Whole Genome Analysis of Pediococcus acidilactici XJ-24 and Its Role in Preventing Listeria monocytogenes ATCC® 19115TM Infection in C57BL/6 Mice
Abstract
1. Introduction
2. Results and Discussion
2.1. Genomic Features of XJ-24
2.2. Evaluation of Adaptability and Stress Response Genes in Human Gastrointestinal Environment
2.3. Safety Assessment of XJ-24
2.4. Bacteriocin of XJ-24
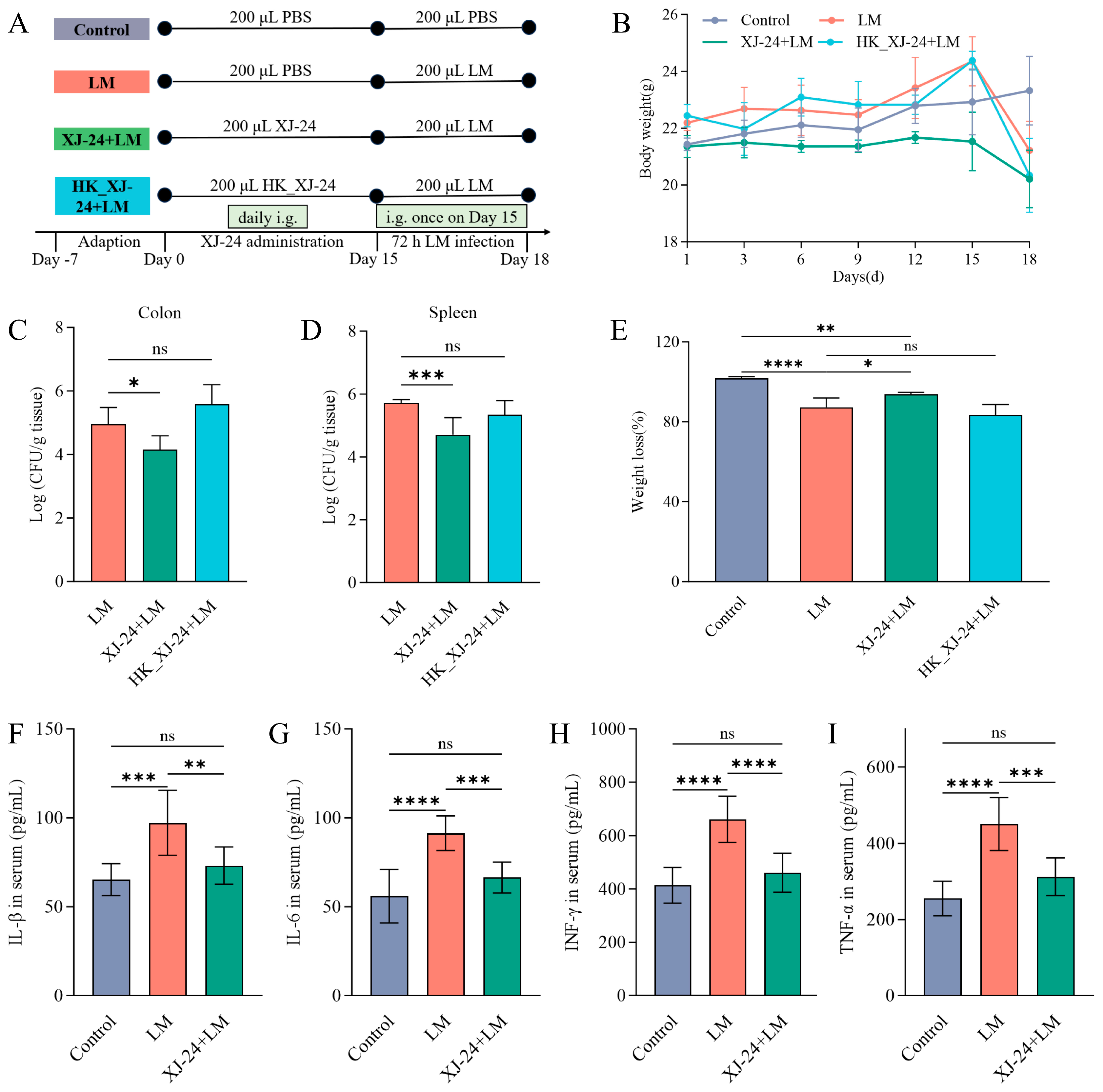
2.5. Effects of XJ-24 Pretreatment on Physiological Indices in Mice Infected with LM
2.6. Effects of XJ-24 Pretreatment on Serum Inflammatory Factors in Mice Infected with LM
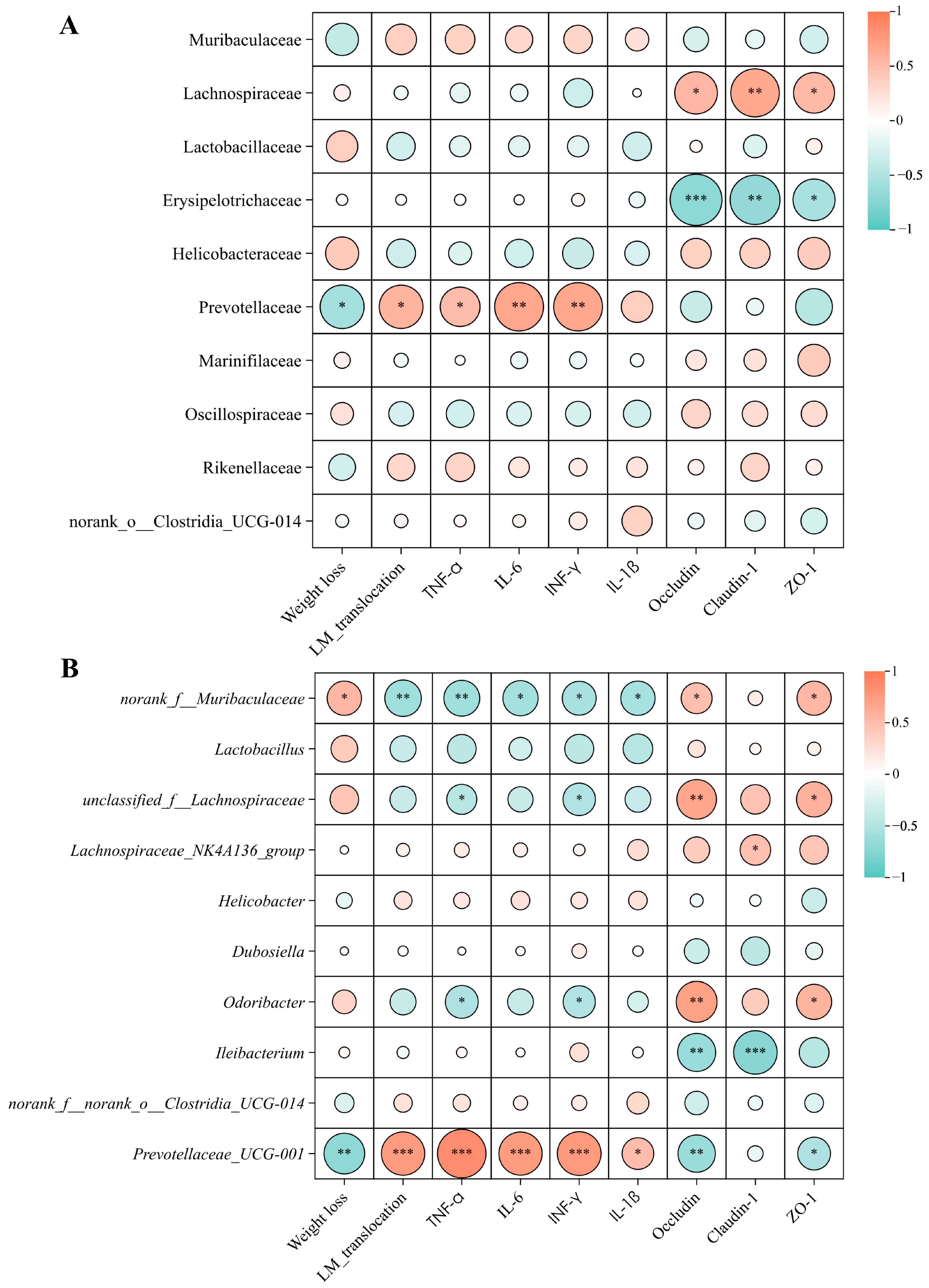
2.7. Effect of XJ-24 Pretreatment on Tissue Damage and Intestinal Barrier Integrity in Mice Infected with LM
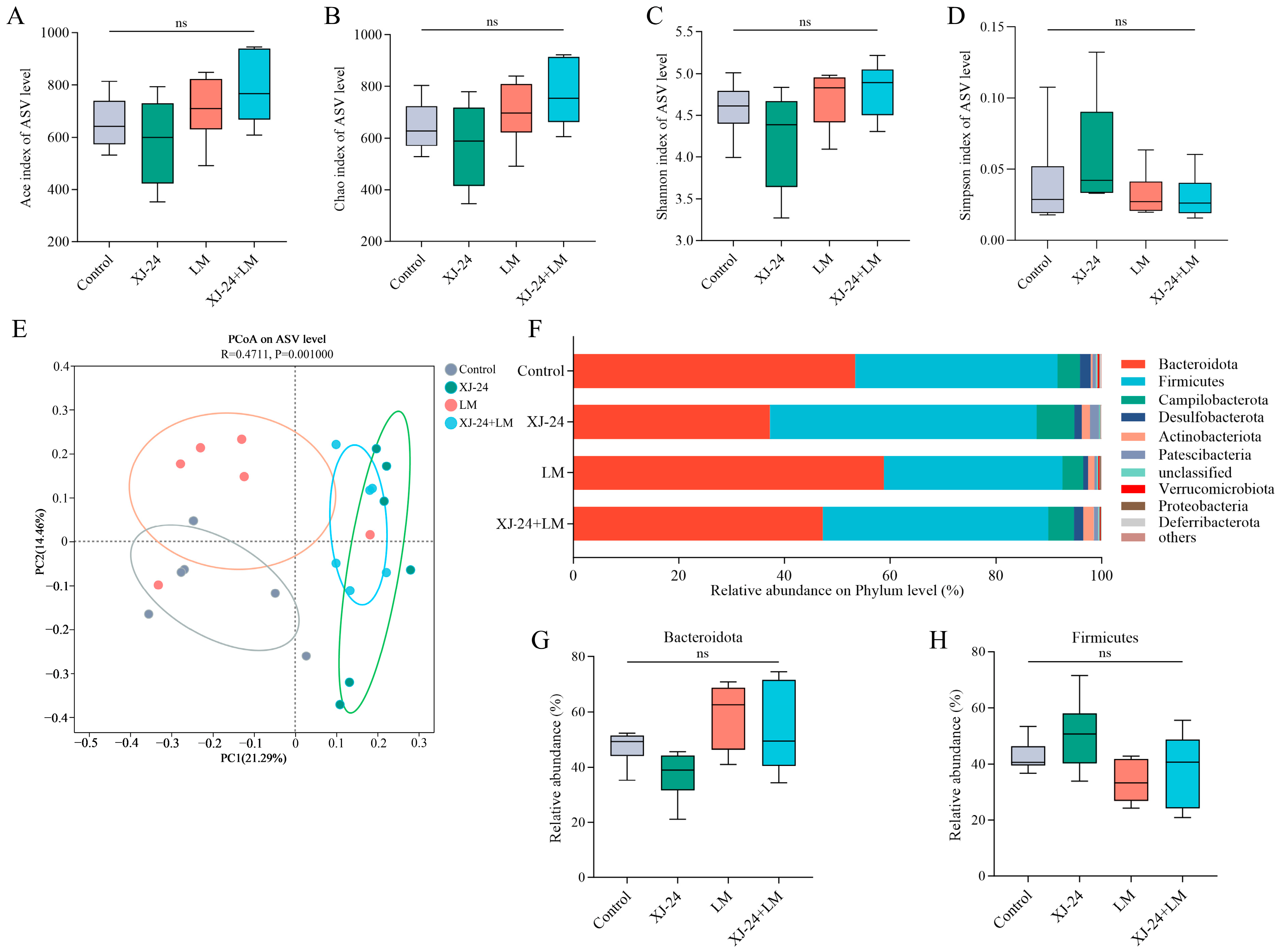
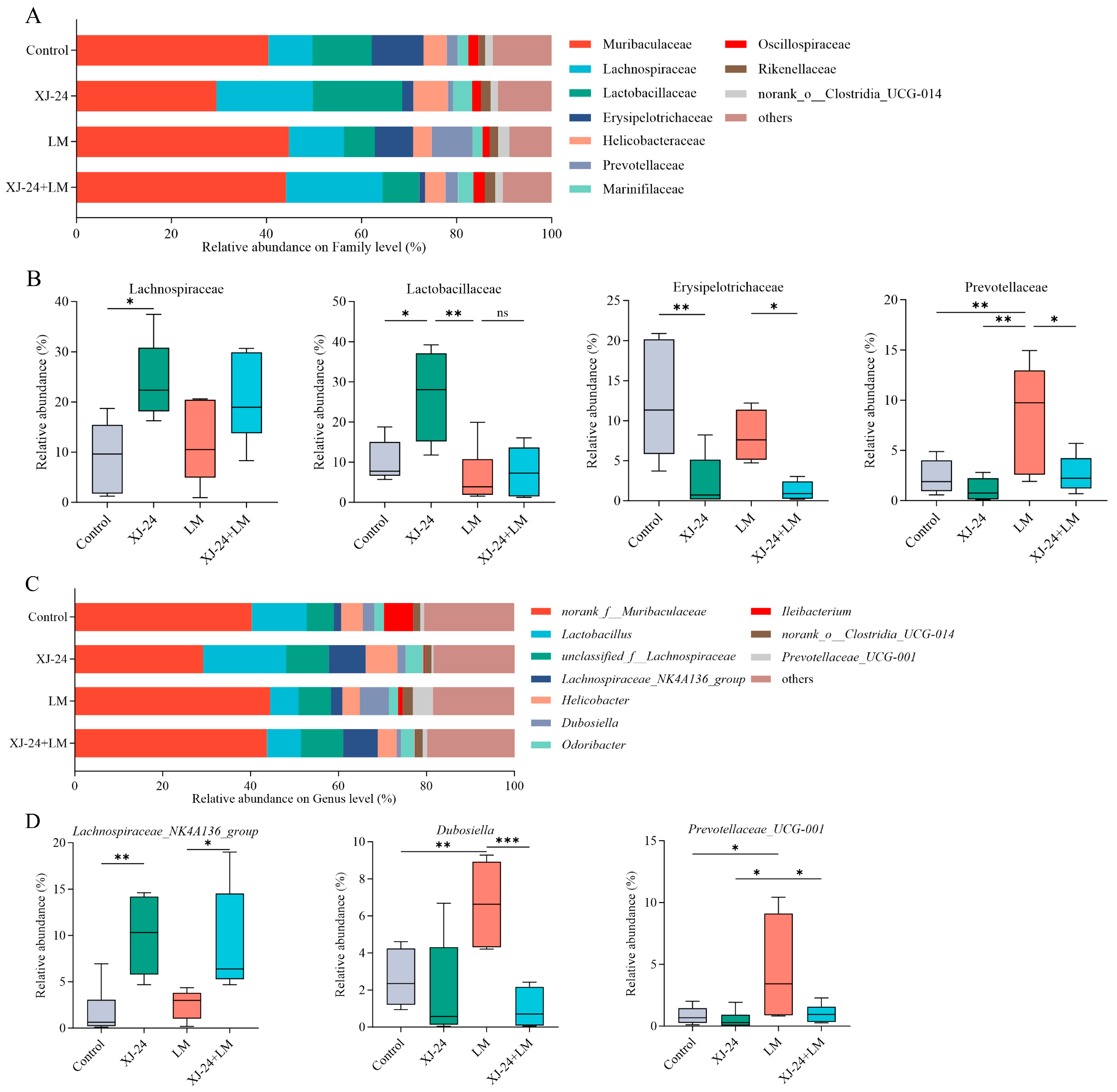
2.8. Effect of XJ-24 Pretreatment on the Gut Microbiota Structure
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
3.2. Whole Genomics Analysis of XJ-24
3.3. Animal Study
3.3.1. Preparation of Strains
3.3.2. Animal Modeling and Grouping
3.3.3. Preparation of Mouse Tissue Samples
3.3.4. Measurement of LM Colonization
3.3.5. Enzyme-Linked Immunosorbent Assay
3.3.6. Histopathological Analysis and Immunohistochemistry
3.3.7. Quantitative Real-Time PCR
3.3.8. Sequencing and Analysis of Gut Microbiota
3.4. Statistical Analysis
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kong, Y.; Yan, H.; Hu, J.; Dang, Y.; Han, Z.; Tian, B.; Wang, P. Antibacterial Activity and Mechanism of Action of Osthole against Listeria monocytogenes. J. Agric. Food Chem. 2024, 72, 10853–10861. [Google Scholar] [CrossRef]
- Li, J.; Yu, J.; Song, Y.; Wang, S.; Mu, G.; Tuo, Y. Exopolysaccharides and Surface-Layer Proteins Expressed by Biofilm-State Lactiplantibacillus plantarum Y42 Play Crucial Role in Preventing Intestinal Barrier and Immunity Dysfunction of Balb/C Mice Infected by Listeria monocytogenes ATCC 19115. J. Agric. Food Chem. 2024, 72, 8581–8594. [Google Scholar] [CrossRef] [PubMed]
- Drolia, R.; Tenguria, S.; Durkes, A.C.; Turner, J.R.; Bhunia, A.K. Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe 2018, 23, 470–484.e7. [Google Scholar] [CrossRef]
