The Invisible Threat of Antibiotic Resistance in Food
Abstract
:1. Introduction
2. Antibiotics and Their Effects on Bacterial Resistance
| Food Type | Bacteria | Antibiotics | Genes | Reference | |
|---|---|---|---|---|---|
| Vegetables, fruits | Lettuce, romaine lettuce | Staphylococcus aureus, Bacillus cereus, E. coli, Enterococcus spp., Aeromonas spp., Clostridium perfringens, Yersinia spp., Campylobacter spp., Salmonella enterica, Listeria spp, Klebsiella pneumoniae | methicillin, macrolide, aminoglycoside, fosfomycin, lincosamide fluoroquinolone, β-lactam, rifampin, tetracycline, sulfonamides, vancomycin, lincosamides, and type B streptogramin (MLSB), oxytetracycline, carbapenem | mecA, mdf(A), aph(3′)-Ia, fosA, lnu(A), lsa(A) and sal(A), oqxA, oqxB and qnrS1, mecA, blaTEM-116, blaACT-15, blaZ, blaLAP-2, blaOXY-1-3, tet(L), tet(M), BLA-1, BLA-2, sul1, str(A), erm(F), str(B), aad(A), int1, IncP oriT, IncQ repB, incW, int3, tet(A), tet(Q), tet(S), str(A), erm(B), blaOXA1, blaVIM, blaTEM tet(B), tet(C), tet(G), tet(L), blaOXA-48 | [45,46,47,48,49,50,51,52] |
| Radish | E. coli, Enterococcus spp., Aeromonas spp., Clostridium perfringens, Yersinia spp., Campylobacter spp., Salmonella enterica, Listeria | aminoglycosides, beta-lactams, macrolides, sulfonamides, tetracyclines, vancomycin, lincosamides, and type B streptogramin (MLSB), | sul1, str(A), erm(F), str(B), aad(A), int1, IncP oriT, IncQ oriV, int2, int3, tet(A), str(A), str(B), erm(B), erm(E), blaCTX-M, blaVIM, blaTEM | [49,50] | |
| Carrot | Staphylococcus aureus, E. coli, Enterococcus spp., Aeromonas spp., Clostridium perfringens, Yersinia spp., Campylobacter spp., Salmonella enterica, Listeria | methicillin, macrolide, aminoglycoside, fosfomycin, lincosamide fluoroquinolone, β-lactam, sulfonamides, tetracyclines, vancomycin, lincosamides, and type B streptogramin (MLSB), colistin | mecA, mdf(A), aph(3′)-Ia, fosA, lnu(A), lsa(A) and sal(A), oqxA, oqxB and qnrS1, mecA, blaTEM-116, blaACT-15, blaZ, blaLAP-2, blaOXY-1-3, sul1, str(A), erm(F), str(B), aad(A), int1, IncP oriT, IncQ oriV, int1, tet(A), tet(S), erm(B), erm(C), erm(E), blaVIM, blaTEM, mcr-1 | [45,49,50,53] | |
| Tomato, cherry tomato | Staphylococcus aureus, E. coli, Clostridium perfringen, Yersinia sp., Campylobacter sp. | methicillin, macrolide, aminoglycoside, fosfomycin, lincosamide fluoroquinolone, β-lactam, lincosamides, and type B streptogramin (MLSB), sulfonamide, tetracycline, | mecA, mdf(A), aph(3′)-Ia, fosA, lnu(A), lsa(A) and sal(A), oqxA, oqxB and qnrS1, mecA, blaTEM-116, blaACT-15, blaZ, blaLAP-2, blaOXY-1-3 IncP oriT, incY, int2, int3, tet(A), tet(T) tet(S), aad(A), str(A), str(B), erm(B), erm(E), blaCTX-M, blaVIM, blaTEM | [45,50] | |
| Pepper | E. coli, Clostridium perfringen, Yersinia sp., Campylobacter sp. | macrolides, lincosamides, and type B streptogramin (MLSB), aminoglycoside, sulfonamide, tetracycline, β-lactam | nt3, tet(T), str(B), sul1, vat(B), blaOXAII | [50] | |
| Cucumber | Staphylococcus aureus, E. coli, Clostridium perfringen, Yersinia sp., Campylobacter sp. | methicillin, macrolide, aminoglycoside, fosfomycin, lincosamide fluoroquinolone, β-lactam, lincosamides, and type B streptogramin (MLSB), sulfonamide, tetracycline, | mecA, mdf(A), aph(3′)-Ia, fosA, lnu(A), lsa(A) and sal(A), oqxA, oqxB and qnrS1, mecA, blaTEM-116, blaACT-15, blaZ, blaLAP-2, blaOXY-1-3, IncP oriT, IncP trfA1, str(B), sul1, erm(B), blaOXAII | [45,50] | |
| Spinach | Pseudomonas teessidea, Morganella morganii | cefotaxime, ceftazidime, carbapenem | blaCTX-M-15, blaKPC | [54,55] | |
| Garlic chives | Bacillus cereus | rifampin, tetracycline, β-lactam | tet(L), tet(M), BLA-1, BLA-2 | [46,47,48] | |
| Perilla leaf | Bacillus cereus | rifampin, tetracycline, β-lactam | tet(L), tet(M), BLA-1, BLA-2 | [46,47,48] | |
| Cabbage | Staphylococcus aureus | methicillin, macrolide, aminoglycoside, fosfomycin, lincosamide fluoroquinolone, β-lactam | mecA, mdf(A), aph(3′)-Ia, fosA, lnu(A), lsa(A) and sal(A), oqxA, oqxB and qnrS1, mecA, blaTEM-116, blaACT-15, blaZ, blaLAP-2 and blaOXY-1-3 | [45] | |
| Watermelon, honeydew melon, peach, grape | Staphylococcus aureus | methicillin, macrolide, aminoglycoside, fosfomycin, lincosamide fluoroquinolone, β-lactam | mecA, mdf(A), aph(3′)-Ia, fosA, lnu(A), lsa(A) and sal(A), oqxA, oqxB and qnrS1, mecA, blaTEM-116, blaACT-15, blaZ, blaLAP-2 and blaOXY-1-3 | [45] | |
| Orange | Klebisiella pneumoniae | colistin, polymyxin B, ampicillin | bla, mcr-1, SHV-110 | [56] | |
| Apple | E. coli | aminoglycoside, colistin, polymyxin B, chloromycetin, sulfonamide, tetracycline, iclaprim | mcr-1, aadA2, aadA1, floR, cmlA1, sul2, sul3, tetA, tetM, dfrA12, mdfA | [56] | |
| Drinking water | Campylobacter spp. Enterococcus spp. Listeria Shigella Staphylococcus aureus Streptococcus pneumoniae Pseudomonas aeruginosa, | erythromycin, aminoglycosides, amphenicols, quinolone, sulfonamides, tetracyclines, β-lactamase, vancomycin | ermB aph(3′)-II cmlA, floR oqxB, qepA sul2 tetO, tetQ, tetW blaTEM vanA | [3,57,58,59,60,61,62,63,64] | |
| Meat/meat-products | Hamburger broiler chicken, poultry | E. coli, Salmonella Enterococcus spp. | amoxicillin, penicillin, cephalexin | blaTEM | [65] |
| erythromycin | tetM, tetL, ermB | [66] | |||
| ciprofloxacin | parC, gyrA | [67,68] | |||
| Dairy products | Cheese | Salmonella enterica | trimethoprim/sulfamethoxazole, ciprofloxacin, cefoxitin, cefuroxime axetil, cefuroxime | aac(6′), mdtK, cat_1, cat_4, golS, mdsA, mdsB, mdsC, rssB+, sdiA, ant(9) | [69] |
| Enterococcus faecalis, E. faecium, E. gallinarum, E. avium, E. casseliflavis | vancomycin, gentamicin, kanamycin, rifampin, tetracycline; erythromycin, lincomycin, linezolid, quinopristine/dalfopristine, chloramphenicol, streptomycin, ciprofloxacin | tetM, ermB, cad, tetL, aph(3)IIIa, acc6-le-aph(2)-la | [70] | ||
| Gram-negative bacteria | cefepime, ertapenem gentamicin, ampicillin ampicillin, sulbactam, chloramphenicol, tetracycline, ciprofloxacin, ceftazidime, sulfamethoxazol, trimethoprim | int 1, tet b, int 2, Shv, tet a, ctx—M, Tem, ctx- M15, oxa—48 | [71] | ||
| Salmonella Typhimurium, S. Typhimurium, S. Infantis, S. Virchow, S. Tsevie, S. Rissen, S. Shubra, S. Anatum | ampicillin, amoxicillin, amoxycillin-clavulanic acid, cefazolin, cephalothin, cefoxitin, ceftazidime, cefepime, imipenem, meropenem, aztreonam, vancomycin, gentamicin, amikacin, neomycin, tetracycline, erythromycin, clindamycin, ciprofloxacin, sulfamethoxazole, trimethoprim/sulfamethoxazole | blaOXA-1, blaOXA-2, blaTEM-1, blaCTX-M, blaCMY-1, blaCMY-2 | [41] | ||
| Cheeses from Bovine, Ovine, and Caprine Milk | Leuconostoc lactis, Leuconostoc mesenteroides, Lactococcus lactis, Lactococcus garviae, Enterococcus faecalis, Lacticaseibacillus plantarum, L. pentosus, L. delbrueckii, L. helveticus, L. brevis, L. casei, L. paracasei | tetracycline, erythromycin, chloramphenicol | tet(M,L,W), ermB, cat-TC | [72] | |
| Raw milk and artisanal cheese | Escherichia cvli | amoxacillin—clavulanate, aztreonam, cefepime, ceftazidime, ceftriaxone, cefotaxime, meropenem, imipenem, cefoxitin, ampicillin, tetracycline, doxycycline | blaTEM | [73] | |
| Raw milk (Bovine) | Escherichia cvli | azithromycin, chloramphenicol, ceftriaxone, penicillin, gentamicin, amoxicillin, tetracycline, cephalexin | blaSHV, blaTEM | [37] | |
| Listeria monocytogenes | ND * | ||||
| Staphylococcus aureus | blaZ, mecA | ||||
| Raw milk (Bovine, Ovine, and Caprine) | Staphylococcus aureus | cefoxitin | SCCmec- Iva | [74] | |
| Mastitis milk (Bovine) | Staphylococcus aureus | cefoxitin, ampicillin, gentamicin, norfloxacin, streptomycin, ciprofloxacin, trimethoprim–Sulfamethoxazole, tetracycline, erythromycin, chloramphenicol | blaZ, tetM, tetK, strB, msrA, ermB, ermC | [75] | |
| Pasteurized milk | Bacillus cereus, B. licheniformis, B. paralicheniformis, B. pumilus, B. safensis, B. Subtilis, B. toyonesis, B. invictae | penicillin, ampicillin, tetracycline, trimethoprim- sulfamethoxazole | tetL | [76] |
| Isolated Genera | Isolated Species/Serotype | Food Source | Resistance Phenotype | Resistance Genes | References |
|---|---|---|---|---|---|
| Salmonella | enterica/Typhimurium | poultry meat, eggs | amoxicillin-clavulanic acid, ampicillin, gentamicin, enrofloxacin, kanamycin, cefixime, cefepime, chloramphenicol, sulfamethoxazole/trimethoprim | blaPSE-1, blaCMY-2, blaTEM, ampC | [77] |
| enterica/Infantis | food from animal origin | tetracycline | tet(A) | [78] | |
| enterica/Dublin | ground beef | ceftriaxone and tetracycline | blaCMY-2, tet(A) | [79] | |
| enterica/Derby and Typhimurium | pork, poultry | cefotaxime | blaTEM, blaSHV, bla CTX-M | [80] | |
| enterica/Heidelberg | pork chop, chicken breast | ampicillin, amoxicillin, clavulanic acid, cefoxitin, ceftiofur | blaCMY | [81] | |
| enterica/Kentucky | cow’s milk | nalidixic acid, ciprofloxacin, amoxicillin–clavulanic acid, cefotaxime | blaTEM, ampC(FOX) | [82] | |
| enterica/Anatum | cow’s milk | nalidixic acid, ciprofloxacin, ofloxacin | qnrB | ||
| enterica/Enteritidis | chicken meat | nalidixic acid, cefotaxime | blaTEM, ampC(EBC) | ||
| Campylobacter | jejuni, coli | chicken, turkey, swine, cattle | tetracycline, quinolone | tet(O), gyrA | [83] |
| chicken | cephalosporin, quinolone, fluoroquinolone | [84] | |||
| jejuni | poultry | ciprofloxacin, nalidixic acid, tetracycline | ND * | [85] | |
| Clostridium | perfringens | fish, shellfish | tetracycline, clindamycin, ampicillin, penicillin, ceftriaxone | ND | [86] |
| duck | gentamicin, bacitracin, lincomycin, tetracycline | ND | [87] | ||
| water | vancomycin, penicillin, erythromycin, tetracycline, trimethoprim, kasugamycin, bacitracin | vanRG, vanRI, bla2, ermQ, tetB(P), dfrK, ksgA, bacA | [88] | ||
| Listeria | monocytogenes | chicken meat | ceftriaxone, cefotetan, amoxicillin, amikacin, ertapenem, erythromycin, ciprofloxacin, trimethoprim | sul1, sul2 | [89] |
| food of animal origin | tetracycline | tetM | [90] | ||
| freshly mixed sausage | cefoxitin, nalidixic acid, streptomycin, erythromycin, clindamycin, rifampicin, meropenem, tetracycline, trimethoprim–sulfamethoxazole | tetM | [91] | ||
| juice | clindamycin, meropenem trimethoprim/sulfamethoxazole | sul1 | [92] | ||
| Yersinia | enterocolitica | pork | neomycin, streptomycin, imipenem, sulfamethoxazole, vancomycin, nitroimidazole, amoxicillin, ampicillin, florfenicol, tiamulin, nalidixic acid | emrD, yfhD, marC | [93] |
| meat | tetracycline, streptomycin, trimethoprim/sulfamethoxazole, cefazolin, chloramphenicol | tetA, aph(6)-Id, aph(3″)-Ib, sul2 | [94] | ||
| chicken meat | ampicilli, ticarcillin, cefoxitin | blaA, blaB | [95] | ||
| Escherchia | coli/verotoxin producing (VTEC) | meat | ampicillin, amoxicillin/clavulanate, caphalothin, streptomycin, tetracycline, nalidixic acid, trimethoprim/sulfamethoxazole | blaTEM, strA, strB, tetB, sul2 | [96] |
| milk | imipenem, meropenem, ampicillin, cephazolin, nalidixic acid, streptomycin, kanamycin, sulfamethoxazole/trimethoprim | blaVIM, blaTEM, | [97] | ||
| meat | ampicillin, cephazolin, cefotaxime | blaTEM, blaCTX | |||
| beef | amoxicillin-clavulanic acid, ampicillin, aztreonam, chloramphenicol, ciprofloxacin, cefpodoxime, ceftriaxone, cefotetan, cefotaxime, cefoxitin, gentamicin, kanamycin, nalidixic acid, oxacillin, spectinomycin, streptomycin, sulfamethoxazole/trimethoprim, tetracycline | blaTEM-1, qnrB, blaCMY-2, blaCTX-M-3, floR | [98] | ||
| chicken | amoxicillin-clavulanic acid, ampicillin, amoxicillin-clavulanic acid, ampicillin, aztreonam, chloramphenicol, ciprofloxacin, cefpodoxime, ceftriaxone, cefotetan, cefotaxime, cefoxitin, kanamycin, nalidixic acid, oxacillin, spectinomycin, streptomycin, sulfamethoxazole/trimethoprim, tetracycline | blaTEM-1, blaCTX-M-15 | |||
| milk | amoxicillin-clavulanic acid, ampicillin, amoxicillin-clavulanic acid, ampicillin, aztreonam, chloramphenicol, cefotetan, ciprofloxacin, cefpodoxime, ceftriaxone, cefotaxime, cefoxitin, gentamicin | blaTEM-1, qnrB, floR | |||
| cheese | amoxicillin-clavulanic acid, ampicillin, amoxicillin-clavulanic acid, ampicillin, aztreonam, chloramphenicol, ciprofloxacin, cefotetan, ciprofloxacin, cefotaxime, cefoxitin, gentamicin, kanamycin, nalidixic acid, oxacillin, spectinomycin, streptomycin, sulfamethoxazole/trimethoprim, tetracycline | blaTEM-1, qnrB, blaCTX-M-15, aac (6′)-Ib-cr |
2.1. Antibiotic Resistance and Tolerance: Adaptation Strategies
2.2. Effect of Different Stressors on Antibiotic Resistance of Foodborne Bacteria
3. Antibiotic Resistance in Traditional Foods
3.1. Antibiotic Resistance in Vegetables and Fruits
3.2. Antibiotic Resistance of Foodborne Pathogenic Bacteria in Meat
3.3. Antibiotic Resistance in Dairy/Fermented Foods
Antibiotic Resistance of LAB in Fermented Dairy Products
4. Antibiotic Resistance in Drinking Water
5. Antibiotic Resistance in Novel Foods
5.1. Antibiotic Resistance Gene Migration Between Microalgae and Bacteria
