Standardization of the Agar Plate Method for Bacteriophage Production
Abstract
1. Introduction
2. Results
2.1. Plaque Morphology
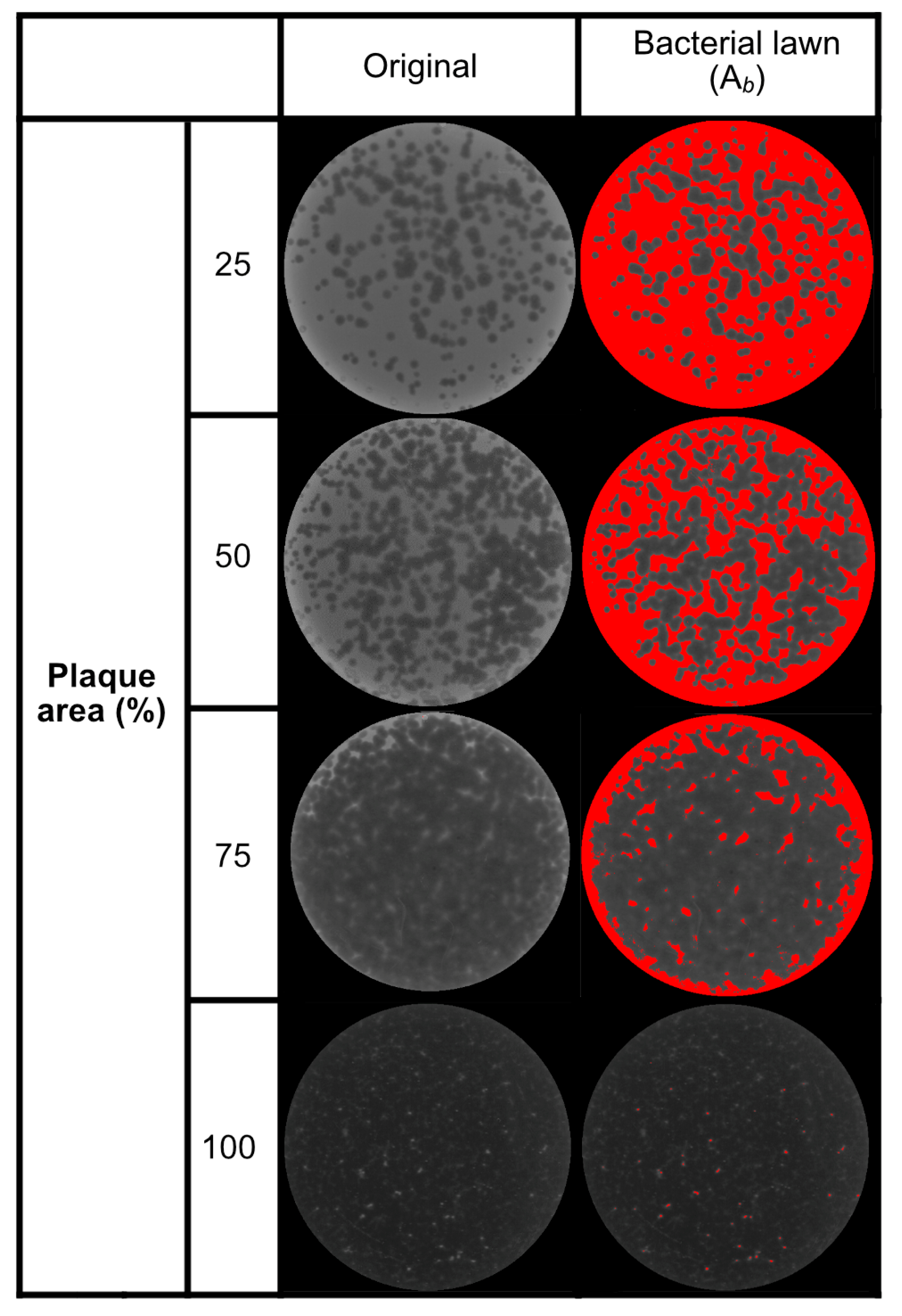
2.2. Analysis of Inoculum and Plaque Area and Regression Model

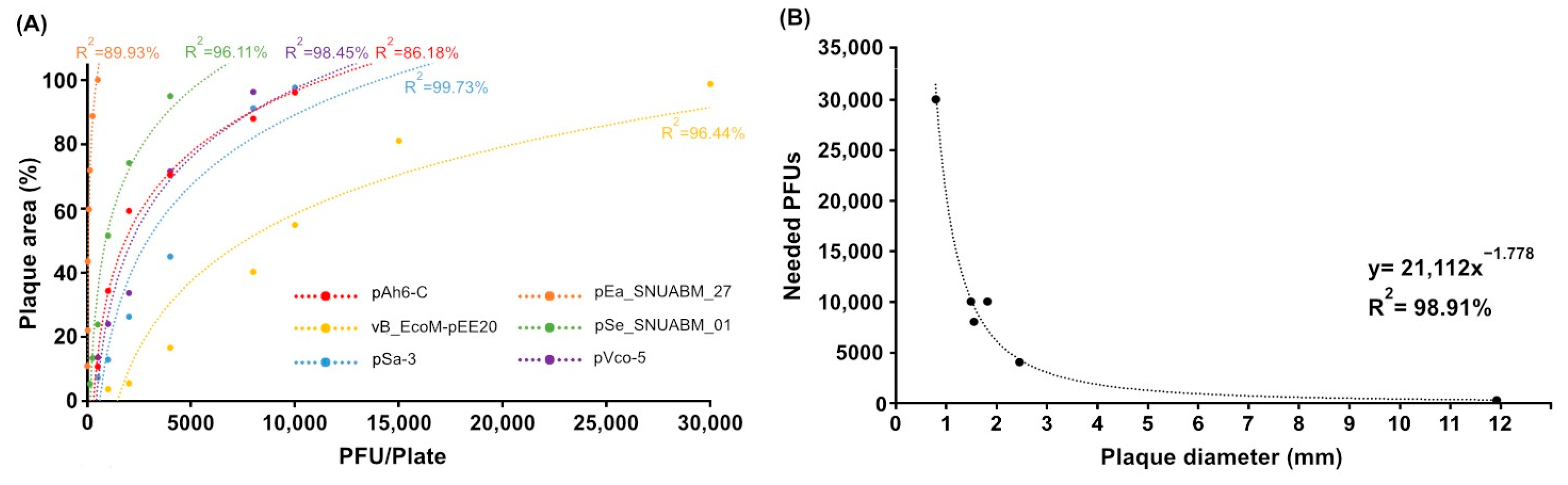
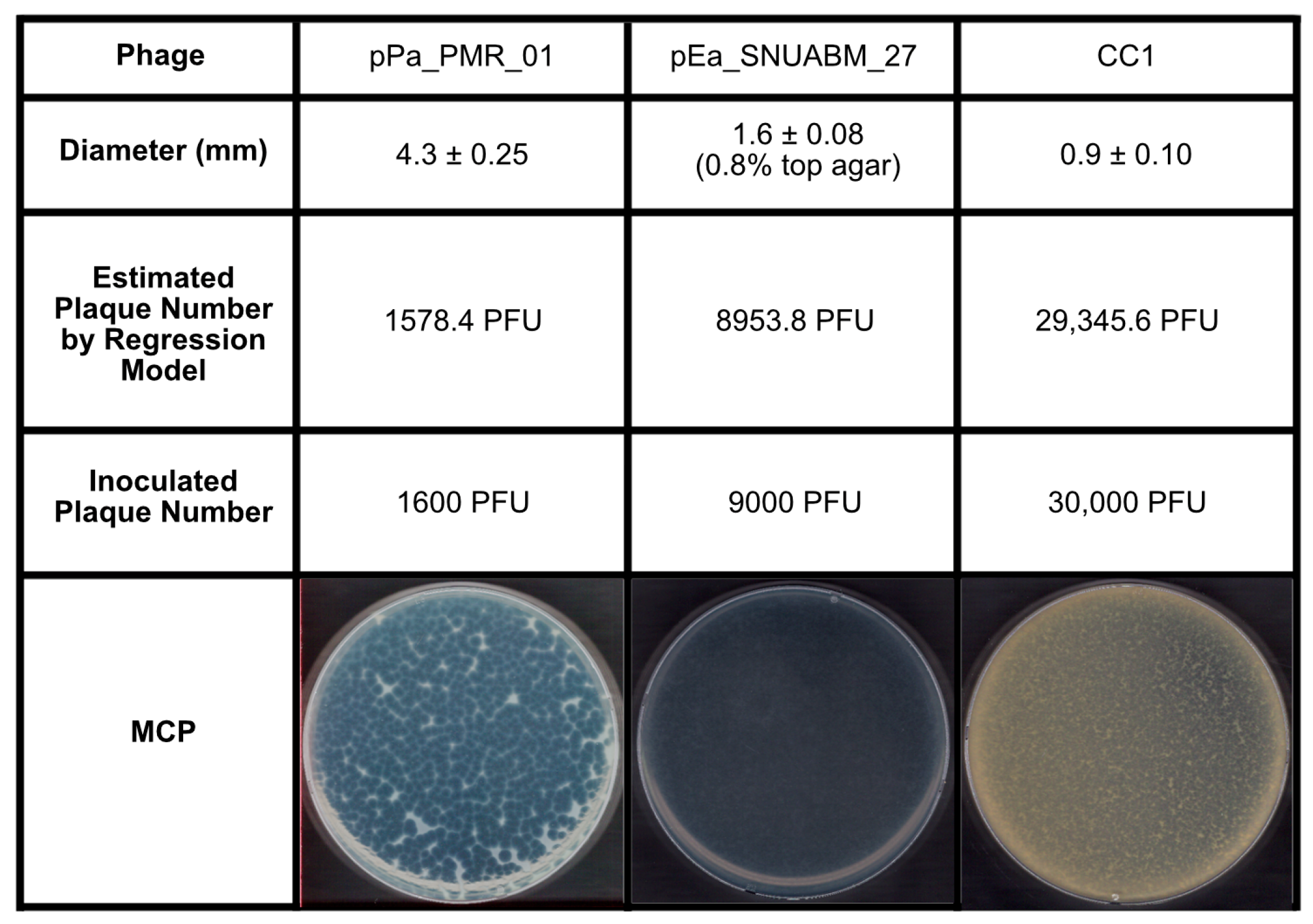
2.3. Validation of the Inoculum by PSC
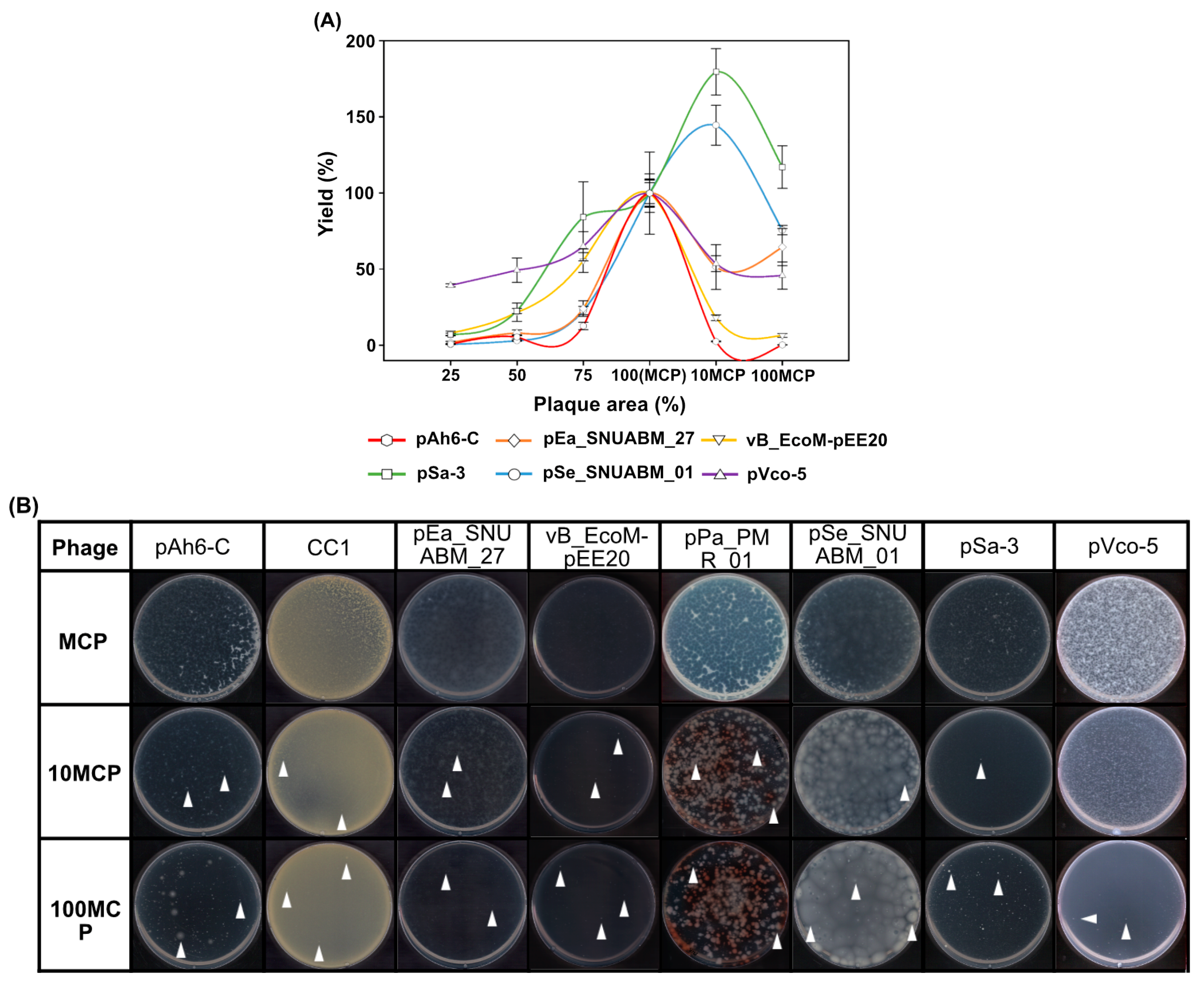
2.4. Optimization of the Phage Inoculum
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Culture Conditions
4.2. Preparation of Bacteriophages
4.3. Bacteriophage Plating, Plaque Area Analysis, and Harvest
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ho, C.S.; Wong, C.T.H.; Aung, T.T.; Lakshminarayanan, R.; Mehta, J.S.; Rauz, S.; McNally, A.; Kintses, B.; Peacock, S.J.; de la Fuente-Nunez, C.; et al. Antimicrobial Resistance: A Concise Update. Lancet Microbe 2024, 5, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.E.; Cadarette, D. Infectious Disease Threats in the Twenty-First Century: Strengthening the Global Response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, F.; Castagna, T.; Pantolini, B.; Campanardi, M.C.; Roperti, M.; Grotto, A.; Fattori, M.; Maso, L.D.; Carrara, F.; Zambarbieri, G.; et al. The challenge of antimicrobial resistance (AMR): Current status and future prospects. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2024, 397, 9603–9615. [Google Scholar] [CrossRef]
- Mantravadi, P.K.; Kalesh, K.A.; Dobson, R.C.J.; Hudson, A.O.; Parthasarathy, A. The Quest for Novel Antimicrobial Compounds: Emerging Trends in Research, Development, and Technologies. Antibiotics 2019, 8, 8. [Google Scholar] [CrossRef]
- Ahmad, M.; Khan, A.U. Global economic impact of antibiotic resistance: A review. J. Glob. Antimicrob. Resist. 2019, 19, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Asghar, R.; Muzammil, S.; Shafique, M.; Siddique, A.B.; Khurshid, M.; Ijaz, M.; Rasool, M.H.; Chaudhry, T.H.; Aamir, A.; et al. AMR and Sustainable Development Goals: At a crossroads. Glob. Health 2024, 20, 73. [Google Scholar] [CrossRef] [PubMed]
- Spellberg, B.; Bartlett, J.; Wunderink, R.; Gilbert, D.N. Novel approaches are needed to develop tomorrow’s antibacterial therapies. Am. J. Respir. Crit. Care Med. 2015, 191, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Murugaiyan, J.; Kumar, P.A.; Rao, G.S.; Iskandar, K.; Hawser, S.; Hays, J.P.; Mohsen, Y.; Adukkadukkam, S.; Awuah, W.A.; Jose, R.A.M.; et al. Progress in Alternative Strategies to Combat Antimicrobial Resistance: Focus on Antibiotics. Antibiotics 2022, 11, 200. [Google Scholar] [CrossRef]