- Pitts, M.G.; D’Orazio, S.E.F. A Comparison of Oral and Intravenous Mouse Models of Listeriosis. Pathogens 2018, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Wilking, H.; Lachmann, R.; Holzer, A.; Halbedel, S.; Flieger, A.; Stark, K. Ongoing High Incidence and Case-Fatality Rates for Invasive Listeriosis, Germany, 2010–2019. Emerg. Infect. Dis. 2021, 27, 2485–2488. [Google Scholar] [CrossRef]
- Mayer, R.L.; Verbeke, R.; Asselman, C.; Aernout, I.; Gul, A.; Eggermont, D.; Boucher, K.; Thery, F.; Maia, T.M.; Demol, H.; et al. Immunopeptidomics-based design of mRNA vaccine formulations against Listeria monocytogenes. Nat. Commun. 2022, 13, 6075. [Google Scholar] [CrossRef]
- Jin, T.; Zhang, Y.; Yang, Y.; Teng, Y.; Yan, C.; Shan, Z.; Meng, J.; Xia, X. Intestinal linoleic acid contributes to the protective effects of Akkermansia muciniphila against Listeria monocytogenes infection in mice. Imeta 2024, 3, e196. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- He, Y.; Yang, Q.; Tian, L.; Zhang, Z.; Qiu, L.; Tao, X.; Wei, H. Protection of surface layer protein from Enterococcus faecium WEFA23 against Listeria monocytogenes CMCC54007 infection by modulating intestinal permeability and immunity. Appl. Microbiol. Biotechnol. 2021, 105, 4269–4284. [Google Scholar] [CrossRef] [PubMed]
- Drolia, R.; Amalaradjou, M.A.R.; Ryan, V.; Tenguria, S.; Liu, D.; Bai, X.; Xu, L.; Singh, A.K.; Cox, A.D.; Bernal-Crespo, V.; et al. Receptor-targeted engineered probiotics mitigate lethal Listeria infection. Nat. Commun. 2020, 11, 6344. [Google Scholar] [CrossRef]
- Keane, J.M.; Las Heras, V.; Pinheiro, J.; FitzGerald, J.A.; Nunez-Sanchez, M.A.; Hueston, C.M.; O’Mahony, L.; Cotter, P.D.; Hill, C.; Melgar, S.; et al. Akkermansia muciniphila reduces susceptibility to Listeria monocytogenes infection in mice fed a high-fat diet. Gut Microbes 2023, 15, 2229948. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, L.M.; Santos, M.M.; de Souza Silva, H.P.; Arantes, R.M.; Nicoli, J.R.; Vieira, L.Q. Monoassociation with probiotic Lactobacillus delbrueckii UFV-H2b20 stimulates the immune system and protects germfree mice against Listeria monocytogenes infection. Med. Microbiol. Immunol. 2011, 200, 29–38. [Google Scholar] [CrossRef]
- Becattini, S.; Littmann, E.R.; Carter, R.A.; Kim, S.G.; Morjaria, S.M.; Ling, L.; Gyaltshen, Y.; Fontana, E.; Taur, Y.; Leiner, I.M.; et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J. Exp. Med. 2017, 214, 1973–1989. [Google Scholar] [CrossRef]