5.2. Microbiota of Edible Insects and Prevalence of Antibiotic Resistance Genes in Their Bacteria
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Das, D.J.; Shankar, A.; Johnson, J.B.; Thomas, S. Critical Insights into Antibiotic Resistance Transferability in Probiotic Lactobacillus. Nutrition 2020, 69, 110567. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mukherjee, M.M.; Varela, F.M. Modulation of Bacterial Multidrug Resistance Efflux Pumps of the Major Facilitator Superfamily. Int. J. Bacteriol. 2013, 2013, 204141. [Google Scholar] [CrossRef] [PubMed]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Flórez, A.B.; Campedelli, I.; Delgado, S.; Alegría, Á.; Salvetti, E.; Felis, G.E.; Mayo, B.; Torriani, S. Antibiotic Susceptibility Profiles of Dairy Leuconostoc, Analysis of the Genetic Basis of Atypical Resistances and Transfer of Genes In Vitro and in a Food Matrix. PLoS ONE 2016, 11, e0145203. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.I.; Joo, I.S.; Choi, J.H.; Jung, K.H.; Choi, E.J.; Han, M.K.; Hwang, I.G. Prevalence and Antimicrobial Resistance of Enterococcus spp. Isolated from Meat and Fishery Production in Korea. Food Sci. Biotechnol. 2013, 22, 161–165. [Google Scholar] [CrossRef]
- Gordoncillo, M.J.N.; Donabedian, S.; Bartlett, P.C.; Perri, M.; Zervos, M.; Kirkwood, R.; Febvay, C. Isolation and Molecular Characterization of Vancomycin-Resistant Enterococcus faecium from Swine in Michigan, USA. Zoonoses Public Health 2013, 60, 319–326. [Google Scholar] [CrossRef]
- Treiber, F.M.; Beranek-Knauer, H. Antimicrobial Residues in Food from Animal Origin—A Review of the Literature Focusing on Products Collected in Stores and Markets Worldwide. Antibiotics 2021, 10, 534. [Google Scholar] [CrossRef]
- Pardon, B.; Catry, B.; Dewulf, J.; Persoons, D.; Hostens, M.; Bleecker, K.D.; Deprez, P. Prospective Study on Quantitative and Qualitative Antimicrobial and Antiinflammatory Drug Use in White Veal Calves. J. Antimicrob. Chemother. 2012, 67, 1027–1038. [Google Scholar] [CrossRef]
- Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. [Google Scholar] [CrossRef]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef] [PubMed]
- Harbarth, S.; Balkhy, H.H.; Goossens, H.; Jarlier, V.; Kluytmans, J.; Laxminarayan, R.; Saam, M.; Van Belkum, A.; Pittet, D.; For the World Healthcare-Associated Infections Resistance Forum Participants. Antimicrobial Resistance: One World, One Fight! Antimicrob. Resist. Infect. Control 2015, 4, 49. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC); European Food Safety Authority (EFSA); European Medicines Agency (EMA). Third Joint Inter-Agency Report on Integrated Analysis of Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals in the EU/EEA. EFSA J. 2021, 19, e06712. [Google Scholar] [CrossRef]
- Food and Drug Administration, U.S., Department of Health and Human Services, Center for Veterinary Medicine. Guidance for Industry: The Judicious Use of Medically Important Antimicrobial Drugs in Food-Producing Animals. 2012. Available online: http://www.fda.gov/downloads/animalveterinary/guidancecomplianceenforcement/guidanceforindustry/ucm216936.pdf (accessed on 25 February 2025).
- Xiao, Y.; Li, L. China’s National Plan to Combat Antimicrobial Resistance. Lancet Infect. Dis. 2016, 16, 1216–1218. [Google Scholar] [CrossRef] [PubMed]
- Rafailidis, P.I.; Kofteridis, D. Proposed amendments regarding the definitions of multidrug-resistant and extensively drug-resistant bacteria. Expert Rev. Anti. Infect. Ther. 2021, 20, 139–146. [Google Scholar] [CrossRef]
- Fu, S.; Wang, Q.; Wang, R.; Zhang, Y.; Lan, R.; He, F.; Yang, Q. Horizontal Transfer of Antibiotic Resistance Genes within the Bacterial Communities in Aquacultural Environment. Sci. Total Environ. 2022, 820, 153286. [Google Scholar] [CrossRef]
- Davison, H.C.; Woolhouse, M.E.J.; Low, J.C. What is antibiotic resistance and how can we measure it? Trends Microbiol. 2000, 8, 554–559. [Google Scholar] [CrossRef]
- Milijasevic, M.; Veskovic-Moracanin, S.; Babic Milijasevic, J.; Petrovic, J.; Nastasijevic, I. Antimicrobial Resistance in Aquaculture: Risk Mitigation within the One Health Context. Foods 2024, 13, 2448. [Google Scholar] [CrossRef]
- Nijsingh, N.; Munthe, C.; Lindblom, A.; Åhrén, C. Screening for Multi-Drug-Resistant Gram-Negative Bacteria: What Is Effective and Justifiable. Monash Bioeth. Rev. 2020, 38, S72–S90. [Google Scholar] [CrossRef]
- Antibiotic Resistance Has Claimed at Least One Million Lives Each Year Since 1990. Available online: https://www.ox.ac.uk/news/2024-09-17-antibiotic-resistance-has-claimed-least-one-million-lives-each-year-1990 (accessed on 20 February 2025).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2015. EFSA J. 2017, 15, 4694. [Google Scholar] [CrossRef]
- Nji, E.; Kazibwe, J.; Hambridge, T.; Joko, C.A.; Larbi, A.A.; Damptey, L.A.O.; Nkansa-Gyamfi, N.A.; Lundborg, C.S.; Lien, L.T.Q. High prevalence of antibiotic resistance in commensal Escherichia coli from healthy human sources in community settings. Sci. Rep. 2021, 11, 3372. [Google Scholar] [CrossRef] [PubMed]
- Yahav, D.; Shepshelovich, D.; Tau, N. Cost analysis of new antibiotics to treat multidrug-resistant bacterial infections: Mind the gap. Infect. Dis. Ther. 2021, 10, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Skov, R.L.; Monnet, D.L. Plasmid-Mediated Colistin Resistance (mcr-1 Gene): Three Months Later, the Story Unfolds. Eurosurveillance 2016, 21, 30155. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards. Scientific Opinion on the Public Health Risks of Bacterial Strains Producing Extended-Spectrum β-Lactamases and/or AmpC β-Lactamases in Food and Food-Producing Animals. EFSA J. 2011, 9, 2322. [Google Scholar] [CrossRef]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; Van der Zwaluw, K.; et al. Extended-Spectrum β-Lactamase Genes of Escherichia coli in Chicken Meat and Humans, the Netherlands. Emerg. Infect. Dis. 2011, 17, 1216. [Google Scholar] [CrossRef]
- Beyene, T. Veterinary Drug Residues in Food-Animal Products: Its Risk Factors and Potential Effects on Public Health. J. Vet. Schi. Technol. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- Bacanli, M.; Basaran, N. Importance of Antibiotic Residues in Animal Food. Food Chem. Toxicol. 2019, 125, 462–466. [Google Scholar] [CrossRef]
- Francino, M.P. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 2016, 6, 1543. [Google Scholar] [CrossRef]
- Wall, B.A.; Mateus, A.; Marshall, L.; Pfeiffer, D.U.; Lubroth, J.; Ormel, H.J.; Otto, P.; Patriarchi, A. Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production; FAO: Rome, Italy, 2016; ISBN 978-92-5-109441-9. [Google Scholar]
- Manie, T.; Brözel, V.S.; Veith, W.J.; Gouws, P.A. Antimicrobial Resistance of Bacterial Flora Associated with Bovine Products in South Africa. J. Food Prot. 1999, 62, 615–618. [Google Scholar] [CrossRef]
- Voidarou, X.; Alexopoulos, A.; Plessas, S.; Bezirtzoglou, E. Antibiotic Profile of Common Pathogens Related to Food Safety and Health. J. Ege Acad. Rev. 2009, 9, 961–967. [Google Scholar] [CrossRef]
- Gao, T.; Ding, Y.; Wu, Q.; Wang, J.; Zhang, J.; Yu, S.; Yu, P.; Liu, C.; Kong, L.; Feng, Z.; et al. Prevalence, Virulence Genes, Antimicrobial Susceptibility, and Genetic Diversity of Bacillus cereus Isolated from Pasteurized Milk in China. Front. Microbiol. 2018, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Dehkordi, F.S.; Yazdani, F.; Mozafari, J.; Valizadeh, Y. Virulence Factors, Serogroups, and Antimicrobial Resistance Properties of Escherichia coli Strains in Fermented Dairy Products. BMC Res. Notes 2014, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hassani, S.; Moosavy, M.H.; Gharajalar, S.N.; Khatibi, S.A.; Hajibemani, A.; Barabadi, Z. High Prevalence of Antibiotic Resistance in Pathogenic Foodborne Bacteria Isolated from Bovine Milk. Sci. Rep. 2022, 12, 3878. [Google Scholar] [CrossRef]
- Brown, K.; Mugoh, M.; Call, D.R.; Omulo, S. Antibiotic Residues and Antibiotic-Resistant Bacteria Detected in Milk Marketed for Human Consumption in Kibera, Nairobi. PLoS ONE 2020, 15, e0233413. [Google Scholar] [CrossRef]
- Tyasningsih, W.; Ramandinianto, S.C.; Ansharieta, R.; Witaningrum, A.M.; Permatasari, D.A.; Wardhana, D.K.; Effendi, M.H.; Ugbo, E.N. Prevalence and Antibiotic Resistance of Staphylococcus aureus and Escherichia coli Isolated from Raw Milk in East Java, Indonesia. Vet. World 2022, 15, 2021–2028. [Google Scholar] [CrossRef]
- Elafify, M.; Khalifa, H.O.; Al-Ashmawy, M.; Elsherbini, M.; El Latif, A.A.; Okanda, T.; Matsumoto, T.; Koseki, S.; Abdelkhalek, A. Prevalence and Antimicrobial Resistance of Shiga Toxin-Producing Escherichia coli in Milk and Dairy Products in Egypt. J. Environ. Sci. Health Part B 2019, 55, 265–272. [Google Scholar] [CrossRef]
- Elzhraa, F.; Al-Ashmawy, M.; El-Sherbini, M.; El-Sebaey, A.M.; Mohácsi-Farkas, C.; Kiskó, G.; Belák, Á. Rumi and Pasteurized Kareish Cheeses Are a Source of β-Lactam-Resistant Salmonella in the Nile Delta Region of Egypt: Insights into Their Incidence, AMR Pattern, Genotypic Determinants of Virulence and β-Lactam Resistance. Antibiotics 2024, 13, 454. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; et al. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Adley, C.C.; Ryan, M.P. The Nature and Extent of Foodborne Disease. In Antimicrobial Food Packaging; Barros-Velázquez, J., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 1–10. [Google Scholar] [CrossRef]
- World Health Organization. WHO Bacterial Priority Pathogens List; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Jia, K.; Qin, X.; Bu, X.; Zhu, H.; Liu, Y.; Wang, X.; Dong, Q. Prevalence, antibiotic resistance and molecular characterization of Staphylococcus aureus in ready-to-eat fruits and vegetables in Shanghai, China. Curr. Res. Food Sci. 2024, 8, 100669. [Google Scholar] [CrossRef]
- Agersø, Y.; Jensen, L.B.; Givskov, M.; Roberts, M.C. The identification of a tetracycline resistance gene tet (M), on a Tn 916-like transposon, in the Bacillus cereus group. FEMS Microbiol. Lett. 2002, 214, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Park, K.M.; Jeong, M.; Park, K.J.; Koo, M. Prevalence, enterotoxin genes, and antibiotic resistance of Bacillus cereus isolated from raw vegetables in Korea. FEMS Microbiol. Lett. 2018, 81, 1590–1597. [Google Scholar] [CrossRef]
- Fraccalvieri, R.; Bianco, A.; Difato, L.M.; Capozzi, L.; Del Sambro, L.; Simone, D.; Parisi, A. Toxigenic genes, pathogenic potential and antimicrobial resistance of Bacillus cereus group isolated from ice cream and characterized by whole genome sequencing. Foods 2022, 11, 2480. [Google Scholar] [CrossRef]
- Tien, Y.C.; Li, B.; Zhang, T.; Scott, A.; Murray, R.; Sabourin, L.; Marti, R.; Topp, E. Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic resistance genes on vegetables at harvest. Sci. Total Environ. 2017, 581, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Marti, R.; Scott, A.; Tien, Y.C.; Murray, R.; Sabourin, L.; Zhang, Y.; Topp, E. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl. Environ. Microbiol. 2013, 79, 5701–5709. [Google Scholar] [CrossRef]
- Rodríguez, C.; Lang, L.; Wang, A.; Altendorf, K.; García, F.; Lipski, A. Lettuce for human consumption collected in Costa Rica contains complex communities of culturable oxytetracycline- and gentamicin-resistant bacteria. Appl. Environ. Microbiol. 2006, 72, 5870–5876. [Google Scholar] [CrossRef] [PubMed]
- Touati, A.; Mairi, A.; Baloul, Y.; Lalaoui, R.; Bakour, S.; Thighilt, L.; Gharout, A.; Rolain, J.M. First detection of Klebsiella pneumoniae producing OXA-48 in fresh vegetables from Béjaïa city, Algeria. J. Glob. Antimicrob. Resist. 2017, 9, 17–18. [Google Scholar] [CrossRef]
- Liu, B.T.; Li, X.; Zhang, Q.; Shan, H.; Zou, M.; Song, F.J. Colistin-resistant mcr-positive Enterobacteriaceae in fresh vegetables, an increasing infectious threat in China. Int. J. Antimicrob. Agents 2019, 54, 89–94. [Google Scholar] [CrossRef]
- Raphael, E.; Wong, L.K.; Riley, L.W. Extended-spectrum beta-lactamase gene sequences in Gram-negative saprophytes on retail organic and nonorganic spinach. Appl. Environ. Microbiol. 2011, 77, 1601–1607. [Google Scholar] [CrossRef]
- Colosi, I.A.; Baciu, A.M.; Opriș, R.V.; Peca, L.; Gudat, T.; Simon, L.M.; Colosi, H.A.; Costache, C. Prevalence of ESBL, AmpC, and carbapenemase-producing Enterobacterales isolated from raw vegetables retailed in Romania. Foods 2020, 9, 1726. [Google Scholar] [CrossRef]
- Yang, F.; Shen, C.; Zheng, X.; Liu, Y.; El-Sayed Ahmed, M.A.E.G.; Zhao, Z.; Liao, K.; Shi, Y.; Guo, X.; Tian, G.B.; et al. Plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli and Klebsiella pneumoniae isolated from market retail fruits in Guangzhou, China. Infect. Drug Resist. 2019, 12, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo, J.R. Tetracyclines. In Antimicrobial Therapy in Veterinary Medicine, 5th ed.; Gigueré, S., Prescott, J., Dowling, P.M., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 256–268. ISBN 9780470963029. [Google Scholar] [CrossRef]