- O’neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Review on Antimicrobial Resistance. 2014. Available online: https://amr-review.org/Publications.html (accessed on 12 August 2024).
- Ghosh, S.; Bornman, C.; Zafer, M.M. Antimicrobial Resistance Threats in the emerging COVID-19 pandemic: Where do we stand? J. Infect. Public. Health. 2021, 14, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Calap, P.; Delgado-Martínez, J. Bacteriophages: Protagonists of a Post-Antibiotic Era. Antibiotics 2018, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Pal, N.; Sharma, P.; Kumawat, M.; Singh, S.; Verma, V.; Tiwari, R.R.; Sarma, D.K.; Nagpal, R.; Kumar, M. Phage therapy: An alternative treatment modality for MDR bacterial infections. Infect. Dis. 2024, 56, 785–817. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.J.; Kwon, J.; Kim, S.G.; Lee, S.-J. The Biotechnological Application of Bacteriophages: What to Do and Where to Go in the Middle of the Post-Antibiotic Era. Microorganisms 2023, 11, 2311. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage Therapy: Various Perspectives on How to Improve the Art. In Host-Pathogen Interactions; Medina, C., López-Baena, F.J., Eds.; Springer: New York, NY, USA, 2018; pp. 113–127. [Google Scholar]
- Zhang, M.; Zhang, T.; Yu, M.; Chen, Y.-L.; Jin, M. The Life Cycle Transitions of Temperate Phages: Regulating Factors and Potential Ecological Implications. Viruses 2022, 14, 1904. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; De Vos, D.; Verbeken, G. Clinical application of bacteriophages in Europe. Microbiol. Aust. 2019, 40, 8–15. [Google Scholar] [CrossRef]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect. Dis. 2020, 7, ofaa389. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Callahan, M.T.L.; Woolston, J.W.; Sharma, M.; Sulakvelidze, A. Phage biocontrol for reducing bacterial foodborne pathogens in produce and other foods. Curr. Opin. Biotechnol. 2022, 78, 102805. [Google Scholar] [CrossRef] [PubMed]
- Kuek, M.; McLean, S.K.; Palombo, E.A. Application of bacteriophages in food production and their potential as biocontrol agents in the organic farming industry. Biol. Control 2022, 165, 104817. [Google Scholar] [CrossRef]