- Amalaradjou, M.A.; Bhunia, A.K. Modern approaches in probiotics research to control foodborne pathogens. Adv. Food Nutr. Res. 2012, 67, 185–239. [Google Scholar] [PubMed]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Nunez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, J.; Troost, F.J.; Konings, I.; Dekker, J.; Kleerebezem, M.; Brummer, R.J.; Wells, J.M. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G851–G859. [Google Scholar] [CrossRef]
- Klaenhammer, T.R.; Kleerebezem, M.; Kopp, M.V.; Rescigno, M. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 2012, 12, 728–734. [Google Scholar] [CrossRef]
- Al-Emran, H.; Moon, J.F.; Miah, M.L.; Meghla, N.S.; Reuben, R.; Uddin, M.J.; Ibnat, H.; Sarkar, S.; Roy, P.; Rahman, M.S. Genomic analysis and in vivo efficacy of Pediococcus acidilactici as a potential probiotic to prevent hyperglycemia, hypercholesterolemia and gastrointestinal infections. Sci. Rep. 2022, 12, 20429. [Google Scholar] [CrossRef]
- Jeong, D.Y.; Ryu, M.S.; Yang, H.J.; Jeong, S.Y.; Zhang, T.; Yang, H.J.; Kim, M.J.; Park, S. Pediococcus acidilactici intake decreases the clinical severity of atopic dermatitis along with increasing mucin production and improving the gut microbiome in NC/Nga mice. Biomed. Pharmacother. 2020, 129, 110488. [Google Scholar] [CrossRef]
- Cabello-Olmo, M.; Oneca, M.; Pajares, M.J.; Jimenez, M.; Ayo, J.; Encio, I.J.; Barajas, M.; Arana, M. Antidiabetic Effects of Pediococcus acidilactici pA1c on HFD-Induced Mice. Nutrients 2022, 14, 692. [Google Scholar] [CrossRef]
- Lin, J.G.; Jiang, W.P.; Tsai, Y.S.; Lin, S.W.; Chen, Y.L.; Chen, C.C.; Huang, G.J. Dietary Probiotic Pediococcus acidilactici GKA4, Dead Probiotic GKA4, and Postbiotic GKA4 Improves Cisplatin-Induced AKI by Autophagy and Endoplasmic Reticulum Stress and Organic Ion Transporters. Nutrients 2024, 16, 3532. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Du, G.; Yan, X.; Ye, H.; Guo, Q.; Wang, Z.; Yuan, Y.; Yue, T. Selenium-Enriched Pediococcus acidilactici MRS-7 Alleviates Patulin-Induced Jejunum Injuries in Mice and Its Possible Mechanisms. J. Agric. Food Chem. 2022, 70, 4755–4764. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Garg, N.; Sachdev, A.; Kumar, B. Effect of the oral intake of probiotic Pediococcus acidilactici BA28 on Helicobacter pylori causing peptic ulcer in C57BL/6 mice models. Appl. Biochem. Biotechnol. 2014, 172, 973–983. [Google Scholar] [CrossRef]
