Spread of Antimicrobial Resistance by Salmonella enterica Serovar Choleraesuis between Close Domestic and Wild Environments
Abstract
:1. Introduction
2. Results
2.1. Clustering of S. Choleraesuis Isolates by PFGE-XbaI
2.2. Resistance Determinants against Clinically Relevant Antimicrobials in the S. Choleraesuis Isolates
2.3. Plasmid Content of S. Choleraesuis Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Animal Sources
4.2. Pulsed-Field Gel Electrophoresis (PFGE) Analysis
4.3. Antibiotic Susceptibility Testing
4.4. Screening for Antibiotic Resistance Genes
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khan, M.A.; Cao, L.; Riaz, A.; Li, Y.; Jiao, Q.; Liu, Z.; Wang, H.; Meng, F.; Ma, Z. The Harm of Salmonella to Pig Industry and Its Control Measures. Int. J. Appl. Agric. Sci. 2019, 5, 24. [Google Scholar]
- Gil Molino, M.; Risco Pérez, D.; Gonçalves Blanco, P.; Fernandez Llario, P.; Quesada Molina, A.; García Sánchez, A.; Cuesta Gerveno, J.M.; Gómez Gordo, L.; Martín Cano, F.E.; Pérez Martínez, R. Outbreaks of antimicrobial resistant Salmonella Choleraesuis in wild boars piglets from central-western Spain. Transbound Emerg. Dis. 2019, 66, 225–233. [Google Scholar] [CrossRef] [Green Version]
- Methner, U.; Heller, M.; Bocklisch, H. Salmonella enterica subspecies enterica serovar Choleraesuis in a wild boar population in Germany. Eur. J. Wildl. Res. 2010, 56, 493–502. [Google Scholar] [CrossRef]
- Fedorka-Cray, P.J.; Gray, J.T.; Wray, C. Salmonella infections in pigs. In Salmonella in Domestic Animals; Wray, C., Wray, A., Eds.; CABI: London, UK, 2000; pp. 191–207. [Google Scholar]
- Asai, T.; Namimatsu, T.; Osumi, T.; Kojima, A.; Harada, K.; Aoki, H.; Sameshima, T.; Takahashi, T. Molecular typing and antimicrobial resistance of Salmonella enterica subspecies enterica serovar Choleraesuis isolates from diseased pigs in Japan. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, 109–119. [Google Scholar] [CrossRef]
- Pedersen, K.; Sørensen, G.; Löfström, C.; Leekitcharoenphon, P.; Nielsen, B.; Wingstrand, A.; Aarestrup, F.M.; Hendriksen, R.S.; Baggesen, D.L. Reappearance of Salmonella serovar Choleraesuis var. Kunzendorf in Danish pig herds. Vet. Microbiol. 2015, 176, 282–291. [Google Scholar] [CrossRef] [Green Version]
- Baggesen, D.L.; Christensen, J.; Jensen, T.K.; Skov, M.; Sørensen, G.; Sørensen, V. Outbreak of salmonellosis caused by Salmonella enterica subsp. enterica serovar. choleraesuis var. Kunzendorf (S. Choleraesuis) on a Danish pig farm. Dan. Veterinærtidsskrift 2000, 83, 6–12. [Google Scholar]
- Conedera, G.; Ustulin, M.; Barco, L.; Bregoli, M.; Re, E.; Vio, D. Outbreak of atypical Salmonella Choleraesuis in wild boar in North Eastern Italy. In Trens in Game Meat Hygiene; Paulsen, P., Bauer, A.F.J.M.S., Eds.; Academic Publishers: Wageningen, The Netherlands, 2014; pp. 151–159. [Google Scholar]
- Perez, J.; Astorga, R.; Carrasco, L.; Mendez, A.; Perea, A.; Sierra, M. Outbreak of salmonellosis in farmed European wild boars (Sus scrofa ferus). Vet. Rec. 1999, 145, 464–465. [Google Scholar] [CrossRef]