- Kakasis, A.; Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 2019, 53, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of bacteriophages in the agro-food sector: A long way toward approval. Front. Cell. Infect. Microbiol. 2018, 8, 296. [Google Scholar] [CrossRef]
- Schmidt, F.R. Optimization and scale up of industrial fermentation processes. Appl. Microbiol. Biotechnol. 2005, 68, 425–435. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Latz, S.; Romero, J.; Higuera, G.; García, K.; Bastías, R. Bacteriophage production models: An overview. Front. Microbiol. 2019, 10, 1187. [Google Scholar] [CrossRef]
- Jurač, K.; Nabergoj, D.; Podgornik, A. Bacteriophage production processes. Appl. Microbiol. Biotechnol. 2019, 103, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Abedon, S.T. Practical Methods for Determining Phage Growth Parameters. In Bacteriophages: Methods and Protocols; Clokie, M.R., Kropinski, A.M., Eds.; Humana Press: New York, NY, USA, 2009; Volume 1, pp. 175–202. [Google Scholar]
- João, J.; Lampreia, J.; Prazeres, D.M.F.; Azevedo, A.M. Manufacturing of bacteriophages for therapeutic applications. Biotechnol. Adv. 2021, 49, 107758. [Google Scholar] [CrossRef]
- González-Menéndez, E.; Arroyo-López, F.N.; Martínez, B.; García, P.; Garrido-Fernández, A.; Rodríguez, A. Optimizing propagation of Staphylococcus aureus infecting bacteriophage vB_sauM-phiIPLA-RODI on Staphylococcus xylosus using response surface methodology. Viruses 2018, 10, 153. [Google Scholar] [CrossRef]
- Kim, B.; Kim, E.S.; Yoo, Y.-J.; Bae, H.-W.; Chung, I.-Y.; Cho, Y.-H. Phage-derived antibacterials: Harnessing the simplicity, plasticity, and diversity of phages. Viruses 2019, 11, 268. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Dąbrowska, K.; Hodyra-Stefaniak, K.; Borysowski, J.; Międzybrodzki, R.; Weber-Dąbrowska, B. Phages targeting infected tissues: Novel approach to phage therapy. Future Microbiol. 2015, 10, 199–204. [Google Scholar] [CrossRef]