- Wu, J.-J.; Zhou, Q.-Y.; Liu, D.-M.; Xiong, J.; Liang, M.-H.; Tang, J.; Xu, Y.-Q. Evaluation of the safety and probiotic properties of Lactobacillus gasseri LGZ1029 based on whole genome analysis. LWT 2023, 184, 114759. [Google Scholar] [CrossRef]
- Wonglapsuwan, M. Genomic Insight into Pediococcus acidilactici HN9, a Potential Probiotic Strain Isolated from the Traditional Thai-Style Fermented Beef Nhang. Microorganisms 2020, 9, 50. [Google Scholar] [CrossRef]
- Xie, G.; Zhu, Y.; Zhong, Z.; Du, Q.; Wu, Y.; Xing, K.; Zhang, M.; Shu, H. Functional genomic characterization unveils probiotic features of Bacillus cereus G1-11 isolated from the gut of the hybrid grouper (Epinephelus fuscoguttatus♀ × E. lanceolatus♂). LWT 2023, 184, 115088. [Google Scholar] [CrossRef]
- Kapse, N.G.; Engineer, A.S.; Gowdaman, V.; Wagh, S.; Dhakephalkar, P.K. Functional annotation of the genome unravels probiotic potential of Bacillus coagulans HS243. Genomics 2019, 111, 921–929. [Google Scholar] [CrossRef]
- Hu, W.; Li, H.; Shi, Z.; Yang, X.; Yi, Z.; Zhou, S.; Kan, J.; Du, M. Screening and in situ inhibitory effects of probiotic lactic acid bacteria against Listeria monocytogenes. Food Ferment. Ind. 2024. [Google Scholar] [CrossRef]
- Cremon, C.; Barbaro, M.R.; Ventura, M.; Barbara, G. Pre-and probiotic overview. Curr. Opin. Pharmacol. 2018, 43, 87–92. [Google Scholar] [CrossRef]
- Cai, Z.; Guo, Y.; Zheng, Q.; Liu, Z.; Zhong, G.; Zeng, L.; Huang, M.; Pan, D.; Wu, Z. Screening of a potential probiotic Lactiplantibacillus plantarum NUC08 and its synergistic effects with yogurt starter. J. Dairy Sci. 2024, 107, 2760–2773. [Google Scholar] [CrossRef]
- Wanna, W.; Surachat, K.; Kaitimonchai, P.; Phongdara, A. Evaluation of probiotic characteristics and whole genome analysis of Pediococcus pentosaceus MR001 for use as probiotic bacteria in shrimp aquaculture. Sci. Rep. 2021, 11, 18334. [Google Scholar] [CrossRef] [PubMed]