- Müller, M.; Weber, A.; Tucher, R.; Naumann, L. Case report: Salmonella choleraesuis as a cause of haematogenous osteomyelitis in a wild boar (Sus scrofa). Fallbericht: Osteomyelitis bei einem wildschwein (Sus scrofa) durch salmonella choleraesuis. Tierarztl. Umsch. 2004, 59, 700–702. [Google Scholar]
- Longo, A.; Petrin, S.; Mastrorilli, E.; Tiengo, A.; Lettini, A.A.; Barco, L.; Ricci, A.; Losasso, C.; Cibin, V. Characterizing Salmonella enterica serovar Choleraesuis, var. Kunzendorf: A comparative case study. Front. Vet. Sci. 2019, 6, 316. [Google Scholar] [CrossRef] [Green Version]
- Rodrigáñez, J.; Silió, L.; Rillo, S.M. El cerdo Ibérico y su sistema de producción. Anim. Genet. Resour. Resour. Génétiques Anim. Recur. Genéticos Anim. 1993, 12, 89–96. [Google Scholar] [CrossRef]
- Rodríguez-Prieto, V.; Kukielka, D.; Martínez-López, B.; de las Heras, A.I.; Barasona, J.Á.; Gortázar, C.; Sánchez-Vizcaíno, J.M.; Vicente, J. Porcine reproductive and respiratory syndrome (PRRS) virus in wild boar and Iberian pigs in south-central Spain. Eur J. Wildl. Res. 2013, 59, 859–867. [Google Scholar] [CrossRef]
- Carrasco García de León, R. Factores de Riesgo de Transmisión de Enfermedades en Ungulados Cinegéticos del Centro y sur de España. Available online: https://ruidera.uclm.es/xmlui/handle/10578/9753 (accessed on 30 December 2016).
- Navarro-Gonzalez, N.; Mentaberre, G.; Porrero, C.M.; Serrano, E.; Mateos, A.; López-Martín, J.M.; Lavín, S.; Domínguez, L. Effect of cattle on Salmonella carriage, diversity and antimicrobial resistance in free-ranging wild boar (Sus scrofa) in northeastern Spain. PLoS ONE 2012, 7, e51614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vittecoq, M.; Godreuil, S.; Prugnolle, F.; Durand, P.; Brazier, L.; Renaud, N.; Arnal, A.; Aberkane, S.; Jean-Pierre, H.; Gauthier-Clerc, M. Antimicrobial resistance in wildlife. J. Appl. Ecol. 2016, 53, 519–529. [Google Scholar] [CrossRef] [Green Version]
- Gil Molino, M.; García Sánchez, A.; Risco Pérez, D.; Gonçalves Blanco, P.; Quesada Molina, A.; Rey Pérez, J.; Martín Cano, F.E.; Cerrato Horrillo, R.; Hermoso-de-Mendoza Salcedo, J.; Fernández Llario, P. Prevalence of Salmonella spp. in tonsils, mandibular lymph nodes and faeces of wild boar from Spain and genetic relationship between isolates. Transbound. Emerg. Dis. 2019, 66, 1218–1266. [Google Scholar] [CrossRef]
- Leekitcharoenphon, P.; Sørensen, G.; Löfström, C.; Battisti, A.; Szabo, I.; Wasyl, D.; Slowey, R.; Zhao, S.; Brisabois, A.; Kornschober, C.; et al. Cross-Border Transmission of Salmonella Choleraesuis var. Kunzendorf in European Pigs and Wild Boar: Infection, Genetics, and Evolution. Front. Microbiol. 2019, 10, 179. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.H.; Su, L.H.; Chu, C. Salmonella enterica Serotype Choleraesuis: Epidemiology, Pathogenesis, Clinical Disease, and Treatment. Clin. Microbiol. Rev. 2004, 17, 311–322. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Wu, C.; Chang, C.; Lee, H.; Lee, N.; Shih, H.; Lee, C.; Ko, N.; Wang, L.; Ko, W. Extraintestinal focal infections in adults with Salmonella enterica serotype Choleraesuis bacteremia. J. Microbiol. Immunol. Infect. 2007, 40, 240–247. [Google Scholar]