- Sanganyado, E.; Gwenzi, W. Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019, 669, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, H.; Xiong, P.; Zhu, Q.; Liao, C.; Jiang, G. Occurrence, fate, and risk assessment of typical tetracycline antibiotics in the aquatic environment: A review. Sci. Total Environ. 2021, 753, 141975. [Google Scholar] [CrossRef]
- Duran, N.; Ozer, B.; Duran, G.G.; Onlen, Y.; Demir, C. Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J. Med. Res. 2012, 135, 389–396. [Google Scholar] [PubMed] [PubMed Central]
- Su, H.C.; Liu, Y.S.; Pan, C.G.; Chen, J.; He, L.Y.; Ying, G.G. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water. Sci. Total Environ. 2018, 616, 453–461. [Google Scholar] [CrossRef]
- Yu, B.; Zhang, Y.; Yang, L.; Xu, J.; Bu, S. Analysis of antibiotic resistance phenotypes and genes of Escherichia coli from healthy swine in Guizhou, China. Onderstepoort J. Vet. Res. 2018, 88, 1880. [Google Scholar] [CrossRef]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef]
- Ullmann, I.F.; Tunsjø, H.S.; Andreassen, M.; Nielsen, K.M.; Lund, V.; Charnock, C. Detection of Aminoglycoside Resistant Bacteria in Sludge Samples from Norwegian Drinking Water Treatment Plants. Front. Microbiol. 2019, 10, 487. [Google Scholar] [CrossRef]
- Rajaei, M.; Moosavy, M.H.; Gharajalar, S.N.; Khatibi, S.A. Antibiotic Resistance in the Pathogenic Foodborne Bacteria Isolated from Raw Kebab and Hamburger: Phenotypic and Genotypic Study. BMC Microbiol. 2021, 21, 272. [Google Scholar] [CrossRef]
- Rehman, M.; Yin, X.; Zaheer, R.; Goji, N.; Amoako, K.; McAllister, T.; Pritchard, J.; Topp, E.; Diarra, M. Genotypes and Phenotypes of Enterococci Isolated from Broiler Chickens. Front. Sustain. Food Syst. 2018, 2, 83. [Google Scholar] [CrossRef]
- Yu, L.; Liu, Y.; Liu, M.; Li, Z.; Li, L.; Wang, F. Research Note: Molecular Characterization of Antimicrobial Resistance and Virulence Gene Analysis of Enterococcus faecalis in Poultry in Tai’an, China. Poult. Sci. 2022, 101, 101763. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Seo, H.J.; Seo, K.W.; Jeon, H.Y.; Kim, D.K.; Kim, S.W.; Lim, S.-K.; Lee, Y.J. Characteristics of High-Level Ciprofloxacin-Resistant Enterococcus faecalis and Enterococcus faecium from Retail Chicken Meat in Korea. J. Food Prot. 2018, 81, 1357–1363. [Google Scholar] [CrossRef]
- Kunadu, A.P.H.; Holmes, M.; Miller, E.L.; Grant, A.J. Microbiological quality and antimicrobial resistance characterization of Salmonella spp. in fresh milk value chains in Ghana. Int. J. Food Microbiol. 2018, 277, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Kürekci, C.; Önen, S.P.; Yipel, M.; Aslantaş, Ö.; Gündoğdu, A. Characterisation of phenotypic and genotypic antibiotic resistance profile of enterococci from cheeses in Turkey. Korean J. Food Sci. An. 2016, 36, 352. [Google Scholar] [CrossRef]
- Silva, C.R.; Okuno, N.T.; de Medeiros Macedo, V.H.L.; da Rocha Freire, I.; Miller, R.M.; Marin, V.A. Resistome in gram-negative bacteria from soft cheese in Brazil. Rev. Ciênc. Méd. Biol. 2020, 19, 430–440. [Google Scholar] [CrossRef]
- Nalepa, B.; Markiewicz, L.H. Microbiological biodiversity of regional cow, goat and ewe milk cheeses produced in Poland and antibiotic resistance of lactic acid bacteria isolated from them. Animals 2022, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Parussolo, L.; Sfaciotte, R.A.P.; Dalmina, K.A.; Melo, F.D.; Costa, U.M.; Ferraz, S.M. Detection of virulence genes and antimicrobial resistance profiles of Escherichia coli isolates from raw milk and artisanal cheese in Southern Brazil. Semin. Ciênc. Agrár. 2019, 40, 163–178. [Google Scholar] [CrossRef]
- Tegegne, H.A.; Florianová, M.; Gelbíčová, T.; Karpíšková, R.; Koláčková, I. Detection and molecular characterization of methicillin-resistant Staphylococcus aureus isolated from bulk tank milk of cows, sheep, and goats. Foodborne Pathog. Dis. 2018, 16, 68–73. [Google Scholar] [CrossRef]
- Mbindyo, C.M.; Gitao, G.C.; Plummer, P.J.; Kulohoma, B.W.; Mulei, C.M.; Bett, R. Antimicrobial Resistance Profiles and Genes of Staphylococci Isolated from Mastitic Cow’s Milk in Kenya. Antibiotics 2021, 10, 772. [Google Scholar] [CrossRef]
- Zhai, Z.; Cui, C.; Li, X.; Yan, J.; Sun, E.; Wang, C.; Guo, H.; Hao, Y. Prevalence, antimicrobial susceptibility, and antibiotic resistance gene transfer of Bacillus strains isolated from pasteurized milk. J. Dairy Sci. 2023, 106, 75–83. [Google Scholar] [CrossRef]
- Siddique, A.; Azim, S.; Ali, A.; Andleeb, S.; Ahsan, A.; Imran, M.; Rahman, A. Antimicrobial resistance profiling of biofilm forming non typhoidal Salmonella enterica isolates from poultry and its associated food products from Pakistan. Antibiotics 2021, 10, 785. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- Schmidt, J.W.; Vikram, A.; Doster, E.; Thomas, K.; Weinroth, M.D.; Parker, J.; Hanes, A.; Geornaras, I.; Morley, P.S.; Belk, K.E.; et al. Antimicrobial resistance in US retail ground beef with and without label claims regarding antibiotic use. J. Food Prot. 2021, 84, 827–842. [Google Scholar] [CrossRef]
- Bacci, C.; Vismarra, A.; Dander, S.; Barilli, E.; Superchi, P. Occurrence and antimicrobial profile of bacterial pathogens in former foodstuff meat products used for pet diets. J. Food Prot. 2019, 82, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Folster, J.P.; Pecic, G.; Singh, A.; Duval, B.; Rickert, R.; Ayers, S.; Abbott, J.; McGlinchey, B.; Bauer-Turpin, J.; Haro, J.; et al. Characterization of extended-spectrum cephalosporin–resistant Salmonella enterica serovar Heidelberg isolated from food animals, retail meat, and humans in the United States 2009. Foodborne Pathog. Dis. 2012, 9, 638–645. [Google Scholar] [CrossRef]
- Hassena, A.B.; Siala, M.; Guermazi, S.; Zormati, S.; Gdoura, R.; Sellami, H. Occurrence and phenotypic and molecular characterization of antimicrobial resistance of Salmonella isolates from food in Tunisia. J. Food Prot. 2019, 82, 1166–1175. [Google Scholar] [CrossRef]
- Hull, D.M.; Harrell, E.; van Vliet, A.H.M.; Correa, M.; Thakur, S. Antimicrobial resistance and interspecies gene transfer in Campylobacter coli and Campylobacter jejuni isolated from food animals, poultry processing, and retail meat in North Carolina, 2018–2019. PLoS ONE 2021, 16, e0246571. [Google Scholar] [CrossRef]
- Silva, W.C.; Targino, B.N.; Mendonça, R.S.; Sant’Ana, A.S.; Hungaro, H.M. Campylobacter: An overview of cases, occurrence in food, contamination sources, and antimicrobial resistance in Brazil. Food Rev. Int. 2017, 34, 364–389. [Google Scholar] [CrossRef]
- Wieczorek, K.; Wołkowicz, T.; Osek, J. Antimicrobial Resistance and Virulence-Associated Traits of Campylobacter jejuni Isolated From Poultry Food Chain and Humans with Diarrhea. Front. Microbiol. 2018, 9, 1508. [Google Scholar] [CrossRef]
- Li, M.; Wang, Y.; Hou, B.; Chen, Y.; Hu, M.; Zhao, X.; Zhang, Q.; Li, L.; Luo, Y.; Liu, Y.; et al. Toxin gene detection and antibiotic resistance of Clostridium perfringens from aquatic sources. Int. J. Food Microbiol. 2024, 415, 110642. [Google Scholar] [CrossRef]
- Xiu, L.; Liu, Y.; Wu, W.; Chen, S.; Zhong, Z.; Wang, H. Prevalence and multilocus sequence typing of Clostridium perfringens isolated from 4 duck farms in Shandong province, China. Poult. Sci. 2020, 99, 5105–5117. [Google Scholar] [CrossRef]
- Fourie, J.C.J.; Bezuidenhout, C.C.; Sanko, T.J.; Mienie, C.; Adeleke, R. Inside environmental Clostridium perfringens genomes: Antibiotic resistance genes, virulence factors and genomic features. J. Water Health 2020, 18, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Kayode, A.J.; Okoh, A.I. Antibiotic resistance profile of Listeria monocytogenes recovered from ready-to-eat foods surveyed in South Africa. J. Food Prot. 2022, 85, 1807–1814. [Google Scholar] [CrossRef]
- Escolar, C.; Gomez, D.; del Carmen Rota García, M.; Conchello, P.; Herrera, A. Antimicrobial resistance profiles of Listeria monocytogenes and Listeria innocua isolated from ready-to-eat products of animal origin in Spain. Foodborne Pathog. Dis. 2017, 14, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; Da Silva, W.P. Listeria monocytogenes isolates from food and food environment harbouring tetM and ermB resistance genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, P.; Chajęcka-Wierzchowska, W.; Zadernowska, A. High-Pressure Processing—Impacts on the Virulence and Antibiotic Resistance of Listeria monocytogenes Isolated from Food and Food Processing Environments. Foods 2023, 12, 3899. [Google Scholar] [CrossRef]
- Martins, B.T.F.; Botelho, C.V.; Silva, D.A.L.; Lanna, F.G.P.A.; Grossi, J.L.; Campos-Galvão, M.E.M.; Ricardo Seiti Yamatogi, R.S.; Falcão, J.P.; dos Santos Bersot, L.; Nero, L.A. Yersinia enterocolitica in a Brazilian pork production chain: Tracking of contamination routes, virulence and antimicrobial resistance. Int. J. Food Microbiol. 2018, 276, 5–9. [Google Scholar] [CrossRef]
- Yue, Y.; Shen, M.; Liu, X.; Hao, Q.; Kang, Y.; Che, Y.; Li, F.; Chen, S.; Xu, S.; Jing, H.; et al. Whole-genome sequencing-based prediction and analysis of antimicrobial resistance in Yersinia enterocolitica from Ningxia, China. Front. Microbiol. 2022, 13, 936425. [Google Scholar] [CrossRef]
- Ozdemır, F.; Arslan, S.; Erturk, H.G. Expression of blaA and blaB and susceptibility to penicillins and cephalosporins in Yersinia enterocolitica from different foods. Eur. J. Biol. 2020, 79, 83–88. [Google Scholar] [CrossRef]
- Krüger, A.; Lucchesi, P.; Sanso, A.M.; Etcheverría, A.I.; Bustamante, A.V.; Burgán, J.; Fernández, L.; Fernández, D.; Leotta, G.; Friedrich, A.W.; et al. Genetic characterization of Shiga toxin-producing Escherichia coli O26: H11 strains isolated from animal, food, and clinical samples. Front. Cell. Infect. Microbiol. 2015, 5, 74. [Google Scholar] [CrossRef]
- Elmonir, W.; Shalaan, S.; Tahoun, A.; Mahmoud, S.F.; Remela, E.M.A.; Eissa, R.; El-Sharkawy, H.; Shukry, M.; Zahran, R.N. Prevalence, antimicrobial resistance, and genotyping of Shiga toxin-producing Escherichia coli in foods of cattle origin, diarrheic cattle, and diarrheic humans in Egypt. Gut Pathog. 2021, 13, 8. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Shimamoto, T. Molecular analysis of multidrug resistance in Shiga toxin-producing Escherichia coli O157: H7 isolated from meat and dairy products. Int. J. Food Microbiol. 2015, 193, 68–73. [Google Scholar] [CrossRef]
- Yen, P.; Papin, J.A. History of antibiotic adaptation influences microbial evolutionary dynamics during subsequent treatment. PLoS Biol. 2017, 15, e2001586. [Google Scholar] [CrossRef]
- Brandon, R.N. Adaptation and Environment; Princeton University Press: Princeton, NJ, USA, 2014. [Google Scholar]
- Tan, Y.S.; Zhang, R.K.; Liu, Z.H.; Li, B.Z.; Yuan, Y.J. Microbial Adaptation to Enhance Stress Tolerance. Front. Microbiol. 2022, 13, 888746. [Google Scholar] [CrossRef] [PubMed]
- Felden, B.; Cattoir, V. Bacterial Adaptation to Antibiotics through Regulatory RNAs. Antimicrob. Agents Chemother. 2018, 62, e02503-17. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.A.; Romesberg, F.E. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 2007, 3, 549–556. [Google Scholar] [CrossRef]
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef] [PubMed]
- Klemm, E.J.; Wong, V.K.; Dougan, G. Emergence of dominant mul-tidrug-resistant bacterial clades: Lessons from history and whole- genome sequencing. Proc. Natl. Acad. Sci. USA 2018, 115, 12872–12877. [Google Scholar] [CrossRef]
- Zhang, F.; Cheng, W. The Mechanism of Bacterial Resistance and Potential Bacteriostatic Strategies. Antibiotics 2022, 11, 1215. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic resistance in bacteria—A review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Chen, L.; Kumar, S.; Wu, H. A review of current antibiotic resistance and promising antibiotics with novel modes of action to combat antibiotic resistance. Arch. Microbiol. 2023, 205, 356. [Google Scholar] [CrossRef] [PubMed]
- Spížek, J.; Řezanka, T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem. Pharmacol. 2017, 133, 20–28. [Google Scholar] [CrossRef]
- Cox, G.; Wright, G.D. Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 2013, 303, 287–292. [Google Scholar] [CrossRef]
- Martinez, J.L. General principles of antibiotic resistance in bacteria. Drug Discov. Today Technol. 2014, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Feng, J. The intrinsic resistance of bacteria. Hereditas 2016, 38, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Selim, S. Mechanisms of Gram-positive vancomycin resistance. Biomed. Rep. 2022, 16, 7. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–569. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Inf. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Hoffman, S.B. Mechanisms of antibiotic resistance. Compendium 2001, 23, 464–473. [Google Scholar]