- Pakroo, S.; Tarrah, A.; Bettin, J.; Corich, V.; Giacomini, A. Genomic and Phenotypic Evaluation of Potential Probiotic Pediococcus Strains with Hypocholesterolemic Effect Isolated from Traditional Fermented Food. Probiotics Antimicrob. Proteins 2022, 14, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.-M.; Huang, Y.-Y.; Liang, M.-H. Analysis of the probiotic characteristics and adaptability of Lactiplantibacillus plantarum DMDL 9010 to gastrointestinal environment by complete genome sequencing and corresponding phenotypes. LWT 2022, 158, 113129. [Google Scholar] [CrossRef]
- Peng, X.; Ed-Dra, A.; Yue, M. Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit. Rev. Food Sci. Nutr. 2023, 63, 11244–11262. [Google Scholar] [CrossRef]
- Jiang, Y.-H.; Liang, M.; Yang, Y.-H.; Xue, J.; Suo, H.-Y. Probiotic Lactobacillus plantarum SHY21-2 protected zebrafish against Aeromonas hydrophila infection by maintaining intestinal barrier integrity, inhibiting inflammatory and oxidative stress responses, and regulating intestinal microbiome. Aquaculture 2024, 582, 740506. [Google Scholar] [CrossRef]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef]
- Jiang, J.; Yang, B.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Comparative genomics of Pediococcus pentosaceus isolated from different niches reveals genetic diversity in carbohydrate metabolism and immune system. Front. Microbiol. 2020, 11, 253. [Google Scholar] [CrossRef]
- Zhang, T.; Pan, Y.; Li, B.; Ou, J.; Zhang, J.; Chen, Y.; Peng, X.; Chen, L. Molecular cloning and antimicrobial activity of enterolysin A and helveticin J of bacteriolysins from metagenome of Chinese traditional fermented foods. Food Control 2013, 31, 499–507. [Google Scholar] [CrossRef]
- Miljkovic, M.; Jovanovic, S.; O’Connor, P.M.; Mirkovic, N.; Jovcic, B.; Filipic, B.; Dinic, M.; Studholme, D.J.; Fira, D.; Cotter, P.D. Brevibacillus laterosporus strains BGSP7, BGSP9 and BGSP11 isolated from silage produce broad spectrum multi-antimicrobials. PLoS ONE 2019, 14, e0216773. [Google Scholar] [CrossRef]
- Zhou, C.; Chang, X.; Zou, Y.; Zhao, F.; Zhou, G.; Ye, K. The mechanism of Enterococcus faecium on the virulence of Listeria monocytogenes during the storage of fermented sausages by whole genome analysis. Int. J. Food Microbiol. 2024, 422, 110826. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13, Erratum in Food Control 2018, 88, 236. [Google Scholar] [CrossRef]