- Jean, S.; Wang, J.; Hsueh, P. Bacteremia caused by Salmonella enterica serotype Choleraesuis in Taiwan. J. Microbiol. Immunol. Infect. 2006, 39, 358. [Google Scholar]
- Chiu, C.-H.; Wu, T.-L.; Su, L.-H.; Chu, C.; Chia, J.-H.; Kuo, A.-J.; Chien, M.-S.; Lin, T.-Y. The Emergence in Taiwan of Fluoroquinolone Resistance in Salmonella enterica Serotype Choleraesuis. N. Engl. J. Med. 2002, 346, 413–419. [Google Scholar] [CrossRef]
- Ferstl, P.G.; Reinheimer, C.; Jozsa, K.; Zeuzem, S.; Kempf, V.A.; Waidmann, O.; Grammatikos, G. Severe infection with multidrug-resistant Salmonella choleraesuis in a young patient with primary sclerosing cholangitis. World J. Gastroenterol. 2017, 23, 2086. [Google Scholar] [CrossRef]
- Goldstein, C.; Lee, M.D.; Sanchez, S.; Hudson, C.; Phillips, B.; Register, B.; Grady, M.; Liebert, C.; Summers, A.O.; White, D.G. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 2001, 45, 723–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass, L.; Liebert, C.A.; Lee, M.D.; Summers, A.O.; White, D.G.; Thayer, S.G.; Maurer, J.J. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avianEscherichia coli. Antimicrob. Agents Chemother. 1999, 43, 2925–2929. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.; Chiu, C.-H.; Wu, W.-Y.; Chu, C.-H.; Liu, T.-P.; Ou, J.T. Large Drug Resistance Virulence Plasmids of Clinical Isolates of Salmonella enterica Serovar Choleraesuis. Antimicrob. Agents Chemother. 2001, 45, 2299–2303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzeng, J.-I.; Chu, C.-H.; Chen, S.-W.; Yeh, C.-M.; Chiu, C.-H.; Chiou, C.-S.; Lin, J.-H.; Chu, C. Reduction of Salmonella enterica serovar Choleraesuis carrying large virulence plasmids after the foot and mouth disease outbreak in swine in southern Taiwan, and their independent evolution in human and pig. J. Microbiol. Immunol. Infect. 2012, 45, 418–425. [Google Scholar] [CrossRef] [Green Version]
- Sirichote, P.; Hasman, H.; Pulsrikarn, C.; Schønheyder, H.C.; Samulioniené, J.; Pornruangmong, S.; Bangtrakulnonth, A.; Aarestrup, F.M.; Hendriksen, R.S. Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar Choleraesuis isolates from patients in Thailand and Denmark. J. Clin. Microbiol. 2010, 48, 883–888. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Su, L.-H.; Janapatla, R.P.; Lin, C.-Y.; Chiu, C.-H. Genetic analysis of virulence and antimicrobial-resistant plasmid pOU7519 in Salmonella enterica serovar Choleraesuis. J. Microbiol. Immunol. Infect. 2020, 53, 49–59. [Google Scholar] [CrossRef]
- Catry, B.; Cavaleri, M.; Baptiste, K.; Grave, K.; Grein, K.; Holm, A.; Jukes, H.; Liebana, E.; Navas, A.L.; Mackay, D. Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): Development of resistance in animals and possible impact on human and animal health. Int. J. Antimicrob. Agents 2015, 46, 297–306. [Google Scholar] [CrossRef]
