11, 1110. [Google Scholar] [CrossRef] [PubMed]
- Campioni, F.; Vilela, F.P.; Cao, G.; Kastanis, G.; Dos Prazeres Rodrigues, D.; Costa, R.G.; Tiba-Casas, M.R.; Yin, L.; Allard, M.; Falcão, J.P. Whole genome sequencing analyses revealed that Salmonella enterica serovar Dublin strains from Brazil belonged to two predominant clades. Sci. Rep. 2022, 12, 10555. [Google Scholar] [CrossRef] [PubMed]
- Afema, J.A.; Mather, A.E.; Sischo, W.M. Antimicrobial Resistance Profiles and Diversity in Salmonella from Humans and Cattle, 2004–2011. Zoonoses Public Health 2015, 62, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rivera, L.D.; Moreno Switt, A.I.; Degoricija, L.; Fang, R.; Cummings, C.A.; Furtado, M.R.; Wiedmann, M.; den Bakker, H.C. Genomic characterization of Salmonella Cerro ST367, an emerging Salmonella subtype in cattle in the United States. BMC Genom. 2014, 15, 427. [Google Scholar] [CrossRef]
- Cummings, K.J.; Warnick, L.D.; Elton, M.; Rodriguez-Rivera, L.D.; Siler, J.D.; Wright, E.M.; Gröhn, Y.T.; Wiedmann, M. Salmonella enterica serotype Cerro among dairy cattle in New York: An emerging pathogen? Foodborne Pathog. Dis. 2010, 7, 659–665. [Google Scholar] [CrossRef]
- Fluit, A.C. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol. Med. Microbiol. 2005, 43, 1–11. [Google Scholar] [CrossRef]
- Mølbak, K. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 2005, 41, 1613–1620. [Google Scholar] [CrossRef]
- Burciaga, S.; Trachsel, J.M.; Sockett, D.; Aulik, N.; Monson, M.S.; Anderson, C.L.; Bearson, S.M.D. Genomic and phenotypic comparison of two variants of multidrug-resistant Salmonella enterica serovar Heidelberg isolated during the 2015–2017 multi-state outbreak in cattle. Front. Microbiol. 2023, 14, 1282832. [Google Scholar] [CrossRef]
- McMillan, E.A.; Weinroth, M.D.; Frye, J.G. Increased Prevalence of Salmonella Infantis Isolated from Raw Chicken and Turkey Products in the United States Is Due to a Single Clonal Lineage Carrying the pESI Plasmid. Microorganisms 2022, 10, 1478. [Google Scholar] [CrossRef]
- Fonseca, M.; Heider, L.C.; Stryhn, H.; McClure, J.T.; Léger, D.; Rizzo, D.; Dufour, S.; Roy, J.P.; Kelton, D.F.; Renaud, D.L.; et al. Frequency of isolation and phenotypic antimicrobial resistance of fecal Salmonella enterica recovered from dairy cattle in Canada. J. Dairy Sci. 2024, 107, 2357–2373. [Google Scholar] [CrossRef] [PubMed]
- Aleri, J.W.; Sahibzada, S.; Harb, A.; Fisher, A.D.; Waichigo, F.K.; Lee, T.; Robertson, I.D.; Abraham, S. Molecular epidemiology and antimicrobial resistance profiles of Salmonella isolates from dairy heifer calves and adult lactating cows in a Mediterranean pasture-based system of Australia. J. Dairy Sci. 2022, 105, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Noyes, N.R.; Yang, X.; Linke, L.M.; Magnuson, R.J.; Cook, S.R.; Zaheer, R.; Yang, H.; Woerner, D.R.; Geornaras, I.; McArt, J.A.; et al. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci. Rep. 2016, 6, 24645. [Google Scholar] [CrossRef]
- Carey, A.M.; Capik, S.F.; Giebel, S.; Nickodem, C.; Piñeiro, J.M.; Scott, H.M.; Vinasco, J.; Norman, K.N. Prevalence and Profiles of Antibiotic Resistance Genes mph(A) and qnrB in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli Isolated from Dairy Calf Feces. Microorganisms 2022, 10, 411. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Pradhan, A.K.; Karns, J.S.; Hovingh, E.; Wolfgang, D.R.; Vinyard, B.T.; Kim, S.W.; Salaheen, S.; Haley, B.J.; Van Kessel, J.A.S. Age-Associated Distribution of Antimicrobial-Resistant Salmonella enterica and Escherichia coli Isolated from Dairy Herds in Pennsylvania, 2013–2015. Foodborne Pathog. Dis. 2019, 16, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.V.; Siler, J.D.; Ng, J.C.; Davis, M.A.; Grohn, Y.T.; Warnick, L.D. Effect of on-farm use of antimicrobial drugs on resistance in fecal Escherichia coli of preweaned dairy calves. J. Dairy Sci. 2014, 97, 7644–7654. [Google Scholar] [CrossRef]
- Hille, K.; Ruddat, I.; Schmid, A.; Hering, J.; Hartmann, M.; von Münchhausen, C.; Schneider, B.; Messelhäusser, U.; Friese, A.; Mansfeld, R.; et al. Cefotaxime-resistant E. coli in dairy and beef cattle farms-Joint analyses of two cross-sectional investigations in Germany. Prev. Vet. Med. 2017, 142, 39–45. [Google Scholar] [CrossRef]