- Ashida, H.; Ogawa, M.; Kim, M.; Mimuro, H.; Sasakawa, C. Bacteria and host interactions in the gut epithelial barrier. Nat. Chem. Biol. 2012, 8, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef]
- Mengaud, J.; Ohayon, H.; Gounon, P.; Mège, R.-M.; Cossart, P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 1996, 84, 923–932. [Google Scholar] [CrossRef]
- Lecuit, M.; Vandormael-Pournin, S.; Lefort, J.; Huerre, M.; Gounon, P.; Dupuy, C.; Babinet, C.; Cossart, P. A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science 2001, 292, 1722–1725. [Google Scholar] [CrossRef]
- Disson, O.; Grayo, S.; Huillet, E.; Nikitas, G.; Langa-Vives, F.; Dussurget, O.; Ragon, M.; Le Monnier, A.; Babinet, C.; Cossart, P. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 2008, 455, 1114–1118. [Google Scholar] [CrossRef]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Chen, Y.; Zhu, X.; Wang, L.; Tian, P.; Jin, X.; Liang, M.; Chen, Z.; Zhang, T.; Qian, L. A randomised double-blind placebo-controlled trial of a probiotic combination for manipulating the gut microbiota and managing metabolic syndrome. Food Biosci. 2024, 59, 104076. [Google Scholar] [CrossRef]
- Becattini, S.; Pamer, E.G. Multifaceted defense against Listeria monocytogenes in the gastro-intestinal lumen. Pathogens 2017, 7, 1. [Google Scholar] [CrossRef]
- Qiao, Y.; Qiu, Z.; Tian, F.; Yu, L.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Effect of bacteriocin-producing Pediococcus acidilactici strains on the immune system and intestinal flora of normal mice. Food Sci. Hum. Wellness 2022, 11, 238–246. [Google Scholar] [CrossRef]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Iljazovic, A.; Roy, U.; Gálvez, E.J.; Lesker, T.R.; Zhao, B.; Gronow, A.; Amend, L.; Will, S.E.; Hofmann, J.D.; Pils, M.C. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol. 2021, 14, 113–124. [Google Scholar] [CrossRef]
- Kumar, P.; Griffen, A.; Barton, J.; Paster, B.; Moeschberger, M.; Leys, E. New bacterial species associated with chronic periodontitis. J. Dent. Res. 2003, 82, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Tett, A.; Pasolli, E.; Masetti, G.; Ercolini, D.; Segata, N. Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 2021, 19, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.-T.; Chan, L.; Chen, K.-Y.; Lee, H.-H.; Huang, L.-K.; Yang, Y.-C.S.; Liu, Y.-R.; Hu, C.-J. Rifaximin modifies gut microbiota and attenuates inflammation in Parkinson’s disease: Preclinical and clinical studies. Cells 2022, 11, 3468. [Google Scholar] [CrossRef]