- Luong, T.; Salabarria, A.C.; Roach, D.R. Phage therapy in the resistance era: Where do we stand and where are we going? Clin. Ther. 2020, 42, 1659–1680. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Chen, K.; Jiang, Z.; Liu, Z.; Wang, S.; Ying, Y.; Zhang, J.; Yuan, S.; Huang, Z.; Gao, R.; et al. Evaluation of the impact of repeated intravenous phage doses on mammalian host–phage interactions. J. Virol. 2024, 98, e01359-23. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, N.; Rojas, M.I.; Netto Flores Cruz, G.; Hung, S.H.; Rohwer, F.; Barr, J.J. Phage on tap-a quick and efficient protocol for the preparation of bacteriophage laboratory stocks. PeerJ 2016, 4, e2261. [Google Scholar] [CrossRef]
- Kim, J.H.; Gomez, D.K.; Nakai, T.; Park, S.C. Isolation and identification of bacteriophages infecting ayu Plecoglossus altivelis altivelis specific Flavobacterium psychrophilum. Vet. Microbiol. 2010, 140, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, M.M.F.; Stimson, J.; Chae, G.T.; Tufenkji, N.; Ptacek, C.J.; Blowes, D.W.; Emelko, M.B. Optimal preparation and purification of PRD1-like bacteriophages for use in environmental fate and transport studies. Water Res. 2010, 44, 1114–1125. [Google Scholar] [CrossRef]
- Payaslian, F.; Gradaschi, V.; Rondón Salazar, L.; Dieterle, M.E.; Urdániz, E.; Di Paola, M.; Cárcamo, J.P.; Zon, F.; Sosa, E.; Porto, D.F.D.; et al. Isolation and characterization of vB_MsmS_Celfi: A new Mycobacterium tuberculosis bacteriophage. PHAGE Ther. Appl. Res. 2021, 2, 43–49. [Google Scholar]
- Skaradzińska, A.; Ochocka, M.; Śliwka, P.; Kuźmińska-Bajor, M.; Skaradziński, G.; Friese, A.; Roschanski, N.; Murugaiyan, J.; Roesler, U. Bacteriophage amplification–A comparison of selected methods. J. Virol. Methods 2020, 282, 113856. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.W.; Kim, H.J.; Yun, S.K.; Chai, J.Y.; Park, S.C. Genomic structure of the Aeromonas bacteriophage pAh6-C and its comparative genomic analysis. Arch. Virol. 2015, 160, 561–564. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, S.B.; Jo, S.J.; Cho, K.; Park, J.K.; Kwon, J.; Giri, S.S.; Kim, S.-W.; Kang, J.-W.; Jung, W.-J.; et al. Phage cocktail in combination with kasugamycin as a potential treatment for fire blight caused by Erwinia amylovora. Antibiotics 2022, 11, 1566. [Google Scholar] [CrossRef]
- Kim, S.G.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Chi, C.; Park, S.C. Complete genome sequence of Staphylococcus aureus bacteriophage pSa-3. Genome Announc. 2017, 5, e00182-17. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Oh, W.T.; Jun, J.W.; Park, S.C. Synergistic phage–surfactant combination clears IgE-promoted Staphylococcus aureus aggregation in vitro and enhances the effect in vivo. Int. J. Antimicrob. Agents 2020, 56, 105997. [Google Scholar] [CrossRef]