- Davis, M.A.; Hancock, D.D.; Besser, T.E.; Daniels, J.B.; Baker, K.N.; Call, D.R. Antimicrobial resistance in Salmonella enterica serovar Dublin isolates from beef and dairy sources. Vet. Microbiol. 2007, 119, 221–230. [Google Scholar] [CrossRef]
- Lipkens, Z.; Piepers, S.; De Vliegher, S. Impact of Selective Dry Cow Therapy on Antimicrobial Consumption, Udder Health, Milk Yield, and Culling Hazard in Commercial Dairy Herds. Antibiotics 2023, 12, 901. [Google Scholar] [CrossRef]
- Stevens, M.; Piepers, S.; Supré, K.; Dewulf, J.; De Vliegher, S. Quantification of antimicrobial consumption in adult cattle on dairy herds in Flanders, Belgium, and associations with udder health, milk quality, and production performance. J. Dairy. Sci. 2016, 99, 2118–2130. [Google Scholar] [CrossRef] [PubMed]
- Tello, M.; Ocejo, M.; Oporto, B.; Hurtado, A. Prevalence of Cefotaxime-Resistant Escherichia coli Isolates from Healthy Cattle and Sheep in Northern Spain: Phenotypic and Genome-Based Characterization of Antimicrobial Susceptibility. Appl. Environ. Microbiol. 2020, 86, e00742-20. [Google Scholar] [CrossRef] [PubMed]
- USDA. Can Antibiotics Be Used in Cattle Raising? Available online: https://ask.usda.gov/s/article/Can-antibiotics-be-used-in-cattle-raising#:~:text=Antibiotics%20may%20be%20given%20to,can%20exit%20the%20animal’s%20system (accessed on 5 May 2024).
- Rovira, P.; McAllister, T.; Lakin, S.M.; Cook, S.R.; Doster, E.; Noyes, N.R.; Weinroth, M.D.; Yang, X.; Parker, J.K.; Boucher, C.; et al. Characterization of the Microbial Resistome in Conventional and “Raised Without Antibiotics” Beef and Dairy Production Systems. Front. Microbiol. 2019, 10, 1980. [Google Scholar] [CrossRef]
- Wang, W.; Wei, X.; Wu, L.; Shang, X.; Cheng, F.; Li, B.; Zhou, X.; Zhang, J. The occurrence of antibiotic resistance genes in the microbiota of yak, beef and dairy cattle characterized by a metagenomic approach. J. Antibiot. 2021, 74, 508–518. [Google Scholar] [CrossRef]
- Warburton, D.W.; Bowen, B.; Konkle, A.; Crawford, C.; Durzi, S.; Foster, R.; Fox, C.; Gour, L.; Krohn, G.; LaCasse, P.; et al. A comparison of six different plating media used in the isolation of Salmonella. Int. J. Food Microbiol. 1994, 22, 277–289. [Google Scholar] [CrossRef] [PubMed]
- British Standards Institution. Methods for Microbiological Examination of Food and Animal Feeding Stuffs: Part 4. Detection of Salmonella; British Standards Institution: London, UK, 1990. [Google Scholar]
- Arroyo, G.; Arroyo, J.A. Efficiency of different enrichment and isolation procedures for the detection of Salmonella serotypes in edible offal. J. Appl. Bacteriol. 1995, 79, 360–367. [Google Scholar] [CrossRef]
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodford, N.; van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J. Antimicrob. Chemother. 2010, 65, 601–604. [Google Scholar] [CrossRef]
- D’Amato, R.F.; Hochstein, L.; Vernaleo, J.R.; Cleri, D.J.; Wallman, A.A.; Gradus, M.S.; Thornsberry, C. Evaluation of the BIOGRAM antimicrobial susceptibility test system. J. Clin. Microbiol. 1985, 22, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Fader, R.C.; Weaver, E.; Fossett, R.; Toyras, M.; Vanderlaan, J.; Gibbs, D.; Wang, A.; Thierjung, N. Multilaboratory study of the Biomic automated well-reading instrument versus MicroScan WalkAway for reading MicroScan antimicrobial susceptibility and identification panels. J. Clin. Microbiol. 2013, 51, 1548–1554. [Google Scholar] [CrossRef]
- Fukasawa, Y.; Ermini, L.; Wang, H.; Carty, K.; Cheung, M.S. LongQC: A Quality Control Tool for Third Generation Sequencing Long Read Data. G3 2020, 10, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Nanoporetech. Medaka. Available online: https://github.com/nanoporetech/medaka (accessed on 7 May 2023).
- Huang, Y.T.; Liu, P.Y.; Shih, P.W. Homopolish: A method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol. 2021, 22, 95. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Seemann, T. Snippy. Available online: https://github.com/tseemann/snippy (accessed on 7 May 2024).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016, 17, 238. [Google Scholar] [CrossRef]
- Lees, J.A.; Galardini, M.; Bentley, S.D.; Weiser, J.N.; Corander, J. pyseer: A comprehensive tool for microbial pangenome-wide association studies. Bioinformatics 2018, 34, 4310–4312. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).