- Zhou, M.; Wu, J.; Wu, L.; Sun, X.; Chen, C.; Huang, L. The utilization of N-acetylgalactosamine and its effect on the metabolism of amino acids in Erysipelotrichaceae strain. BMC Microbiol. 2024, 24, 397. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.; Yeoh, Y.K.; Li, A.Y.; Zhan, H.; Wan, Y.; Chung, A.C.; Cheung, C.P.; Chen, N. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef]
- Zhang, N.; Ju, Z.; Zuo, T. Time for food: The impact of diet on gut microbiota and human health. Nutrition 2018, 51, 80–85. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.J.; Beiko, R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Huynh, U.; Zastrow, M.L. Metallobiology of Lactobacillaceae in the gut microbiome. J. Inorg. Biochem. 2023, 238, 112023. [Google Scholar] [CrossRef] [PubMed]


| Attributes | Values |
|---|---|
| Genome Size (bp) | 2,069,896 |
| G + C Content (%) | 42.18 |
| Coding Gene Number | 2010 |
| Coding Gene Average Length (bp) | 886.43 |
| Genes Assigned to COGs | 1620 |
| rRNA | 15 |
| 5S rRNA | 5 |
| 16S rRNA | 5 |
| 23S rRNA | 5 |
| tRNA | 58 |
| sRNA | 27 |
| Plasmids | 1 |
| Gene Locus | Gene Name | Gene Function | |
|---|---|---|---|
| Universal stress family protein | ACG4EF_00285 | - | universal stress protein |
| ACG4EF_01025 | - | universal stress protein | |
| ACG4EF_01030 | - | universal stress protein | |
| ACG4EF_01595 | - | universal stress protein | |
| ACG4EF_03305 | - | universal stress protein | |
| ACG4EF_07140 | - | universal stress protein | |
| Proteases and chaperones | ACG4EF_02365 | clpP | ATP-dependent Clp endopeptidase proteolytic |
| ACG4EF_05205 | hslU | ATP-dependent protease ATPase subunit HslU | |
| ACG4EF_05210 | hslV | ATP-dependent protease subunit HslV | |
| ACG4EF_05335 | - | ATP-dependent Clp protease ATP-binding subunit | |
| ACG4EF_05700 | clpB | ATP-dependent chaperone ClpB | |
| ACG4EF_06340 | clpX | ATP-dependent Clp protease ATP-binding subunit ClpX | |
| ACG4EF_07860 | - | ATP-dependent Clp protease ATP-binding subunit | |
| Temperature stress | ACG4EF_02195 | groES | co-chaperone GroES |
| ACG4EF_02200 | groL | chaperonin GroEL | |
| ACG4EF_04750 | hrcA | heat-inducible transcriptional repressor HrcA | |