- Grant, S.S.; Hung, D.T. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 2013, 4, 273–283. [Google Scholar] [CrossRef]
- Verstraete, L.; Van den Bergh, B.; Verstraeten, N.; Michiels, J. Ecology and evolution of antibiotic persistence. Trends Microbiol. 2022, 30, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ning, Q.; Deng, Z.; Zhang, M.; You, J. Role of environmental stresses in elevating resistance mutations in bacteria: Phenomena and mechanisms. Environ. Pollut. 2022, 307, 119603. [Google Scholar] [CrossRef] [PubMed]
- Dalbanjan, N.P.; Kadapure, A.J.; Praveen Kumar, S.K. A comprehensive review on latent role of stress proteins in antibiotic resistance. Microbe 2024, 4, 100151. [Google Scholar] [CrossRef]
- Luo, D.; Wu, Z.; Bai, Q.; Zhang, Y.; Huang, M.; Huang, Y.; Li, X. Universal Stress Proteins: From Gene to Function. Int. J. Mol. Sci. 2023, 24, 4725. [Google Scholar] [CrossRef]
- Zhou, Y.; Liao, H.; Pei, L.; Pu, Y. Combatting persister cells: The daunting task in post-antibiotics era. Cell Insight 2023, 2, 100104. [Google Scholar] [CrossRef]
- Kratz, J.C.; Banerjee, S. Gene expression tradeoffs determine bacterial survival and adaptation to antibiotic stress. PRX Life 2024, 2, 013010. [Google Scholar] [CrossRef]
- Maeda, T.; Furusawa, C. Laboratory Evolution of Antimicrobial Resistance in Bacteria to Develop Rational Treatment Strategies. Antibiotics 2024, 13, 94. [Google Scholar] [CrossRef]
- Melo, J.; Andrew, P.W.; Faleiro, M.L. Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: The role of stress responses. Food Res. Int. 2015, 67, 75–90. [Google Scholar] [CrossRef]
- Sharma, M.; Taormina, P.J.; Beuchat, L.R. Habituation of foodborne pathogens exposed to extreme pH conditions: Genetic basis and implications in foods and food processing environments. Food Sci. Technol. Res. 2003, 9, 115–127. [Google Scholar] [CrossRef]
- Komora, N.; Bruschi, C.; Magalhães, R.; Ferreira, V.; Teixeira, P. Survival of Listeria monocytogenes with different antibiotic resistance patterns to food-associated stresses. Int. J. Food Microbiol. 2017, 245, 79–87. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Stanton, P.R.C.; Turroni, F.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837. [Google Scholar] [CrossRef]
- Kovacevic, J.; Sagert, J.; Wozniak, A.; Gilmour, M.W.; Allen, K.J. Antimicrobial resistance and co-selection phenomenon in Listeria spp. recovered from food and food production environments. Food Microbiol. 2013, 34, 319–327. [Google Scholar] [CrossRef]
- Rakic-Martinez, M.; Drevets, D.A.; Dutta, V.; Katic, V.; Kathariou, S. Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl. Environ. Microbiol. 2011, 77, 8714–8721. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.J.; Wałecka-Zacharska, E.; Chen, J.C.; Katarzyna, K.P.; Devlieghere, F.; Van Meervenne, E.; Osek, J.; Wieczorek, K.; Bania, J. Listeria monocytogenes—An examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiol. 2016, 54, 178–189. [Google Scholar] [CrossRef]
- Zarzecka, U.; Zadernowska, A.; Chajęcka-Wierzchowska, W. Starter cultures as a reservoir of antibiotic resistant microorganisms. LWT 2020, 127, 109424. [Google Scholar] [CrossRef]
- Amund, O.D. Exploring the relationship between exposure to technological and gastrointestinal stress and probiotic functional properties of lactobacilli and bifidobacteria. Can. J. Microbiol. 2016, 62, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Natt, N.K.; Garcha, S. Antibiotic sensitivity of acid-stressed probiotic Lactobacillus acidophilus NCDC 291. Int. J. Microbiol. 2011, 9, 134. [Google Scholar] [CrossRef]
- Casado Muñoz, M.d.C.; Benomar, N.; Lavilla Lerma, L.; Knapp, C.W.; Gálvez, A.; Abriouel, H. Biocide tolerance, phenotypic and molecular response of lactic acid bacteria isolated from naturally-fermented Aloreña table olives to different physicochemical stresses. Food Microbiol. 2016, 60, 1–12. [Google Scholar] [CrossRef]
- Kovács, M.; Wojnárovits, L.; Homlok, R.; Tegze, A.; Mohácsi-Farkas, C.; Takács, E.; Belák, Á. Changes in the behavior of Staphylococcus aureus strains in the presence of oxacillin under the effect of gamma radiation. Environ. Pollut. 2024, 340, 122843. [Google Scholar] [CrossRef] [PubMed]
- Dorrian, J.M.; Briggs, D.A.; Ridley, M.L.; Layfield, R.; Kerr, I.D. Induction of a stress response in Lactococcus lactis is associated with resistance to ribosomally active antibiotics. FEBS J. 2011, 278, 4015–4024. [Google Scholar] [CrossRef]
- Van Hoek, A.H.; Veenman, C.; Van Overbeek, W.M.; Lynch, G.; De Roda Husman, A.M.; Blaak, H. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 2015, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ramya, V.; Patel, P. Health benefits of vegetables. Int. J. Chem. Stud. 2019, 7, 82–87. [Google Scholar]
- Heng, Y.; House, L.A. Cluster analysis for fruit consumption patterns: An international study. Br. Food J. 2018, 120, 1942–1952. [Google Scholar] [CrossRef]
- Yu, H.; Neal, J.A.; Sirsat, S.A. Consumers’ Food Safety Risk Perceptions and Willingness to Pay for Fresh-Cut Produce with Lower Risk of Foodborne Illness. Food Control 2018, 86, 83–89. [Google Scholar] [CrossRef]
- Zhou, S.Y.; Wei, M.Y.; Giles, M.; Neilson, R.; Zheng, F.; Zhang, Q.; Zhu, Y.-G.; Yang, X.-R. Prevalence of antibiotic resistome in ready-to-eat salad. Front. Public Health 2020, 8, 92. [Google Scholar] [CrossRef] [PubMed]
- Machado-Moreira, B.; Richards, K.; Brennan, F.; Abram, F.; Burgess, C.M. Microbial contamination of fresh produce: What, where, and how? Compr. Rev. Food Sci. Food Saf. 2019, 18, 1727–1750. [Google Scholar] [CrossRef]
- Xylia, P.; Botsaris, G.; Chrysargyris, A.; Skandamis, P.; Tzortzakis, N. Variation of microbial load and biochemical activity of ready-to-eat salads in Cyprus as affected by vegetable type, season, and producer. Food Microbiol. 2019, 83, 200–210. [Google Scholar] [CrossRef]
- Richter, L.; Du Plessis, E.M.; Duvenage, S.; Korsten, L. Occurrence, identification, and antimicrobial resistance profiles of Extended-Spectrum and AmpC beta-Lactamase-producing Enterobacteriaceae from fresh vegetables retailed in Gauteng Province, South Africa. Foodborne Pathog. Dis. 2019, 16, 421–427. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.; He, J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019, 130, 104912. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Amoah, I.D.; Stenström, T.A.; Verbyla, M.E.; Mihelcic, J.R. Epidemiological evidence and health risks associated with agricultural reuse of partially treated and untreated wastewater: A review. Front. Public Health 2018, 6, 337. [Google Scholar] [CrossRef] [PubMed]
- Scaccia, N.; Vaz-Moreira, I.; Manaia, C.M. The risk of transmitting antibiotic resistance through endophytic bacteria. Trends Plant Sci. 2021, 26, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Chen, Q.; Chen, S.; Zhu, Y.G. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 2017, 98, 152–159. [Google Scholar] [CrossRef]
- Araujo, S.; Silva, I.A.; Tacão, M.; Patinha, C.; Alves, A.; Henriques, I. Characterization of antibiotic-resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms. Int. J. Food Microbiol. 2017, 257, 192–200. [Google Scholar] [CrossRef]
- Blau, K.; Bettermann, A.; Jechalke, S.; Fornefeld, E.; Vanrobaeys, Y.; Stalder, T.; Top, E.M.; Smalla, K. The transferable resistome of produce. mBio 2018, 9, e01300-18. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: A need for quantitative risk assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- Alegbeleye, O.O.; Singleton, I.; Sant’Ana, A.S. Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiol. 2018, 73, 177–208. [Google Scholar] [CrossRef]
- Schierstaedt, J.; Grosch, R.; Schikora, A. Agricultural production systems can serve as reservoirs for human pathogens. FEMS Microbiol. Lett. 2019, 366, fnaa016. [Google Scholar] [CrossRef]
- Yang, X.; Wu, Q.; Huang, J.; Wu, S.; Zhang, J.; Chen, L.; Wei, X.; Ye, Y.; Li, Y.; Wang, J.; et al. Prevalence and characterization of salmonella isolated from raw vegetables in China. Food Control 2020, 109, 106915. [Google Scholar] [CrossRef]
- Campos, J.; Mourão, J.; Pestana, N.; Peixe, L.; Novais, C.; Antunes, P. Microbiological quality of ready-to-eat salads: An underestimated vehicle of bacteria and clinically relevant antibiotic resistance genes. Int. J. Food Microbiol. 2013, 166, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, N.; Muraleedharan, C.; Talreja, D.; Rana, S.W.; Walia, S.; Kumar, A.; Walia, S.K. Occurrence of multidrug-resistant extended-spectrum beta-lactamase-producing bacteria on iceberg lettuce retailed for human consumption. Biomed. Res. Int. 2015, 2015, 547547. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.T.; Song, F.J. Emergence of two Escherichia coli strains co-harboring mcr-1 and bla NDM in fresh vegetables from China. Infect. Drug Resist. 2019, 12, 2627–2635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Q.; Song, J.; Zhang, Z.; Chen, S.; Long, Z.; Wang, M.; Yu, Y.; Fang, H. Tracking resistomes, virulence genes, and bacterial pathogens in long-term manure-amended greenhouse soils. J. Hazard. Mater. 2020, 396, 122618. [Google Scholar] [CrossRef]
- Sun, Y.; Snow, D.; Walia, H.; Li, X. Transmission routes of the microbiome and resistome from manure to soil and lettuce. Environ. Sci. Technol. 2021, 55, 11102–11112. [Google Scholar] [CrossRef]
- Sanseverino, I.; Navarro Cuenca, A.; Loos, R.; Marinov, D.; Lettieri, T. State of the Art on the Contribution of Water to Antimicrobial Resistance; Publications Office of the European Union: Luxembourg, 2018; ISBN 9789279984785. [Google Scholar]
- Kolpin, D.W.; Furlong, E.T.; Meyer, M.T.; Thurman, E.M.; Zaugg, S.D.; Barber, L.B.; Buxton, H.T. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: A national reconnaissance. Environ. Sci. Technol. 2002, 36, 1202–1211. [Google Scholar] [CrossRef]
- Faleye, A.C.; Adegoke, A.A.; Ramluckan, K.; Fick, J.; Bux, F.; Stenström, T.A. Concentration and reduction of antibiotic residues in selected wastewater treatment plants and receiving waterbodies in Durban, South Africa. Sci. Total Environ. 2019, 678, 10–20. [Google Scholar] [CrossRef]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P. Antibiotic-resistance genes in waste water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef]
- Fillinger, U.; Lindsay, S.W. Suppression of exposure to malaria vectors by an order of magnitude using microbial larvicides in rural Kenya. Trop. Med. Int. Health 2006, 11, 1629–1642. [Google Scholar] [CrossRef]
- Boyce, R.; Lenhart, A.; Kroeger, A.; Velayudhan, R.; Roberts, B.; Horstick, O. Bacillus Thuringiensis Israelensis (Bti) for the control of dengue vectors: Systematic literature review. Trop. Med. Int. Health 2013, 18, 564–577. [Google Scholar] [CrossRef]
- Cerqueira, F.; Matamoros, V.; Bayona, J.; Pina, B. Antibiotic resistance genes distribution in microbiomes from the soil-plant-fruit continuum in commercial Lycopersicon esculentum fields under different agricultural practices. Sci. Total Environ. 2019, 652, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Huang, C.; Xi, B.; Tang, Z.; Tan, W.; Li, W.; Zhang, Y.; Li, W. The maturity period is the main stage of antibiotic resistance genes reduction in aerobic composting process of swine manure in sub-scale farms. Bioresour. Technol. 2021, 319, 124139. [Google Scholar] [CrossRef]
- Burch, T.R.; Sadowsky, M.J.; LaPara, T.M. Effect of different treatment technologies on the fate of antibiotic resistance genes and class 1 integrons when residual municipal wastewater solids are applied to soil. Environ. Sci. Technol. 2017, 51, 14225–14232. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.L.; An, X.L.; Zheng, B.X.; Ma, Y.B.; Su, J.Q. Long-term organic fertilization increased antibiotic resistome in phyllosphere of maize. Sci. Total Environ. 2018, 645, 1230–1237. [Google Scholar] [CrossRef]
- Blaak, H.; Van Hoek, A.H.; Veenman, C.; Docters Van Leeuwen, A.E.; Lynch, G.; Van Overbeek, W.M.; De Roda Husman, A.M. Extended spectrum β -lactamase- and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment. Int. J. Food Microbiol. 2014, 168, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Nuesch-Inderbinen, M.; Morach, M.; Zihler, B.A.; Hachler, H.; Stephan, R. Extended-spectrum-beta-lactamaseproducing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 2015, 81, 3115–3120. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Zurfluh, K.; Peterhans, S.; Hächler, H.; Stephan, R. Assessment of the prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in ready-to-eat salads, fresh-cut fruit, and sprouts from the Swiss market. J. Food Prot. 2015, 78, 1178–1181. [Google Scholar] [CrossRef]
- Song, J.; Oh, S.S.; Kim, J.; Shin, J. Extended-spectrum β-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea. Sci. Rep. 2020, 10, 19721. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Incidence and genetic diversity of multidrug-resistant Listeria monocytogenes isolates recovered from fruits and vegetables in the Eastern Cape Province, South Africa. Int. J. Food Microbiol. 2022, 363, 109513. [Google Scholar] [CrossRef]