- Kim, H.J.; Giri, S.S.; Kim, S.G.; Kim, S.W.; Kwon, J.; Lee, S.B.; Park, S.C. Isolation and characterization of two bacteriophages and their preventive effects against pathogenic Vibrio coralliilyticus causing mortality of Pacific oyster (Crassostrea gigas) larvae. Microorganisms 2020, 8, 926. [Google Scholar] [CrossRef] [PubMed]
- Regulski, K.; Champion-Arnaud, P.; Gabard, J. Bacteriophage manufacturing: From early twentieth-century processes to current GMP. In Bacteriophages; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer: New York, NY, USA, 2021; pp. 699–729. [Google Scholar]
- Wiebe, K.G.; Cook, B.W.; Lightly, T.J.; Court, D.A.; Theriault, S.S. Investigation into scalable and efficient enterotoxigenic Escherichia coli bacteriophage production. Sci. Rep. 2024, 14, 3618. [Google Scholar] [CrossRef]
- Rohde, C.; Resch, G.; Pirnay, J.-P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Górski, A.; Hazan, R.; Huys, I.; Kakabadze, E.; et al. Expert opinion on three phage therapy related topics: Bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 2018, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; De Vos, D.; Verbeken, G.; Merabishvili, M.; Chanishvili, N.; Vaneechoutte, M.; Zizi, M.; Laire, G.; Lavigne, R.; Huys, I.; et al. The phage therapy paradigm: Prêt-à-porter or sur-mesure? Pharm. Res. 2011, 28, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.B.; Carvalho, C.; Azeredo, J.; Ferreira, E.C. Correction: Population Dynamics of a Salmonella Lytic Phage and Its Host: Implications of the Host Bacterial Growth Rate in Modelling. PLoS ONE 2015, 10, e0136007. [Google Scholar] [CrossRef] [PubMed]
- Cairns, B.J.; Timms, A.R.; Jansen, V.A.; Connerton, I.F.; Payne, R.J. Quantitative models of in vitro bacteriophage–host dynamics and their application to phage therapy. PLoS Pathog. 2009, 5, e1000253. [Google Scholar] [CrossRef]