| ACG4EF_04760 | dnaK | molecular chaperone DnaK | |
| ACG4EF_04765 | dnaJ | molecular chaperone DnaJ | |
| ACG4EF_06525 | cspA | cold-shock protein | |
| ACG4EF_07950 | - | cold-shock protein | |
| Bile tolerance | ACG4EF_01095 | arcD | arginine-ornithine antiporter |
| ACG4EF_01470 | nagB | glucosamine-6-phosphate deaminase | |
| ACG4EF_08565 | pyrG | CTP synthase | |
| Acid tolerance | ACG4EF_07220 | atpC | ATP synthase subunit epsilon |
| ACG4EF_07225 | atpD | ATP synthase subunit beta | |
| ACG4EF_07230 | atpG | ATP synthase subunit gamma | |
| ACG4EF_07235 | atpA | ATP synthase subunit alpha | |
| ACG4EF_07240 | atpH | ATP synthase F1 subunit delta | |
| ACG4EF_07245 | atpF | ATP synthase subunit B | |
| ACG4EF_07250 | atpE | ATP synthase subunit C | |
| ACG4EF_07255 | atpB | ATP synthase subunit A | |
| ACG4EF_06250 | - | Na+/H+ antiporter NhaC family protein | |
| Alkaline stress | ACG4EF_09110 | amaP | alkaline shock response membrane anchor protein AmaP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; Zhou, S.; Ibrahim, A.; Li, G.; Awad, S.; Ramos-Vivas, J.; Kan, J.; Du, M. Whole Genome Analysis of Pediococcus acidilactici XJ-24 and Its Role in Preventing Listeria monocytogenes ATCC® 19115TM Infection in C57BL/6 Mice. Antibiotics 2025, 14, 323. https://doi.org/10.3390/antibiotics14030323
Hu W, Zhou S, Ibrahim A, Li G, Awad S, Ramos-Vivas J, Kan J, Du M. Whole Genome Analysis of Pediococcus acidilactici XJ-24 and Its Role in Preventing Listeria monocytogenes ATCC® 19115TM Infection in C57BL/6 Mice. Antibiotics. 2025; 14(3):323. https://doi.org/10.3390/antibiotics14030323
Chicago/Turabian StyleHu, Weizhong, Shuxin Zhou, Amel Ibrahim, Guannan Li, Sameh Awad, José Ramos-Vivas, Jianquan Kan, and Muying Du. 2025. "Whole Genome Analysis of Pediococcus acidilactici XJ-24 and Its Role in Preventing Listeria monocytogenes ATCC® 19115TM Infection in C57BL/6 Mice" Antibiotics 14, no. 3: 323. https://doi.org/10.3390/antibiotics14030323
APA StyleHu, W., Zhou, S., Ibrahim, A., Li, G., Awad, S., Ramos-Vivas, J., Kan, J., & Du, M. (2025). Whole Genome Analysis of Pediococcus acidilactici XJ-24 and Its Role in Preventing Listeria monocytogenes ATCC® 19115TM Infection in C57BL/6 Mice. Antibiotics, 14(3), 323. https://doi.org/10.3390/antibiotics14030323