- Usui, M.; Ozeki, K.; Komatsu, T.; Fukuda, A.; Tamura, Y. Prevalence of Extended-Spectrum β-Lactamase–Producing Bacteria on Fresh Vegetables in Japan. J. Food Prot. 2019, 82, 1663–1666. [Google Scholar] [CrossRef]
- Salmanov, A.G.; Ushkalov, V.O.; Shunko, Y.Y.; Piven, N.; Vygovska, L.M.; Verner, O.M.; Kushnirenko, S. One health: Antibiotic-resistant bacteria contamination in fresh vegetables sold at retail markets in Kyiv. Wiad. Lek. 2021, 74, 116. [Google Scholar] [CrossRef]
- Trocado, N.D.; de Moraes, M.S.; Aveleda, L.; Silva, C.R.; Marin, V.A. Phenotypic and genotypic detection of antibiotic-resistant bacteria in fresh fruit juices from a public hospital in Rio de Janeiro. Arch. Microbiol. 2021, 203, 1471–1475. [Google Scholar] [CrossRef]
- Freitag, C.; Michael, G.B.; Li, J.; Kadlec, K.; Wang, Y.; Hassel, M.; Schwarz, S. Occurrence and characterisation of ESBL-encoding plasmids among Escherichia coli isolates from fresh vegetables. Vet. Microbiol. 2018, 219, 63–69. [Google Scholar] [CrossRef]
- Mesbah, Z.F.; Granier, S.A.; Touati, A.; Millemann, Y. Occurrence of third-generation cephalosporins-resistant Klebsiella pneumoniae in fresh fruits and vegetables purchased at markets in Algeria. Microb. Drug Resist. 2020, 26, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Dai, J.; Chen, J.; He, Y.; Su, L.; Gong, M.; Cao, M.; Wei, K.; You, Y.; Liu, L.; et al. Antibiotic susceptibility and genomic analysis of ciprofloxacin-resistant and ESBLs-producing Escherichia coli in vegetables and their irrigation water and growing soil. Int. J. Food Microbiol. 2024, 414, 110629. [Google Scholar] [CrossRef]
- Hu, F.; Guo, Y.; Zhu, D. Surveillance of bacterial drug resistance in China in 2021. J. Chin. Infect. Chemother. 2022, 22, 521–530. [Google Scholar] [CrossRef]
- Iseppi, R.; de Niederhäusern, S.; Bondi, M.; Messi, P.; Sabia, C. Extended-spectrum β-lactamase, AmpC, and MBL-producing gram-negative bacteria on fresh vegetables and ready-to-eat salads sold in local markets. Microb. Drug Resist. 2018, 24, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Poirel, L.; Nordmann, P.; Nüesch-Inderbinen, M.; Hächler, H.; Stephan, R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob. Agents Chemother. 2016, 60, 2594–2595. [Google Scholar] [CrossRef]
- Manageiro, V.; Jones-Dias, D.; Ferreira, E.; Caniça, M. Plasmid-mediated colistin resistance (mcr-1) in Escherichia coli from non-imported fresh vegetables for human consumption in Portugal. Microorganisms 2020, 8, 429. [Google Scholar] [CrossRef]
- Xedzro, C.; Shimamoto, T.; Yu, L.; Zuo, H.; Sugawara, Y.; Sugai, M.; Shimamoto, T. Emergence of colistin-resistant Enterobacter cloacae and Raoultella ornithinolytica carrying the phosphoethanolamine transferase gene, mcr-9, derived from vegetables in Japan. Microbiol. Spectr. 2023, 11, e01063-23. [Google Scholar] [CrossRef]
- Oh, S.S.; Song, J.; Kim, J.; Shin, J. Increasing prevalence of multidrug-resistant mcr-1-positive Escherichia coli isolates from fresh vegetables and healthy food animals in South Korea. Int. J. Inf. Dis. 2020, 92, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Chelaghma, W.; Loucif, L.; Bendjama, E.; Cherak, Z.; Bendahou, M.; Rolain, J.M. Occurrence of extended-spectrum cephalosporin-, carbapenem-, and colistin-resistant gram-negative bacteria in fresh vegetables, an increasing human health concern in Algeria. Antibiotics 2022, 11, 988. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Poirel, L.; Nordmann, P.; Klumpp, J.; Stephan, R. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob. Resist. Infect. Control 2015, 4, 38. [Google Scholar] [CrossRef]
- Yigrem, C.; Fisseha, R.; Eribo, B. Characterization of carbapenem and β-lactam resistance in Klebsiella pneumonia associated with leafy vegetables and clinical isolates from Gondar, Ethiopia. J. Microbiol. Res. 2021, 11, 21–32. [Google Scholar] [CrossRef]
- Nketiah, A.; Quansah, J.K.; Kunadu, A.P.H. Presence of carbapenem resistance in hybrid Escherichia coli pathovars from ready-to-eat fresh-cut fruits in Accra, Ghana. J. Appl. Microbiol. 2024, 135, lxae239. [Google Scholar] [CrossRef]
- Abreu, R.; Semedo-Lemsaddek, T.; Cunha, E.; Tavares, L.; Oliveira, M. Antimicrobial Drug Resistance in Poultry Production: Current Status and Innovative Strategies for Bacterial Control. Microorganisms 2023, 11, 953. [Google Scholar] [CrossRef] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Holman, D.B.; Klima, C.L.; Gzyl, K.E.; Zaheer, R.; Service, C.; Jones, T.H.; McAllister, T.A. Antimicrobial Resistance in Enterococcus spp. Isolated from a Beef Processing Plant and Retail Ground Beef. Microbiol. Spectr. 2021, 9, e0198021. [Google Scholar] [CrossRef]
- Kim, C.; Goodwyn, B.; Albukhaytan, S.; Nartea, T.; Ndegwa, E.; Dhakal, R. Microbiological Survey and Antimicrobial Resistance of Foodborne Bacteria in Select Meat Products and Ethnic Food Products Procured from Food Desert Retail Outlets in Central Virginia, USA. Pathogens 2023, 12, 965. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, Y.; Peng, W.; Huang, B.; Ma, L.; Zheng, M.; Ding, S.; Zhu, K. Prevalence of Pathogens Harbouring Mobile Antimicrobial Resistance Genes and Virulence Factors in Retail Beef and Mutton. FEMS Microbiol. Lett. 2020, 367, fnaa089. [Google Scholar] [CrossRef]
- Gutema, F.D.; Rasschaert, G.; Agga, G.E.; Jufare, A.; Duguma, A.B.; Abdi, R.D.; Duchateau, L.; Crombe, F.; Gabriël, S.; De Zutter, L. Occurrence, Molecular Characteristics, and Antimicrobial Resistance of Escherichia coli O157 in Cattle, Beef, and Humans in Bishoftu Town, Central Ethiopia. Foodborne Pathog. Dis. 2021, 18, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Obaidat, M.M. Prevalence and Antimicrobial Resistance of Listeria monocytogenes, Salmonella enterica and Escherichia coli O157:H7 in Imported Beef Cattle in Jordan. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101447. [Google Scholar] [CrossRef]
- Zamil, S.; Ferdous, J.; Zannat, M.M.; Biswas, P.K.; Gibson, J.S.; Henning, J.; Hoque, M.A.; Barua, H. Isolation and Antimicrobial Resistance of Motile Salmonella enterica from the Poultry Hatchery Environment. Vet. Res. Commun. 2021, 45, 277–284. [Google Scholar] [CrossRef]
- Li, Z.; Jia, C.; Hu, Z.; Jin, Y.; Li, T.; Zhang, X.; Peng, Z.; Yang, R.; Chen, H.; Wang, X. Antimicrobial Resistance and Genomic Characteristics of Escherichia coli Strains Isolated from the Poultry Industry in Henan Province, China. Microorganisms 2024, 12, 575. [Google Scholar] [CrossRef] [PubMed]
- Marotta, F.; Garofolo, G.; di Marcantonio, L.; Di Serafino, G.; Neri, D.; Romantini, R.; Sacchini, L.; Alessiani, A.; Di Donato, G.; Nuvoloni, R.; et al. Antimicrobial Resistance Genotypes and Phenotypes of Campylobacter jejuni Isolated in Italy from Humans, Birds from Wild and Urban Habitats, and Poultry. PLoS ONE 2019, 14, e0223804. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global Trends in Antimicrobial Resistance in Animals in Low- and Middle-Income Countries. Science 2019, 36, eaaw1944. [Google Scholar] [CrossRef] [PubMed]
- Raut, R.; Maharjan, P.; Fouladkhah, A.C. Practical Preventive Considerations for Reducing the Public Health Burden of Poultry-Related Salmonellosis. Int. J. Environ. Res. Public Health 2023, 20, 6654. [Google Scholar] [CrossRef]
- Diarra, M.S.; Malouin, F. Antibiotics in Canadian Poultry Productions and Anticipated Alternatives. Front. Microbiol. 2014, 5, 282. [Google Scholar] [CrossRef]
- Saleem, G.N.; Gu, R.; Qu, H.; Bahar Khaskheli, G.; Rashid Rajput, I.; Qasim, M.; Chen, X. Therapeutic Potential of Popular Fermented Dairy Products and Its Benefits on Human Health. Front. Nutr. 2024, 11, 1328620. [Google Scholar] [CrossRef]
- Sharma, H.; Ozogul, F.; Bartkiene, E.; Rocha, J.M. Impact of Lactic Acid Bacteria and Their Metabolites on the Techno-Functional Properties and Health Benefits of Fermented Dairy Products. Crit. Rev. Food Sci. Nutr. 2023, 63, 4819–4841. [Google Scholar] [CrossRef]
- García-Burgos, M.; Moreno-Fernández, J.; Alférez, M.J.; Díaz-Castro, J.; López-Aliaga, I. New Perspectives in Fermented Dairy Products and Their Health Relevance. J. Funct. Foods 2020, 72, 104059. [Google Scholar] [CrossRef]
- Engelhardt, T.; Albano, H.; Kiskó, G.; Mohácsi-Farkas, C.; Teixeira, P. Antilisterial Activity of Bacteriocinogenic Pediococcus acidilactici HA6111-2 and Lactobacillus plantarum ESB 202 Grown Under pH and Osmotic Stress Conditions. Food Microbiol. 2015, 48, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, T.; Szakmár, K.; Kiskó, G.; Mohácsi-Farkas, C.; Reichart, O. Combined Effect of NaCl and Low Temperature on Antilisterial Bacteriocin Production of Lactobacillus plantarum ST202Ch. LWT-Food Sci. Technol. 2018, 89, 104–109. [Google Scholar] [CrossRef]
- Ladha, G.; Jeevaratnam, K. Characterization of Purified Antimicrobial Peptide Produced by Pediococcus pentosaceus LJR1 and Its Application in Preservation of White Leg Shrimp. World J. Microbiol. Biotechnol. 2020, 36, 72. [Google Scholar] [CrossRef]
- Sonbol, F.I.; Abdel Aziz, A.A.; El-Banna, T.E.; Al-Fakhrany, O.M. Antimicrobial Activity of Bacteriocins Produced by Enterococcus Isolates Recovered from Egyptian Homemade Dairy Products Against Some Foodborne Pathogens. Int. Microbiol. 2020, 23, 533–547. [Google Scholar] [CrossRef]
- Gálvez, A.; Abriouel, H.; Benomar, N.; Lucas, R. Microbial Antagonists to Foodborne Pathogens and Biocontrol. Curr. Opin. Biotechnol. 2010, 21, 142–148. [Google Scholar] [CrossRef]
- Castellano, P.; Pérez Ibarreche, M.; Blanco Massani, M.; Fontana, C.; Vignolo, G. Strategies for Pathogen Biocontrol Using Lactic Acid Bacteria and Their Metabolites: A Focus on Meat Ecosystems and Industrial Environments. Microorganisms 2017, 5, 38. [Google Scholar] [CrossRef]
- Rodríguez-Alonso, P.; Fernández-Otero, C.; Centeno, J.A.; Garabal, J.I. Antibiotic Resistance in Lactic Acid Bacteria and Micrococcaceae/Staphylococcaceae Isolates from Artisanal Raw Milk Cheeses, and Potential Implications on Cheese Making. J. Food Sci. 2009, 74, M284–M293. [Google Scholar] [CrossRef]
- Devirgiliis, C.; Zinno, P.; Perozzi, G. Update on Antibiotic Resistance in Foodborne Lactobacillus and Lactococcus Species. Front. Microbiol. 2013, 4, 301. [Google Scholar] [CrossRef]
- Tóth, A.G.; Csabai, I.; Krikó, E.; Tőzsér, D.; Maróti, G.; Patai, Á.V.; Makrai, L.; Szita, G.; Solymosi, N. Antimicrobial Resistance Genes in Raw Milk for Human Consumption. Sci. Rep. 2020, 10, 7464. [Google Scholar] [CrossRef]
- Duche, R.T.; Singh, A.; Wandhare, A.G.; Sangwan, V.; Sihag, M.K.; Nwagu, T.N.; Panwar, H.; Ezeogu, L.I. Antibiotic Resistance in Potential Probiotic Lactic Acid Bacteria of Fermented Foods and Human Origin from Nigeria. BMC Microbiol. 2023, 23, 142. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP). Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Wang, H.H.; Schaffner, D.W. Antibiotic Resistance: How Much Do We Know and Where Do We Go from Here? Appl. Environ. Microbiol. 2011, 77, 7093–7095. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, H.; Feng, J.; Ma, L.; de la Fuente-Núñez, C.; Wang, S.; Lu, X. Antibiotic Resistance of Lactic Acid Bacteria Isolated from Dairy Products in Tianjin, China. J. Agric. Food Res. 2019, 1, 100006. [Google Scholar] [CrossRef]
- Feld, L.; Schjørring, S.; Hammer, K.; Licht, T.R.; Danielsen, M.; Krogfelt, K.; Wilcks, A. Selective Pressure Affects Transfer and Establishment of a Lactobacillus plantarum Resistance Plasmid in the Gastrointestinal Environment. J. Antimicrob. Chemother. 2008, 61, 845–852. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Millar, B.C.; Xu, J. Characterization and Transfer of Antibiotic Resistance in Lactic Acid Bacteria from Fermented Food Products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef] [PubMed]
- EFSA-FEEDAP. Guidance on the Characterisation of Microorganisms Used as Feed Additives or as Production Organisms. EFSA J. 2018, 16, 5206. [Google Scholar] [CrossRef]
- Zarzecka, U.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Microorganisms from Starter and Protective Cultures—Occurrence of Antibiotic Resistance and Conjugal Transfer of tet Genes In Vitro and During Food Fermentation. LWT 2022, 153, 112490. [Google Scholar] [CrossRef]
- Luo, H.; Wan, K.; Wang, H.H. High-Frequency Conjugation System Facilitates Biofilm Formation and pAMβ1 Transmission by Lactococcus lactis. Appl. Environ. Microbiol. 2005, 71, 2970–2978. [Google Scholar] [CrossRef]
- Rozman, V.; Mohar Lorbeg, P.; Accetto, T.; Bogovič Matijašič, B. Characterization of Antimicrobial Resistance in Lactobacilli and Bifidobacteria Used as Probiotics or Starter Cultures Based on Integration of Phenotypic and In Silico Data. Int. J. Food Microbiol. 2020, 314, 108388. [Google Scholar] [CrossRef]