- Storms, Z.J.; Brown, T.; Cooper, D.G.; Sauvageau, D.; Leask, R.L. Impact of the cell life-cycle on bacteriophage T4 infection. FEMS Microbiol. Lett. 2014, 353, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P. Phage therapy in the year 2035. Front. Microbiol. 2020, 11, 1171. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Djebara, S.; Steurs, G.; Griselain, J.; Cochez, C.; De Soi, S.; Glonti, T.; Spiessens, A.; Berghe, E.V.; Green, S.; et al. Personalized bacteriophage therapy outcomes for 100 consecutive cases: A multicentre, multinational, retrospective observational study. Nat. Microbiol. 2024, 9, 1434–1453. [Google Scholar] [CrossRef]
- Romero-Calle, D.; Guimarães Benevides, R.; Góes-Neto, A.; Billington, C. Bacteriophages as Alternatives to Antibiotics in Clinical Care. Antibiotics 2019, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Mattila, S.; Ruotsalainen, P.; Jalasvuori, M. On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front. Microbiol. 2015, 6, 1271. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Chen, H.; Zhang, M.; Zhao, Y.; Jiang, Y.; Liu, X.; Huang, W.; Ma, Y. Clinical experience of personalized phage therapy against carbapenem-resistant Acinetobacter baumannii lung infection in a patient with chronic obstructive pulmonary disease. Front. Cell. Infect. Microbiol. 2021, 11, 631585. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.R.; Oliveira, H.; Pires, D.P.; Dabrowska, K.; Azeredo, J. Phage therapy efficacy: A review of the last 10 years of preclinical studies. Crit. Rev. Microbiol. 2020, 46, 78–99. [Google Scholar] [CrossRef] [PubMed]
- Gencay, Y.E.; Birk, T.; Sørensen, M.C.H.; Brøndsted, L. Methods for isolation, purification, and propagation of bacteriophages of Campylobacter jejuni. In Methods in Molecular Biology; Butcher, J., Stintzi, A., Eds.; Humana Press: New York, NY, USA, 2017; pp. 19–28. [Google Scholar]
- Lavigne, R.; Burkal’tseva, M.V.; Robben, J.; Sykilinda, N.N.; Kurochkina, L.P.; Grymonprez, B.; Jonckx, B.; Krylov, V.N.; Mesyanzhinov, V.V.; Volckaert, G. The genome of bacteriophage φKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 2003, 312, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Kim, S.G.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Han, S.J.; Jeong, D.; Park, S.C. Isolation and characterisation of pVa-21, a giant bacteriophage with anti-biofilm potential against Vibrio alginolyticus. Sci. Rep. 2019, 9, 6284. [Google Scholar] [CrossRef] [PubMed]
- Supply, Storage, Propagation and Purification of Pahges (DSMZ). Available online: https://www.dsmz.de/fileadmin/user_upload/Collection_allg/DSMZ_Cultivation_of_Phages.pdf (accessed on 20 November 2024).