- Darwich, L.; Vidal, A.; Seminati, C.; Albamonte, A.; Casado, A.; López, F.; Molina-López, R.A.; Migura-Garcia, L. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 2019, 14, e0210686. [Google Scholar] [CrossRef] [Green Version]
- Guenther, S.; Falgenhauer, L.; Semmler, T.; Imirzalioglu, C.; Chakraborty, T.; Roesler, U.; Roschanski, N. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 2017, 72, 1289–1292. [Google Scholar]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-mediated colistin resistance in Salmonella enterica: A review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, C.A.d.; Cunha, M.P.V.; Bertani, A.M.d.J.; de Almeida, E.A.; Gonçalves, C.R.; Sacchi, C.T.; de Paiva, J.B.; Camargo, C.H.; Tiba-Casas, M.R. Detection of multidrug-and colistin-resistant Salmonella Choleraesuis causing bloodstream infection. J. Antimicrob. Chemother. 2020, 75, 2009–2010. [Google Scholar] [CrossRef]
- Sánchez-Benito, R.; Iglesias, M.R.; Quijada, N.M.; Campos, M.J.; Ugarte-Ruiz, M.; Hernández, M.; Pazos, C.; Rodríguez-Lázaro, D.; Garduño, E.; Domínguez, L. Escherichia coli ST167 carrying plasmid mobilisable mcr-1 and blaCTX-M-15 resistance determinants isolated from a human respiratory infection. Int. J. Antimicrob. Agents 2017, 50, 285. [Google Scholar] [CrossRef] [PubMed]
- Zottola, T.; Montagnaro, S.; Magnapera, C.; Sasso, S.; De Martino, L.; Bragagnolo, A.; D’Amici, L.; Condoleo, R.; Pisanelli, G.; Iovane, G. Prevalence and antimicrobial susceptibility of Salmonella in European wild boar (Sus scrofa); Latium Region–Italy. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 161–168. [Google Scholar] [PubMed]
- Chiari, M.; Zanoni, M.; Tagliabue, S.; Lavazza, A.; Alborali, L.G. Salmonella serotypes in wild boars (Sus scrofa) hunted in northern Italy. Acta Vet. Scand. 2013, 55, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-C.; Lin, Y.-H.; Chang, C.-F.; Yeh, K.-S.; Chiu, C.-H.; Chu, C.; Chien, M.-S.; Hsu, Y.-M.; Tsai, L.-S.; Chiou, C.-S. Epidemiologic relationship between fluoroquinolone-resistant Salmonella enterica serovar Choleraesuis strains isolated from humans and pigs in Taiwan (1997 to 2002). J. Clin. Microbiol. 2005, 43, 2798–2804. [Google Scholar] [CrossRef] [Green Version]
- Donazzolo, C.; Turchetto, S.; Ustulin, M.; Citterio, C.; Conedera, G.; Vio, D.; Cocchi, M. Antimicrobial susceptibility of Salmonella enterica subsp. enterica serovar Choleraesuis strains forum wild boar (Sus scrofa) in Italy. In Game Meat Hygiene; Paulsen, P.B.A., Smulders, F.J.M., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2017; p. 307. [Google Scholar]
- Hsu, Y.-M.; Tang, C.-Y.; Lin, H.; Chen, Y.-H.; Chen, Y.-L.; Su, Y.-H.; Chen, D.S.; Lin, J.-H.; Chang, C.-C. Comparative study of class 1 integron, ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline (ACSSuT) and fluoroquinolone resistance in various Salmonella serovars from humans and animals. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 9–16. [Google Scholar] [CrossRef]
- Chiu, C.-H.; Su, L.-H. Salmonella, Non-Typhoidal Species (S. choleraesuis, S. enteritidis, S. hadar, S. typhimurium). Available online: http://www.antimicrobe.org/b258.asp (accessed on 28 October 2020).