- Jacobsen, L.; Wilcks, A.; Hammer, K.; Huys, G.; Gevers, D.; Andersen, S.R. Horizontal Transfer of tet(M) and erm(B) Resistance Plasmids from Food Strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the Gastrointestinal Tract of Gnotobiotic Rats. FEMS Microbiol. Ecol. 2007, 59, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Thumu, S.C.R.; Halami, P.M. Conjugal Transfer of erm(B) and Multiple tet Genes from Lactobacillus spp. to Bacterial Pathogens in Animal Gut, In Vitro, and During Food Fermentation. Food Res. Int. 2019, 116, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Toomey, N.; Monaghan, Á.; Fanning, S.; Bolton, D. Transfer of Antibiotic Resistance Marker Genes Between Lactic Acid Bacteria in Model Rumen and Plant Environments. Appl. Environ. Microbiol. 2009, 75, 3146–3152. [Google Scholar] [CrossRef]
- Toomey, N.; Monaghan, A.; Fanning, S.; Bolton, D.J. Assessment of Antimicrobial Resistance Transfer Between Lactic Acid Bacteria and Potential Foodborne Pathogens Using In Vitro Methods and Mating in a Food Matrix. Foodborne Pathog. Dis. 2009, 6, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Ojha, A.K.; Shah, N.P.; Mishra, V. Conjugal Transfer of Antibiotic Resistances in Lactobacillus spp. Curr. Microbiol. 2021, 78, 2839–2849. [Google Scholar] [CrossRef]
- Tosi, L.; Berruti, G.; Danielsen, M.; Wind, A.; Huys, G.; Morelli, L. Susceptibility of Streptococcus thermophilus to antibiotics. Antonie Van Leeuwenhoek 2007, 92, 21–28. [Google Scholar] [CrossRef]
- Morandi, S.; Brasca, M. Safety aspects, genetic diversity and technological characterisation of wild-type Streptococcus thermophilus strains isolated from north Italian traditional cheeses. Food Cont. 2012, 23, 203–209. [Google Scholar] [CrossRef]
- Yang, C.; Yu, T. Characterization and transfer of antimicrobial resistance in lactic acid bacteria from fermented dairy products in China. J. Infect. Dev. Ctries. 2019, 13, 137–148. [Google Scholar] [CrossRef]
- Nunziata, L.; Brasca, M.; Morandi, S.; Silvetti, T. Antibiotic Resistance in Wild and Commercial Non-Enterococcal Lactic Acid Bacteria and Bifidobacteria Strains of Dairy Origin: An Update. Food Microbiol. 2022, 104, 103999. [Google Scholar] [CrossRef]
- Erginkaya, Z.; Turhan, E.U.; Tatlı, D. Determination of Antibiotic Resistance of Lactic Acid Bacteria Isolated from Traditional Turkish Fermented Dairy Products. Iran. J. Vet. Res. 2018, 19, 53–56. [Google Scholar] [CrossRef]
- Morandi, S.; Brasca, M.; Andrighetto, C.; Lombardi, A.; Lodi, R. Technological and Molecular Characterisation of Enterococci Isolated from Northwest Italian Dairy Products. Int. Dairy J. 2006, 16, 867–875. [Google Scholar] [CrossRef]
- Ogier, J.C.; Seror, P. Safety Assessment of Dairy Microorganisms: The Enterococcus Genus. Int. J. Food Microbiol. 2008, 126, 291–301. [Google Scholar] [CrossRef]
- Jamet, E.; Akary, E.; Poisson, M.; Chamba, J.; Bertrand, X.; Serror, P. Prevalence and Characterization of Antibiotic-Resistant Enterococcus faecalis in French Cheeses. Food Microbiol. 2012, 31, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Hammad, A.M.; Hassan, H.A.; Shimamoto, T. Prevalence, Antibiotic Resistance, and Virulence of Enterococcus spp. in Egyptian Fresh Raw Milk Cheese. Food Control 2015, 50, 815–820. [Google Scholar] [CrossRef]
- Výrostková, J.; Regecová, I.; Dudriková, E.; Marcincak, S.; Vargová, M.; Kovacová, M.; Mal’ová, J. Antimicrobial Resistance of Enterococcus sp. Isolated from Sheep and Goat Cheeses. Foods 2021, 10, 1844. [Google Scholar] [CrossRef] [PubMed]
- Claeys, W.L.; Cardoen, S.; Daube, G.; De Block, J.; Dewettinck, K.; Dierick, K.; De Zutter, L.; Huyghebaert, A.; Imberechts, H.; Thiange, P.; et al. Raw or Heated Cow Milk Consumption: Review of Risks and Benefits. Food Control 2013, 31, 251–262. [Google Scholar] [CrossRef]
- Lucey, J.A. Raw Milk Consumption. Nutr. Today 2015, 50, 189–193. [Google Scholar] [CrossRef]
- Law, B.A.; Tamime, A.Y. (Eds.) Technology of Cheesemaking, 2nd ed.; Wiley-Blackwell: Oxford, UK, 2010. [Google Scholar] [CrossRef]
- Salazar, J.K.; Carstens, C.K.; Ramachandran, P.; Shazer, A.G.; Narula, S.S.; Reed, E.; Ottesen, A.; Schill, K.M. Metagenomics of Pasteurized and Unpasteurized Gouda Cheese Using Targeted 16S rDNA Sequencing. BMC Microbiol. 2018, 18, 189. [Google Scholar] [CrossRef]
- Alexa, E.A.; Walsh, C.J.; Coughlan, L.M.; Awad, A.; Simon, C.A.; Ruiz, L.; Crispie, F.; Cotter, P.D.; Alvarez-Ordóñez, A. Dairy Products and Dairy-Processing Environments as a Reservoir of Antibiotic Resistance and Quorum-Quenching Determinants as Revealed Through Functional Metagenomics. mSystems 2020, 5, e00723-19. [Google Scholar] [CrossRef]
- Guo, H.; Pan, L.; Li, L.; Lu, J.; Kwok, L.; Menghe, B.; Zhang, H.; Zhang, W. Characterization of Antibiotic Resistance Genes from Lactobacillus Isolated from Traditional Dairy Products. J. Food Sci. 2017, 82, 724–730. [Google Scholar] [CrossRef]
- Colombo, M.; Nero, L.A.; Todorov, S.D. Safety Profiles of Beneficial Lactic Acid Bacteria Isolated from Dairy Systems. Braz. J. Microbiol. 2020, 51, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Akpınar Kankaya, D.; Tuncer, Y. Antibiotic Resistance in Vancomycin-Resistant Lactic Acid Bacteria (VRLAB) Isolated from Foods of Animal Origin. J. Food Process. Preserv. 2020, 44, e14468. [Google Scholar] [CrossRef]
- Haryani, Y.; Halid, N.A.; Guat, G.S.; Nor-Khaizura, M.A.R.; Hatta, M.A.M.; Sabri, S.; Hasan, H. High Prevalence of Multiple Antibiotic Resistance in Fermented Food-Associated Lactic Acid Bacteria in Malaysia. Food Control 2023, 147, 109558. [Google Scholar] [CrossRef]
- Blandino, G.; Milazzo, I.; Fazio, D. Antibiotic susceptibility of bacterial isolates from probiotic products available in Italy. Microb. Ecol. Health Dis. 2008, 20, 199–203. [Google Scholar] [CrossRef]
- Sharma, C.; Gulati, S.; Thakur, N.; Singh, B.P.; Gupta, S.; Kaur, S.; Mishra, S.K.; Puniya, A.K.; Gill, J.P.S.; Panwar, H. Antibiotic Sensitivity Pattern of Indigenous Lactobacilli Isolated from Curd and Human Milk Samples. 3 Biotech 2017, 7, 53. [Google Scholar] [CrossRef]
- Hoxha, R.; Nikolova, D.; Evstatieva, Y. Antibiotic Resistance Profile of the Newly Isolated Lactic Acid Bacteria Strains from Traditional Fermented Foods. Curr. Trends Nat. Sci. 2022, 11, 247–253. [Google Scholar] [CrossRef]
- Hummel, A.S.; Hertel, C.; Holzapfel, W.H.; Franz, C.M. Antibiotic Resistances of Starter and Probiotic Strains of Lactic Acid Bacteria. Appl. Environ. Microbiol. 2007, 73, 730–739. [Google Scholar] [CrossRef]
- Tavsanli, H.; Elal Mus, T.; Cetinkaya, F.; Ayanoglu, E.; Cibik, R. Isolation of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophiles from Nature: Technological Characterisation and Antibiotic Resistance. Czech J. Food Sci. 2021, 39, 305–311. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Kromann, S.; Chen, M.; Shi, L.; Meng, H. Antibiotic Resistance of Lactobacillus spp. and Streptococcus thermophiles Isolated from Chinese Fermented Milk Products. Foodborne Pathog. Dis. 2019, 16, 221–228. [Google Scholar] [CrossRef]
- Shin, E.; Paek, J.J.; Lee, Y. Antimicrobial Resistance of Seventy Lactic Acid Bacteria Isolated from Commercial Probiotics in Korea. Microbiol. Biotechnol. 2023, 33, 500. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic Resistance in Non-Enterococcal Lactic Acid Bacteria and Bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Knapp, C.W.; Gálvez, A.; Benomar, N. Antibiotic Resistance Profile of Microbes from Traditional Fermented Foods. In Fermented Foods in Health and Disease Prevention; Frias, J., Martinez-Villaluenga, C., Peñas, E., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 675–704. [Google Scholar] [CrossRef]
- Tóth, A.G.; Csabai, I.; Judge, M.F.; Maróti, G.; Becsei, Á.; Spisák, S.; Solymosi, N. Mobile Antimicrobial Resistance Genes in Probiotics. Antibiotics 2021, 10, 1287. [Google Scholar] [CrossRef]
- Fatahi-Bafghi, M.; Naseri, S.; Alizehi, A. Genome Analysis of Probiotic Bacteria for Antibiotic Resistance Genes. Antonie Van Leeuwenhoek 2022, 115, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Leuschner, R.G.K.; Robinson, T.P.; Hugas, M.; Cocconcelli, P.S.; Richard-Forget, F.; Klein, G.; Licht, T.R.; Nguyen-The, C.; Querol, A.; Richardson, M.; et al. Qualified Presumption of Safety (QPS): A Generic Risk Assessment Approach for Biological Agents Notified to the European Food Safety Authority (EFSA). Trends Food Sci. Technol. 2010, 21, 425–435. [Google Scholar] [CrossRef]
- Obioha, P.I.; Anyogu, A.; Awamaria, B.; Ghoddusi, H.B.; Ouoba, L.I.I. Antimicrobial Resistance of Lactic Acid Bacteria from Nono, a Naturally Fermented Milk Product. Antibiotics 2023, 12, 843. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.M.; Prathan, R.; Hein, S.T.; Chuanchuen, R. Microbiological Quality and Antimicrobial Resistance of Commercial Probiotic Products for Food-Producing Animals. Antibiotics 2024, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.S.; Pillidge, C.J.; Gopal, P.K.; Gill, H.S. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 2005, 98, 211–217. [Google Scholar] [CrossRef]
- Coton, M.; Lebreton, M.; Salas, M.L.; Garnier, L.; Navarri, M.; Pawtowski, A.; Mounier, J. Biogenic amine and antibiotic resistance profiles determined for lactic acid bacteria and a propionibacterium prior to use as antifungal bioprotective cultures. Int. Dairy J. 2018, 85, 21–26. [Google Scholar] [CrossRef]
- Anisimova, E.A.; Yarullina, D.R. Antibiotic Resistance of Lactobacillus Strains. Curr. Microbiol. 2019, 76, 1407–1416. [Google Scholar] [CrossRef]
- Zhou, N.; Zhang, J.X.; Fan, M.T.; Wang, J.; Guo, G.; Wei, X.Y. Antibiotic resistance of lactic acid bacteria isolated from Chinese yogurts. J. Dairy Sci. 2012, 95, 4775–4783. [Google Scholar] [CrossRef]
- Comunian, R.; Daga, E.; Dupré, I.; Paba, A.; Devirgiliis, C.; Piccioni, V.; Giraffa, G. Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int. J. Food Microbiol. 2010, 138, 151–156. [Google Scholar] [CrossRef]
- Fortina, M.G.; Nicastro, G.; Carminati, D.; Neviani, E.; Manachini, P.L. Lactobacillus helveticus heterogeneity in natural cheese starters: The diversity in phenotypic characteristics. J. Appl. Microbiol. 1998, 84, 72–80. [Google Scholar] [CrossRef]
- Cho, G.-S.; Cappello, C.; Schrader, K.; Fagbemigum, O.; Oguntoyinbo, F.A.; Csovcsics, C.; Rösch, N.; Kabisch, J.; Neve, H.; Bockelmann, W.; et al. Isolation and characterization of lactic acid bacteria from fermented goat milk in Tajikistan. J. Microbiol. Biotechnol. 2018, 28, 1834–1845. [Google Scholar] [CrossRef]
- Majhenič, C.; Mohar Lorberg, A.; Rogelj, P.I. Characterisation of the Lactobacillus community in traditional Karst ewe’s cheese. Int. J. Dairy Technol. 2007, 60, 182–190. [Google Scholar] [CrossRef]
- Fortina, M.G.; Ricci, G.; Foschino, R.; Picozzi, C.; Dolci, P.; Zeppa, G.; Manachini, P.L. Phenotypic typing, technological properties and safety aspects of Lactococcus garvieae strains from dairy environments. J. Appl. Microbiol. 2007, 103, 445–453. [Google Scholar] [CrossRef]
- Morandi, S.; Silvetti, T.; Miranda Lopez, J.M.; Brasca, M. Antimicrobial Activity, Antibiotic Resistance and the Safety of Lactic Acid Bacteria in Raw Milk V altellina C asera Cheese. J. Food Saf. 2015, 35, 193–205. [Google Scholar] [CrossRef]
- Coppola, R.; Succi, M.; Tremonte, P.; Reale, A.; Salzano, G.; Sorrentino, E. Antibiotic susceptibility of Lactobacillus rhamnosus strains isolated from Parmigiano Reggiano cheese. Lait 2005, 85, 193–204. [Google Scholar] [CrossRef]
- Quaresma, L.S.; Santos, R.C.V.; Gomes, G.C.; Américo, M.F.; Campos, G.M.; Laguna, J.G.; de Jesus, L.C.L. Multidrug resistance profile in Lactobacillus delbrueckii: A food industry species with probiotic properties. World J. Microbiol. Biotechnol. 2024, 40, 235. [Google Scholar] [CrossRef]
- Fernández, E.; Alegría, Á.; Delgado, S.; Martín, M.C.; Mayo, B. Comparative phenotypic and molecular genetic profiling of wild Lactococcus lactis subsp. lactis strains of the L. lactis subsp. lactis and L. lactis subsp. cremoris genotypes, isolated from starter-free cheeses made of raw milk. Appl. Environ. Microbiol. 2011, 77, 5324–5335. [Google Scholar] [CrossRef]
- Morandi, S.; Cremonesi, P.; Silvetti, T.; Brasca, M. Technological characterisation, antibiotic susceptibility and antimicrobial activity of wild-type Leuconostoc strains isolated from north Italian traditional cheeses. J. Dairy Res. 2013, 80, 457–466. [Google Scholar] [CrossRef]
- Flórez, A.B.; Delgado, S.; Mayo, B. Antimicrobial susceptibility of lactic acid bacteria isolated from a cheese environment. Can. J. Microbiol. 2005, 51, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Akpinar, A.; Yerlikaya, O. Some potential beneficial properties of Lacticaseibacillus paracasei subsp. paracasei and Leuconostoc mesenteroides strains originating from raw milk and kefir grains. J. Food Proc. Preserv. 2021, 45, e15986. [Google Scholar] [CrossRef]