- Kim, S.G.; Giri, S.S.; Yun, S.; Kim, S.W.; Han, S.J.; Kwon, J.; Oh, W.T.; Lee, S.B.; Park, Y.H.; Park, S.C. Two novel bacteriophages control multidrug-and methicillin-resistant Staphylococcus pseudintermedius biofilm. Front. Med. 2021, 8, 524059. [Google Scholar] [CrossRef]
- Dennehy, J.J.; Abedon, S.T. Phage infection and lysis. In Bacteriophages: Biology, Technology, Therapy; Harper, D.R., Abedon, S.T., Burrowes, B.H., McConville, M.L., Eds.; Springer: New York, NY, USA, 2021; pp. 341–383. [Google Scholar]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.J.; Park, D.H.; Lee, Y.K.; Han, S.W.; Kwak, Y.S.; Oh, C.S. Complete genome sequence of Erwinia amylovora strain TS3128, a Korean strain isolated in an Asian pear orchard in 2015. Microbiol. Resour. Announc. 2021, 10, e0069421. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, J.H.; Jun, J.W.; Giri, S.S.; Chi, C.; Yun, S.; Kim, S.G.; Kim, S.W.; Kang, J.W.; Jeong, D.G.; et al. Complete genome sequence of Vibrio coralliilyticus 58, Isolated from Pacific oyster (Crassostrea gigas) larvae. Genome Announc. 2017, 5, e00437-17. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Giri, S.S.; Kim, S.W.; Kwon, J.; Lee, S.B.; Park, S.C. First isolation and characterization of Chryseobacterium cucumeris SKNUCL01, isolated from diseased pond loach (Misgurnus anguillicaudatus) in Korea. Pathogens 2020, 9, 397. [Google Scholar] [CrossRef]
- Pulverer, G.; Schütt-Gerowitt, H.; Schaal, K.P. Bacteriophages of Nocardia asteroides. Med. Microbiol. Immunol. 1975, 161, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Tanji, Y.; Tanaka, A.; Tani, K.; Kurimoto, M.; Miyanaga, K. IgG-dependent aggregation of Staphylococcus aureus inhibits bacteriophage attack. Biochem. Eng. J. 2015, 97, 17–24. [Google Scholar] [CrossRef]
- Tzipilevich, E.; Pollak-Fiyaksel, O.; Shraiteh, B.; Ben-Yehuda, S. Bacteria elicit a phage tolerance response subsequent to infection of their neighbors. EMBO J. 2022, 41, e109247. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.; Fusco, D. Superinfection exclusion: A viral strategy with short-term benefits and long-term drawbacks. PLoS Comput. Biol. 2022, 18, e1010125. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophage secondary infection. Virol. Sin. 2015, 30, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Bucher, M.J.; Czyż, D.M. Phage against the machine: The SIE-ence of superinfection exclusion. Viruses 2024, 16, 1348. [Google Scholar] [CrossRef] [PubMed]
- Millman, A.; Melamed, S.; Leavitt, A.; Doron, S.; Bernheim, A.; Hör, J.; Garb, J.; Bechon, N.; Brandis, A.; Lopatina, A.; et al. An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 2022, 30, 1556–1569. [Google Scholar] [CrossRef]
- Huiting, E.; Bondy-Denomy, J. Defining the expanding mechanisms of phage-mediated activation of bacterial immunity. Curr. Opin. Microbiol. 2023, 74, 102325. [Google Scholar] [CrossRef] [PubMed]