- Su, L.H.; Chiu, C.H.; Chu, C.; Ou, J.T. Antimicrobial resistance in nontyphoid Salmonella serotypes: A global challenge. Clin. Infect. Dis. 2004, 39, 546–551. [Google Scholar] [CrossRef] [Green Version]
- Prescott, J.F. Sulfonamides, Diaminopyrimidines, and Their Combinations. In Antimicrobial Therapy in Veterinary Medicine, 5th ed.; Giguère, S., Prescott, J.F., Dowling, P.M., Eds.; John Wiley & Sons, Inc.: Ames, IA, USA, 2013. [Google Scholar]
- Nature, E. The antibiotic alarm. Nature 2013, 495, 141. [Google Scholar]
- Sato, T.; Okubo, T.; Usui, M.; Yokota, S.-I.; Izumiyama, S.; Tamura, Y. Association of Veterinary Third-Generation Cephalosporin Use with the Risk of Emergence of Extended-Spectrum-Cephalosporin Resistance in Escherichia coli from Dairy Cattle in Japan. PLoS ONE 2014, 9, e96101. [Google Scholar] [CrossRef] [Green Version]
- Olesen, S.W.; Barnett, M.L.; MacFadden, D.R.; Brownstein, J.S.; Hernández-Díaz, S.; Lipsitch, M.; Grad, Y.H. The distribution of antibiotic use and its association with antibiotic resistance. Elife 2018, 7, e39435. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gonzalez, N.; Casas-Díaz, E.; Porrero, C.M.; Mateos, A.; Domínguez, L.; Lavín, S.; Serrano, E. Food-borne zoonotic pathogens and antimicrobial resistance of indicator bacteria in urban wild boars in Barcelona, Spain. Vet. Microbiol. 2013, 167, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gonzalez, N.; Castillo-Contreras, R.; Casas-Díaz, E.; Morellet, N.; Porrero, M.C.; Molina-Vacas, G.; Torres, R.T.; Fonseca, C.; Mentaberre, G.; Domínguez, L. Carriage of antibiotic-resistant bacteria in urban versus rural wild boars. Eur. J. Wildl. Res. 2018, 64, 60. [Google Scholar] [CrossRef]
- Fluit, A.; Schmitz, F.J. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 2004, 10, 272–288. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.-F.; Chen, Y.-H.; Peng, C.-F. Molecular characterisation of class 1 integrons in Salmonella enterica serovar Choleraesuis isolates from southern Taiwan. Int. J. Antimicrob. Agents 2009, 33, 216–222. [Google Scholar] [CrossRef]
- Sköld, O. Resistance to trimethoprim and sulfonamides. Vet. Res. 2001, 32, 261–273. [Google Scholar] [CrossRef] [Green Version]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef] [Green Version]