- De Paula, A.T.; Jeronymo-Ceneviva, A.B.; Silva, L.F.; Todorov, S.D.; Franco, B.D.G.M.; Penna, A.L.B. Leuconostoc mesenteroides SJRP55: A potential probiotic strain isolated from Brazilian water buffalo mozzarella cheese. Ann. Microbiol. 2015, 65, 899–910. [Google Scholar] [CrossRef]
- Lüdin, P.; Roetschi, A.; Wüthrich, D.; Bruggmann, R.; Berthoud, H.; Shani, N. Update on tetracycline susceptibility of Pediococcus acidilactici based on strains isolated from swiss cheese and whey. J. Food Prot. 2018, 81, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, V.Q.; Camargo, A.C.; Todorov, S.D.; Nero, L.A. Potential control of Listeria monocytogenes by bacteriocinogenic Enterococcus hirae ST57ACC and Pediococcus pentosaceus ST65ACC strains isolated from artisanal cheese. Probiotics Antimicrob. Proteins 2019, 11, 696–704. [Google Scholar] [CrossRef]
- Basbülbül, G.; Özteber, M.; Biyik, H.H. Antibiotic resistance in lactic acid bacteria isolated from fermented dairy products and boza. J. Microb. Biotech. Food Sci. 2015, 4, 513. [Google Scholar] [CrossRef]
- Shi, Y.; Cui, X.; Gu, S.; Yan, X.; Li, R.; Xia, S.; Ge, J. Antioxidative and probiotic activities of lactic acid bacteria isolated from traditional artisanal milk cheese from Northeast China. Probiotics Antimicrob. Proteins 2019, 11, 1086–1099. [Google Scholar] [CrossRef]
- Xu, F.; Wang, J.; Guo, Y.; Fu, P.; Zeng, H.; Li, Z.; Wang, S. Antibiotic resistance, biochemical typing, and PFGE typing of Bifidobacterium strains commonly used in probiotic health foods. Food Sci. Biotechnol. 2018, 27, 467–477. [Google Scholar] [CrossRef]
- van Hoek, A.H.; Mayrhofer, S.; Domig, K.J.; Aarts, H.J. Resistance determinant erm (X) is borne by transposon Tn5432 in Bifidobacterium thermophilum and Bifidobacterium animalis subsp. lactis. Int. J. Antimicrob. Agents 2008, 31, 544–548. [Google Scholar] [CrossRef]
- Delgado, S.; Flórez, A.B.; Mayo, B. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr. Microbiol. 2005, 50, 202–207. [Google Scholar] [CrossRef]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tanweer, S. In vitro probiotic potential and safety evaluation (hemolytic, cytotoxic activity) of Bifidobacterium strains isolated from raw camel milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef]
- Chen, J.; Li, W.; Zhang, J.; Qi, W.; Li, Y.; Chen, S.; Zhou, W. Prevalence of antibiotic resistance genes in drinking water and biofilms: The correlation with the microbial community and opportunistic pathogens. Chemosphere 2020, 259, 127483. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Niu, Z.; Wang, M.; Zhang, Y. The occurrence and variations of extracellular antibiotic resistance genes in drinking water supply system: A potential risk to our health. J. Clean. Prod. 2023, 402, 136714. [Google Scholar] [CrossRef]
- Oberle, K.; Capdeville, M.J.; Berthe, T.; Budzinski, H.; Petit, F. Evidence for a complex relationship between antibiotics and antibiotic-resistant Escherichia coli: From medical center patients to a receiving environment. Environ. Sci. Technol. 2012, 46, 1859–1868. [Google Scholar] [CrossRef]
- Jia, S.; Shi, P.; Hu, Q.; Li, B.; Zhang, T.; Zhang, X.X. Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ. Sci. Technol. 2015, 49, 12271–12279. [Google Scholar] [CrossRef]
- Liu, S.S.; Qu, H.M.; Yang, D.; Hu, H.; Liu, W.L.; Qiu, Z.G.; Hou, A.M.; Guo, J.; Li, J.W.; Shen, Z.Q.; et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136. [Google Scholar] [CrossRef]
- Larson, A.J.; Haver, S.; Hattendorf, J.; Salmon-Mulanovich, G.; Riveros, M.; Verastegui, H.; Hartinger, S.M. Household-Level Risk Factors for Water Contamination and Antimicrobial Resistance in Drinking Water Among Households with Children Under 5 in Rural San Marcos, Cajamarca, Peru. One Health 2023, 16, 100482. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yu, H.; Liang, Y.; Zhang, X.; Yang, D.; Wang, L.; Jin, M. Extended chloramination significantly enriched intracellular antibiotic resistance genes in drinking water treatment plants. Water Res. 2023, 232, 119689. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ondon, B.S.; Ho, S.H.; Zhou, Q.; Li, F. Drinking water sources as hotspots of Antibiotic-Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARGs): Occurrence, spread, and mitigation strategies. J. Water Process Eng. 2023, 53, 103907. [Google Scholar] [CrossRef]
- Erdei-Tombor, P.; Kiskó, G.; Taczman-Brückner, A. Biofilm Formation in Water Distribution Systems. Processes 2024, 12, 280. [Google Scholar] [CrossRef]
- Krol, J.E.; Wojtowicz, A.J.; Rogers, L.M.; Heuer, H.; Smalla, K.; Krone, S.M.; Top, E.M. Invasion of E. coli Biofilms by Antibiotic Resistance Plasmids. Plasmid 2013, 70, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, T.; Chen, H. Antibiotic resistome promotion in drinking water during biological activated carbon treatment: Is it influenced by quorum sensing? Sci. Total Environ. 2018, 612, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.; Feng, M.; Chen, J.; Shen, W.; Zhang, S. Occurrence of antibiotics and antibiotic resistance genes and their correlations in river-type drinking water source, China. Environ. Sci. Pollut. Res. 2021, 28, 42339–42352. [Google Scholar] [CrossRef]
- Zhong, D.; Zhou, Z.; Ma, W.; Ma, J.; Feng, W.; Li, J.; Du, X. Antibiotic enhances the spread of antibiotic resistance among chlorine-resistant bacteria in drinking water distribution system. Environ. Res. 2022, 211, 113045. [Google Scholar] [CrossRef]
- Yan, Y.; Xu, J.; Huang, W.; Fan, Y.; Li, Z.; Tian, M.; Ma, J.; Lu, X.; Liang, J. Metagenomic and culturomics analysis of microbial communities within surface sediments and the prevalence of antibiotic resistance genes in a pristine river: The Zaqu River in the Lancang River Source Region, China. Microorganisms 2024, 12, 911. [Google Scholar] [CrossRef] [PubMed]
- Root, H.; Daniels, L.; Marx, A.; Bartelt, L.A.; Lachiewicz, A.M.; van Duin, D. Sulfonamides without trimethoprim in the treatment of Nocardia infections: A case report and literature review. Transpl. Infect. Dis. 2020, 23, e13452. [Google Scholar] [CrossRef]
- Gholipour, S.; Shamsizadeh, Z.; Gwenzi, W.; Nikaeen, M. The Bacterial Biofilm Resistome in Drinking Water Distribution Systems: A Systematic Review. Chemosphere 2023, 329, 138642. [Google Scholar] [CrossRef]
- Regulation (EU) 2015/2283 of the European Parliament and of the Council of 25 November 2015 on Novel Foods, Amending Regulation (EU) No 1169/2011 of the European Parliament and of the Council and Repealing Regulation (EC) No 258/97 of the European Parliament and of the Council and Commission Regulation (EC) No 1852/2001 (Text with EEA Relevance). Available online: https://eur-lex.europa.eu/eli/reg/2015/2283/oj (accessed on 24 October 2024).
- What Are the Novel Foods? Available online: https://www.foodchainid.com/resources/what-are-the-novel-foods/ (accessed on 2 January 2025).
- Food and Feed Information Portal Database Novel Food Catalogue. Available online: https://ec.europa.eu/food/food-feed-portal/screen/novel-food-catalogue/search (accessed on 24 November 2024).
- Novel Food Legislation. Available online: https://vb.nweurope.eu/media/17698/presentatie-novel-food-fod-volksgezondheid.pdf (accessed on 2 January 2025).
- Novel Foods. Available online: https://www.ages.at/en/human/nutrition-food/food-information/novel-foods (accessed on 2 January 2025).
- Ameta, S.K.; Rai, A.K.; Hiran, D.; Ameta, R.; Ameta, S.C. Use of Nanomaterials in Food Science. In Biogenic Nano-Particles and Their Use in Agro-Ecosystems; Ghorbanpour, M., Bhargava, P., Varma, A., Choudhary, D., Eds.; Springer: Singapore, 2020; Volume 21, pp. 457–488. [Google Scholar] [CrossRef]
- Barsanti, L.; Gualtieri, P. Is exploitation of microalgae economically and energetically sustainable? Algal Res. 2018, 31, 107–115. [Google Scholar] [CrossRef]
- Vieira, M.V.; Pastrana, L.M.; Fuciños, P. Microalgae encapsulation systems for food, pharmaceutical and cosmetics applications. Mar. Drugs 2020, 18, 644. [Google Scholar] [CrossRef]
- Villarruel-López, A.; Ascencio, F.; Nuño, K. Microalgae, a potential natural functional food source–a review. Pol. J. Food Nutr. Sci. 2017, 67, 251–263. [Google Scholar] [CrossRef]
- Niccolai, A.; Zittelli, G.C.; Rodolfi, L.; Biondi, N.; Tredici, M.R. Microalgae of interest as food source: Biochemical composition and digestibility. Algal Res. 2019, 42, 101617. [Google Scholar] [CrossRef]
- Matos, J.; Cardoso, C.; Bandarra, N.M.; Afonso, C. Microalgae as healthy ingredients for functional food: A review. Food Funct. 2017, 8, 2672–2685. [Google Scholar] [CrossRef] [PubMed]
- da Silva Vaz, B.; Moreira, J.B.; de Morais, M.G.; Costa, J.A.V. Microalgae as a new source of bioactive compounds in food supplements. Curr. Opin. Food Sci. 2016, 7, 73–77. [Google Scholar] [CrossRef]
- Torres-Tiji, Y.; Fields, F.J.; Mayfield, S.P. Microalgae as a future food source. Biotechnol. Adv. 2020, 41, 107536. [Google Scholar] [CrossRef]
- Koller, M.; Muhr, A.; Braunegg, G. Microalgae as versatile cellular factories for valued products. Algal Res. 2014, 6, 52–63. [Google Scholar] [CrossRef]
- Wells, M.L.; Potin, P.; Craigie, J.S.; Raven, J.A.; Merchant, S.S.; Helliwell, K.E.; Smith, A.G.; Camire, M.E.; Brawley, S.H. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol. 2017, 29, 949–982. [Google Scholar] [CrossRef]
- Vigani, M.; Parisi, C.; Rodríguez-Cerezo, E.; Barbosa, M.J.; Sijtsma, L.; Ploeg, M.; Enzing, C. Food and feed products from micro-algae: Market opportunities and challenges for the EU. Trends Food Sci. Technol. 2015, 42, 81–92. [Google Scholar] [CrossRef]
- Su, M.; Bastiaens, L.; Verspreet, J.; Hayes, M. Applications of microalgae in foods, pharma and feeds and their use as fertilizers and biostimulants: Legislation and regulatory aspects for consideration. Foods 2023, 12, 3878. [Google Scholar] [CrossRef]
- Chen, C.; Tang, T.; Shi, Q.; Zhou, Z.; Fan, J. The potential and challenge of microalgae as promising future food sources. Trends Food Sci. Technol. 2022, 126, 99–112. [Google Scholar] [CrossRef]
- Bazarnova, J.; Nilova, L.; Trukhina, E.; Bernavskaya, M.; Smyatskaya, Y.; Aktar, T. Use of microalgae biomass for fortification of food products from grain. Foods 2021, 10, 3018. [Google Scholar] [CrossRef] [PubMed]
- Amaro, H.M.; Guedes, A.C.; Malcata, F.X. Antimicrobial activities of microalgae: An invited review. In Science Against Microbial Pathogens: Communicating Current Research and Technological Advances; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2011; Volume 1, pp. 1272–1280. [Google Scholar]
- Chacón-Lee, T.L.; González-Mariño, G.E. Microalgae for “healthy” foods—Possibilities and challenges. Comp. Rev. Food Sci. Food Saf. 2010, 9, 655–675. [Google Scholar] [CrossRef]
- Pagels, F.; Amaro, H.M.; Tavares, T.G.; Amil, B.F.; Guedes, A.C. Potential of microalgae extracts for food and feed supplementation—A promising source of antioxidant and anti-inflammatory compounds. Life 2022, 12, 1901. [Google Scholar] [CrossRef]
- Guedes, A.C.; Barbosa, C.R.; Amaro, H.M.; Pereira, C.I.; Malcata, F.X. Microalgal and cyanobacterial cell extracts for use as natural antibacterial additives against food pathogens. Int. J. Food Sci. Technol. 2011, 46, 862–870. [Google Scholar] [CrossRef]
- Finney, K.F.; Pomeranz, Y.; Bruinsma, B.L. Use of algae Dunaliella as a protein supplement in bread. Cereal Chem. 1984, 61, 402–406. [Google Scholar]
- Matos, Â.P. The impact of microalgae in food science and technology. J. Am. Oil Chem. Soc. 2017, 94, 1333–1350. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Ali, S.S.; Ramadan, H.; El-Aswar, E.I.; Eltawab, R.; Ho, S.H.; Elsamahy, T.; Li, S.; El-Sheekh, M.M.; Schagerl, M.; et al. Microalgae-based wastewater treatment: Mechanisms, challenges, recent advances, and future prospects. Environ. Sci. Ecotechnol. 2022, 8, 100205. [Google Scholar] [CrossRef]
- Inuwa, A.B.; Mahmood, Q.; Iqbal, J.; Widemann, E.; Shafiq, S.; Irshad, M.; Irshad, U.; Iqbal, A.; Hafeez, F.; Nazir, R. Removal of antibiotic resistance genes, class 1 integrase gene and Escherichia coli indicator gene in a microalgae-based wastewater treatment system. Antibiotics 2022, 11, 1531. [Google Scholar] [CrossRef]
- Liu, L.; Yu, X.; Wu, D.; Su, J. Antibiotic resistance gene profile in aerobic granular reactor under antibiotic stress: Can eukaryotic microalgae act as inhibiting factor? Environ. Pollut. 2022, 304, 119221. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Z.; Lu, T.; Peijnenburg, W.J.G.M.; Gillings, M.; Yang, X.; Chen, J.; Penuelas, J.; Zhu, Y.-G.; Zhou, N.-Y.; et al. Cyanobacterial blooms contribute to the diversity of antibiotic-resistance genes in aquatic ecosystems. Commun. Biol. 2020, 3, 737. [Google Scholar] [CrossRef]
- Zourou, A.C. Evaluation of Horizontal Gene Transfer Between Genetically Engineered Cyanobacteria and Gram-Negative Bacteria. Master’s Thesis, Old Dominion University, Norfolk, VA, USA, 2021. Available online: https://www.proquest.com/openview/b455e969ac78c1c994946a639005ed15/1?pq-origsite=gscholar&cbl=18750&diss=y (accessed on 24 November 2024).