- Laub, M.T.; Typas, A. Principles of bacterial innate immunity against viruses. Curr. Opin. Immunol. 2024, 89, 102445. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Kwon, J.; Giri, S.S.; Yun, S.; Kim, H.J.; Kin, S.W.; Kang, J.W.; Bin Lee, S.; Jung, W.J.; Park, S.C. Strategy for mass production of lytic Staphylococcus aureus bacteriophage pSa-3: Contribution of multiplicity of infection and response surface methodology. Microb. Cell Fact. 2021, 20, 56. [Google Scholar] [CrossRef] [PubMed]
| Plaque Area (%) | ||||||
|---|---|---|---|---|---|---|
| 25 | 50 | 75 | 100 (=MCP) | 10 MCP | 100 MCP | |
| pAh6-C | 5.17 ± 0.38 × 109 PFU/mL | 4.50 ± 0.11 × 1010 PFU/mL | 1.05 ± 0.20 × 1010 PFU/mL | 8.33 ± 0.22 × 1011 PFU/mL | 1.97 ± 0.26 × 1010 PFU/mL | 1.43 ± 0.32 × 109 PFU/mL |
| pEa_27 | 4.33 ± 1.88 × 107 PFU/mL | 2.03 ± 0.46 × 108 PFU/mL | 6.10 ± 0.12 × 108 PFU/mL | 2.53 ± 0.23 × 109 PFU/mL | 1.30 ± 0.37 × 109 PFU/mL | 1.63 ± 0.30 × 109 PFU/mL |
| vB_EcoM-pEE20 | 1.02 ± 0.17 × 109 PFU/mL | 2.80 ± 0.77 × 109 PFU/mL | 7.20 ± 0.98 × 109 PFU/mL | 1.29 ± 0.16 × 1010 PFU/mL | 2.33 ± 0.24 × 109 PFU/mL | 8.23 ± 0.16 × 108 PFU/mL |
| pSe_01 | 5.30 ± 0.72 × 108 PFU/mL | 1.72 ± 0.12 × 109 PFU/mL | 6.40 ± 0.17 × 109 PFU/mL | 7.60 ± 0.64 × 109 PFU/mL | 1.37 ± 0.11 × 1010 PFU/mL | 8.90 ± 0.10 × 109 PFU/mL |
| pSa-3 | 1.07 ± 0.23 × 108 PFU/mL | 7.40 ± 0.73 × 108 PFU/mL | 5.86 ± 0.67 × 109 PFU/mL | 2.58 ± 0.17 × 1010 PFU/mL | 3.73 ± 0.33 × 1010 PFU/mL | 1.95 ± 0.77 × 1010 PFU/mL |
| pVco-5 | 1.83 ± 0.47 × 109 PFU/mL | 2.30 ± 0.37 × 109 PFU/mL | 3.03 ± 0.45 × 109 PFU/mL | 4.67 ± 0.41 × 109 PFU/mL | 2.50 ± 0.24 × 109 PFU/mL | 2.13 ± 0.41 × 109 PFU/mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, S.J.; Lee, Y.M.; Cho, K.; Park, S.Y.; Kwon, H.; Giri, S.S.; Lee, S.B.; Jung, W.J.; Park, J.H.; Hwang, M.H.; et al. Standardization of the Agar Plate Method for Bacteriophage Production. Antibiotics 2025, 14, 2. https://doi.org/10.3390/antibiotics14010002
Jo SJ, Lee YM, Cho K, Park SY, Kwon H, Giri SS, Lee SB, Jung WJ, Park JH, Hwang MH, et al. Standardization of the Agar Plate Method for Bacteriophage Production. Antibiotics. 2025; 14(1):2. https://doi.org/10.3390/antibiotics14010002
Chicago/Turabian StyleJo, Su Jin, Young Min Lee, Kevin Cho, Seon Young Park, Hyemin Kwon, Sib Sankar Giri, Sung Bin Lee, Won Joon Jung, Jae Hong Park, Mae Hyun Hwang, and et al. 2025. "Standardization of the Agar Plate Method for Bacteriophage Production" Antibiotics 14, no. 1: 2. https://doi.org/10.3390/antibiotics14010002
APA StyleJo, S. J., Lee, Y. M., Cho, K., Park, S. Y., Kwon, H., Giri, S. S., Lee, S. B., Jung, W. J., Park, J. H., Hwang, M. H., Park, D. S., Park, E. J., Kim, S. W., Jun, J. W., Kim, S. G., Kim, J. H., & Park, S. C. (2025). Standardization of the Agar Plate Method for Bacteriophage Production. Antibiotics, 14(1), 2. https://doi.org/10.3390/antibiotics14010002