- Nguyen, T.H.; Barnes, C.L.; Agola, J.P.; Sherazi, S.; Greene, L.H.; Lee, J.W. Demonstration of horizontal gene transfer from genetically engineered Thermosynechococcus elongatus BP1 to wild-type E. coli DH5α. Gene 2019, 704, 49–58. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Q.; Zhang, J.; Guan, T.; Chen, Y.; Shi, W. Critical roles of cyanobacteria as reservoir and source for antibiotic resistance genes. Environ. Int. 2020, 144, 106034. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Chang, H.; Zhong, N.; Ren, N.; Ho, S.H. Comprehensive insights into antibiotic resistance gene migration in microalgal-bacterial consortia: Mechanisms, factors, and perspectives. Sci. Total Environ. 2023, 901, 166029. [Google Scholar] [CrossRef] [PubMed]
- Inuwa, A.B.; Pervez, A.; Nazir, R. Microalgaebased wastewater treatment system: Current state, antibiotic resistant bacteria and antibiotic resistance genes reduction potentials. Int. J. Environ. Sci. Technol. 2023, 20, 14053–14072. [Google Scholar] [CrossRef]
- Cao, M.; Wang, F.; Zhou, B.; Chen, H.; Yuan, R.; Ma, S.; Geng, H.; Li, J.; Lv, W.; Wang, Y.; et al. Nanoparticles and antibiotics stress proliferated antibiotic resistance genes in microalgae-bacteria symbiotic systems. J. Hazard. Mater. 2023, 443, 130201. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Milanović, V.; Cardinali, F.; Aquilanti, L.; Clementi, F.; Osimani, A. Current knowledge on the microbiota of edible insects intended for human consumption: A state-of-the-art review. Food Res. Int. 2019, 125, 108527. [Google Scholar] [CrossRef]
- Commission Implementing Regulation (EU) 2021/1975 of 12 November 2021 Authorising the Placing on the Market of Frozen, Dried and Powder Forms of Locusta migratoria as a Novel Food Under Regulation (EU) 2015/2283 of the European Parliament and of the Council and Amending Commission Implementing Regulation (EU) 2017/2470. Off. J. Eur. Union L 2021, 402, 10–16. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32021R1975 (accessed on 24 October 2024).
- Commission Implementing Regulation (EU) 2022/169 of 8 February 2022 Authorising the Placing on the Market of Frozen, Dried and Powder Forms of Yellow Mealworm (Tenebrio molitor Larva) as a Novel Food under Regulation (EU) 2015/2283 of the European Parliament and of the Council and Amending Commission Implementing Regulation (EU) 2017/2470. Off. J. Eur. Union L 2022, 28, 10–16. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32022R0169 (accessed on 23 October 2024).
- Commission Implementing Regulation (EU) 2022/188 of 10 February 2022 Authorising the Placing on the Market of Frozen, Dried and Powder Forms of Acheta domesticus as a Novel Food under Regulation (EU) 2015/2283 of the European Parliament and of the Council, and Amending Commission Implementing Regulation (EU) 2017/2470. Off. J. Eur. Union L 2022, 30, 108–113. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32022R0188 (accessed on 28 October 2024).
- Commission Implementing Regulation (EU) 2023/58 of 5 January 2023 Authorising the Placing on the Market of the Frozen, Paste, Dried and Powder Forms of Alphitobius diaperinus Larvae (Lesser Mealworm) as a Novel Food and Amending Implementing Regulation (EU) 2017/2470. Off. J. Eur. Union L 2023, 5, 10–15. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32023R0058 (accessed on 29 October 2024).
- Mudalungu, C.M.; Mokaya, H.O.; Tanga, C.M. Beneficial sterols in selected edible insects and their associated antibacterial activities. Sci Rep. 2023, 13, 10786. [Google Scholar] [CrossRef] [PubMed]
- Gałęcki, R.; Bakuła, T.; Gołaszewski, J. Foodborne diseases in the edible insect industry in Europe—New challenges and old problems. Foods 2023, 12, 770. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Mann, A.; den Bakker, H.C. Analysis of microbial composition of edible insect products available for human consumption within the United States using traditional microbiological methods and whole genome sequencing. J. Food Prot. 2024, 87, 100277. [Google Scholar] [CrossRef]
- Imathiu, S. Benefits and food safety concerns associated with consumption of edible insects. NFS J. 2020, 18, 1–11. [Google Scholar] [CrossRef]
- Gałęcki, R.; Sokół, R. A parasitological evaluation of edible insects and their role in the transmission of parasitic diseases to humans and animals. PLoS ONE 2019, 14, e0219303. [Google Scholar] [CrossRef]
- Zurek, L.; Ghosha, A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl. Environ. Microbiol. 2014, 80, 3562–3567. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Anjali; Shreyata; Sabu, B.; Jamwal, R.; Devi, P.P.; Yadav, K.; Raina, H.S.; Rajagopal, R. Understanding the role of insects in the acquisition and transmission of antibiotic resistance. Sci. Total Environ. 2023, 858, 159805. [Google Scholar] [CrossRef]
- Milanović, V.; Osimani, V.; Pasquini, M.; Aquilanti, L.; Garofalo, C.; Taccari, M.; Cardinali, F.; Riolo, P.; Clementi, F. Getting insight into the prevalence of antibiotic resistance genes in specimens of marketed edible insects. Int. J. Food Microbiol. 2016, 227, 22–28. [Google Scholar] [CrossRef]
- Milanović, V.; Roncolini, A.; Cardinali, F.; Garofalo, C.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Corsi, L.; Isidoro, N.; Zarantoniello, M.; et al. Occurrence of antibiotic resistance genes in Hermetia illucens larvae fed coffee silverskin enriched with Schizochytrium limacinum or Isochrysis galbana microalgae. Genes 2021, 12, 213. [Google Scholar] [CrossRef]
- Vandeweyer, D.; Milanović, V.; Garofalo, C.; Osimani, A.; Clementi, F.; Van Campenhout, L.; Aquilanti, L. Real-time PCR detection and quantification of selected transferable antibiotic resistance genes in fresh edible insects from Belgium and the Netherlands. Int. J. Food Microbiol. 2019, 290, 288–295. [Google Scholar] [CrossRef]
- Osimani, A.; Milanović, V.; Cardinali, F.; Garofalo, C.; Clementi, F.; Ruschioni, S.; Riolo, P.; Isidoro, N.; Loreto, N.; Galarini, R.; et al. Distribution of transferable antibiotic resistance genes in laboratory-reared edible mealworms (Tenebrio molitor L.). Front. Microbiol. 2018, 9, 2702. [Google Scholar] [CrossRef] [PubMed]
- WHO. Clinically Important Antimicrobials for Human Medicine, 5th ed.; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-151222-0. Available online: https://iris.who.int/bitstream/handle/10665/255027/9789241512220-eng.pdf (accessed on 11 November 2024).
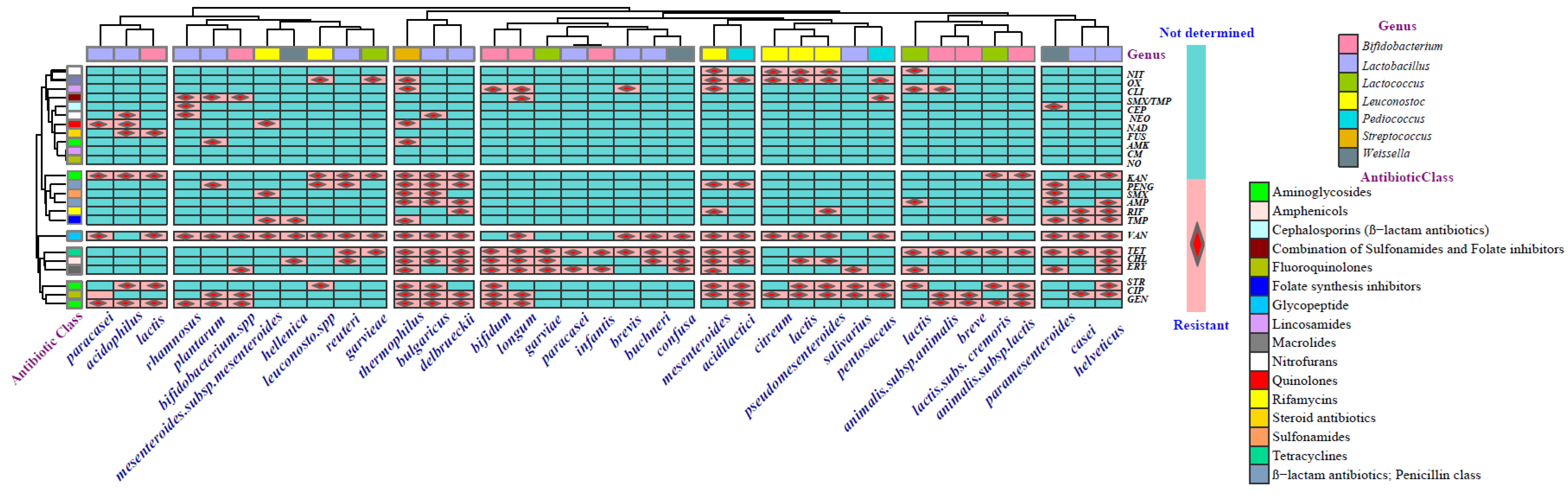
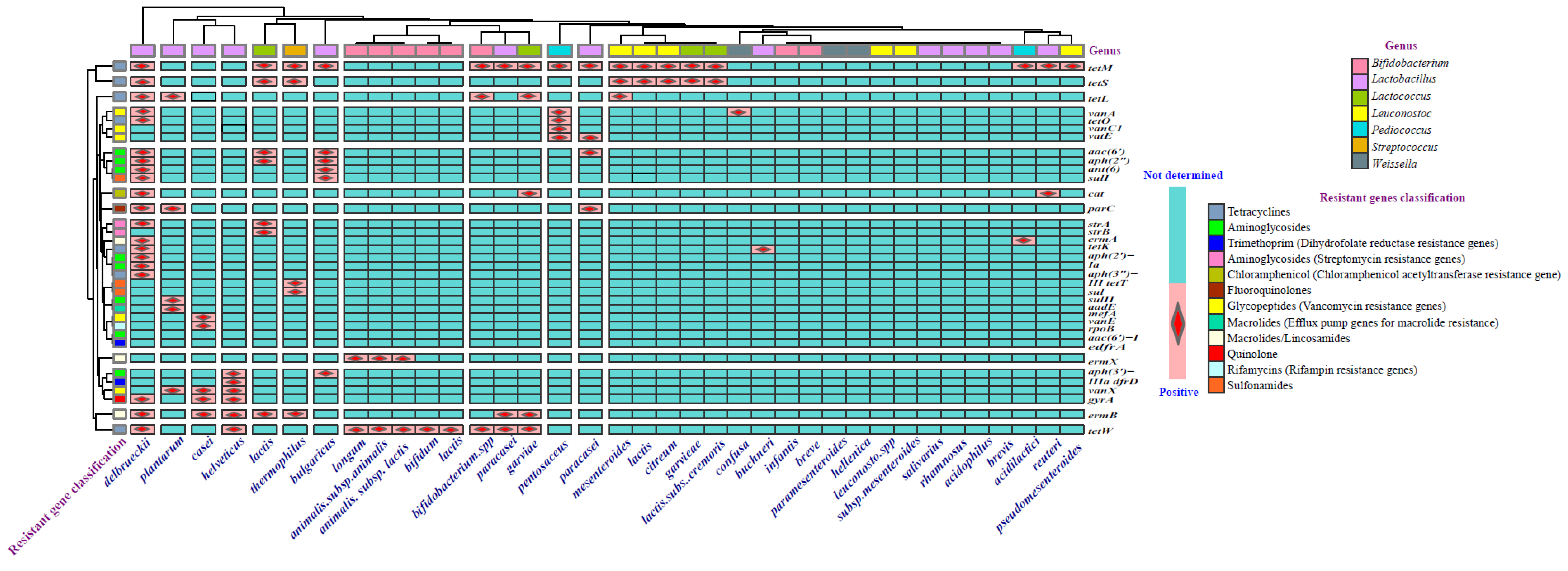
| Categories of Novel Food | Examples |
|---|---|
| Foods with new or modified molecular structure | D-Tagatose, salatrim |
| Foods consisting of, isolated from or produced from material of mineral origin | clinoptilolite (zeolite) |
| Foods consisting of, isolated from or produced from microorganisms, fungi, algae | algae oil from the microalgae Ulkenia sp. |
| Foods consisting of, isolated from or produced from plants or their parts | noni juice (Morinda citrifolia), chia seeds (Salvia hispanica) |
| Foods consisting of, isolated from or produced from animals or their parts | insects, oil from Antarctic krill (Euphasia superba), peptides from the fish Sardinops sagax |
| Food consisting of, isolated from or produced from cell culture or tissue culture derived from animals, plants, micro-organisms, fungi, or algae | extract from cell cultures of Echinacea angustifolia, in vitro meat |
| Food resulting from a production process not used for food production within the Union before 15 May 1997 | high pressure pasteurized fruit preparations, UV-treated food: mushrooms (Agaricus bisporus), baker’s yeast (Saccharomyces cerevisiae), bread, milk |
| Food consisting of engineered nanomaterials | nanosilver provides antimicrobial properties to food packaging, nanocapsules (containing flavor or color enhancers, or added vitamins) |
| Vitamins, minerals and other substances used in accordance with Directive 2002/46/EC, Regulation (EC) No 1925/2006 or Regulation (EU) No 609/2013 | iron (II) ammonium phosphate, vitamin K2 (menaquinone), chromium picolinate |
| Food used exclusively in food supplements within the Union before 15 May 1997 | maqui berry (Aristotelia chilensis), rose root (Rhodiola rosea) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiskó, G.; Bajramović, B.; Elzhraa, F.; Erdei-Tombor, P.; Dobó, V.; Mohácsi-Farkas, C.; Taczman-Brückner, A.; Belák, Á. The Invisible Threat of Antibiotic Resistance in Food. Antibiotics 2025, 14, 250. https://doi.org/10.3390/antibiotics14030250
Kiskó G, Bajramović B, Elzhraa F, Erdei-Tombor P, Dobó V, Mohácsi-Farkas C, Taczman-Brückner A, Belák Á. The Invisible Threat of Antibiotic Resistance in Food. Antibiotics. 2025; 14(3):250. https://doi.org/10.3390/antibiotics14030250
Chicago/Turabian StyleKiskó, Gabriella, Belma Bajramović, Fatma Elzhraa, Patrícia Erdei-Tombor, Viktória Dobó, Csilla Mohácsi-Farkas, Andrea Taczman-Brückner, and Ágnes Belák. 2025. "The Invisible Threat of Antibiotic Resistance in Food" Antibiotics 14, no. 3: 250. https://doi.org/10.3390/antibiotics14030250
APA StyleKiskó, G., Bajramović, B., Elzhraa, F., Erdei-Tombor, P., Dobó, V., Mohácsi-Farkas, C., Taczman-Brückner, A., & Belák, Á. (2025). The Invisible Threat of Antibiotic Resistance in Food. Antibiotics, 14(3), 250. https://doi.org/10.3390/antibiotics14030250