- Chiu, T.H.; Pang, J.C.; Hwang, W.Z.; Tsen, H.Y. Development of PCR primers for the detection of Salmonella enterica serovar Choleraesuis based on the fliC gene. J. Food Prot. 2005, 68, 1575–1580. [Google Scholar] [CrossRef]
- Ribot, E.M.; Fair, M.; Gautom, R.; Cameron, D.; Hunter, S.; Swaminathan, B.; Barrett, T.J. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7, Salmonella, and Shigella for PulseNet. Foodbourne Pathog. Dis. 2006, 3, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Briñas, L.; Zarazaga, M.; Sáenz, Y.; Ruiz-Larrea, F.; Torres, C. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob. Agents Chemother. 2002, 46, 3156–3163. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Gao, S.; Jiao, X.; Liu, X.F. Prevalence of serogroups and virulence factors of Escherichia coli strains isolated from pigs with postweaning diarrhoea in eastern China. Vet. Microbiol. 2004, 103, 13–20. [Google Scholar] [CrossRef]
- Sengeløv, G.; Agersø, Y.; Halling-Sørensen, B.; Baloda, S.B.; Andersen, J.S.; Jensen, L.B. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 2003, 28, 587–595. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Lertworapreecha, M.; Evans, M.C.; Bangtrakulnonth, A.; Chalermchaikit, T.; Hendriksen, R.S.; Wegener, H.C. Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J. Antimicrob. Chemother. 2003, 52, 715–718. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Bangtrakulnonth, A.; Pulsrikarn, C.; Pornreongwong, S.; Hasman, H.; Song, S.W.; Aarestrup, F.M. Antimicrobial resistance and molecular epidemiology of Salmonella Rissen from animals, food products, and patients in Thailand and Denmark. Foodborne Pathog. Dis. 2008, 5, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, M.; Peighambari, S.M.; Svendsen, C.A.; Cavaco, L.M.; Agersø, Y.; Hendriksen, R.S. Molecular clonality and antimicrobial resistance in Salmonella enterica serovars Enteritidis and Infantis from broilers in three Northern regions of Iran. BMC Vet. Res. 2013, 9, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Leverstein-van Hall, M.; Paauw, A.; Box, A.; Blok, H.; Verhoef, J.; Fluit, A. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Microbiol. 2002, 40, 3038–3040. [Google Scholar] [CrossRef] [Green Version]
- Levesque, C.; Piche, L.; Larose, C.; Roy, P.H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 1995, 39, 185–191. [Google Scholar] [CrossRef] [Green Version]
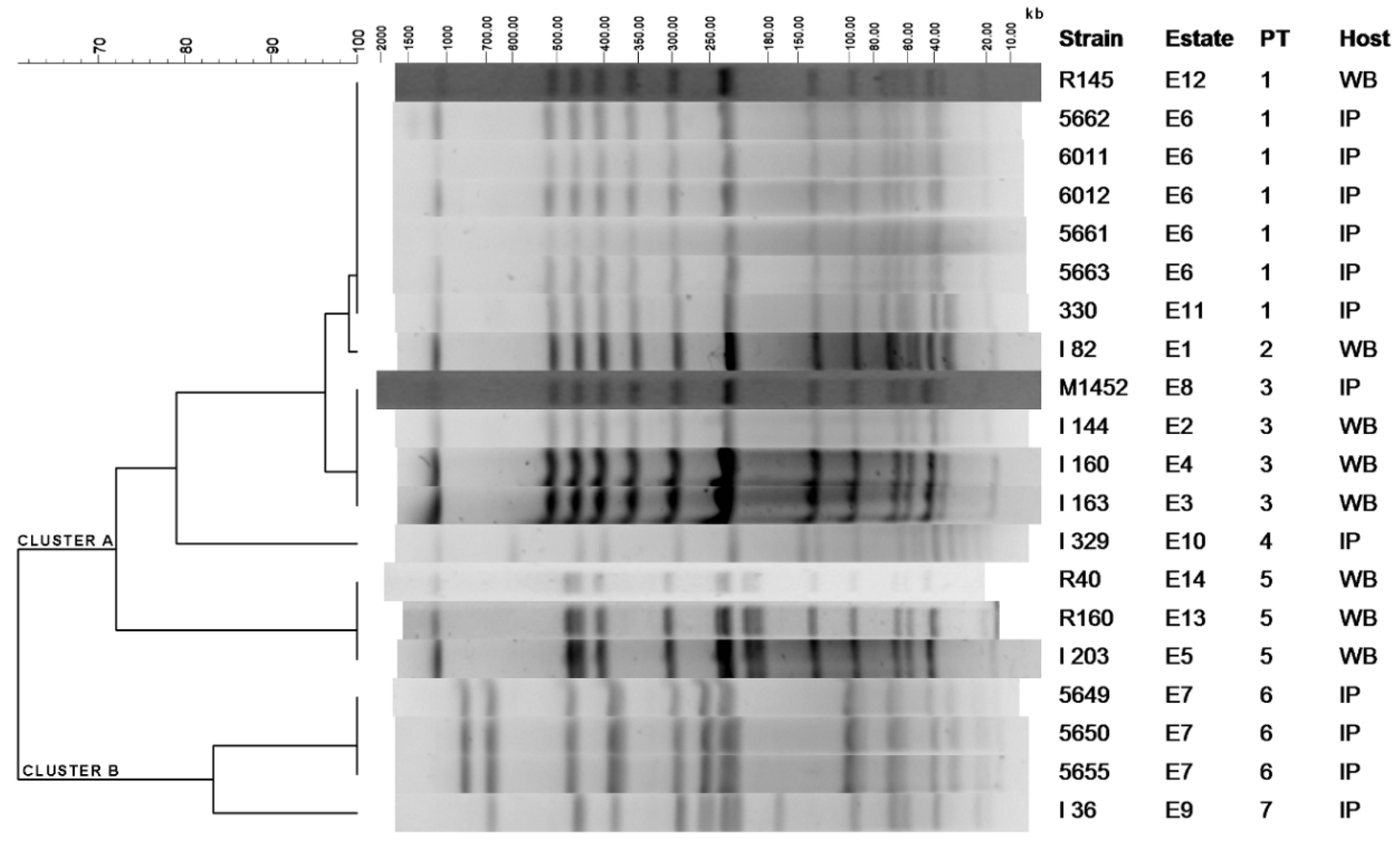
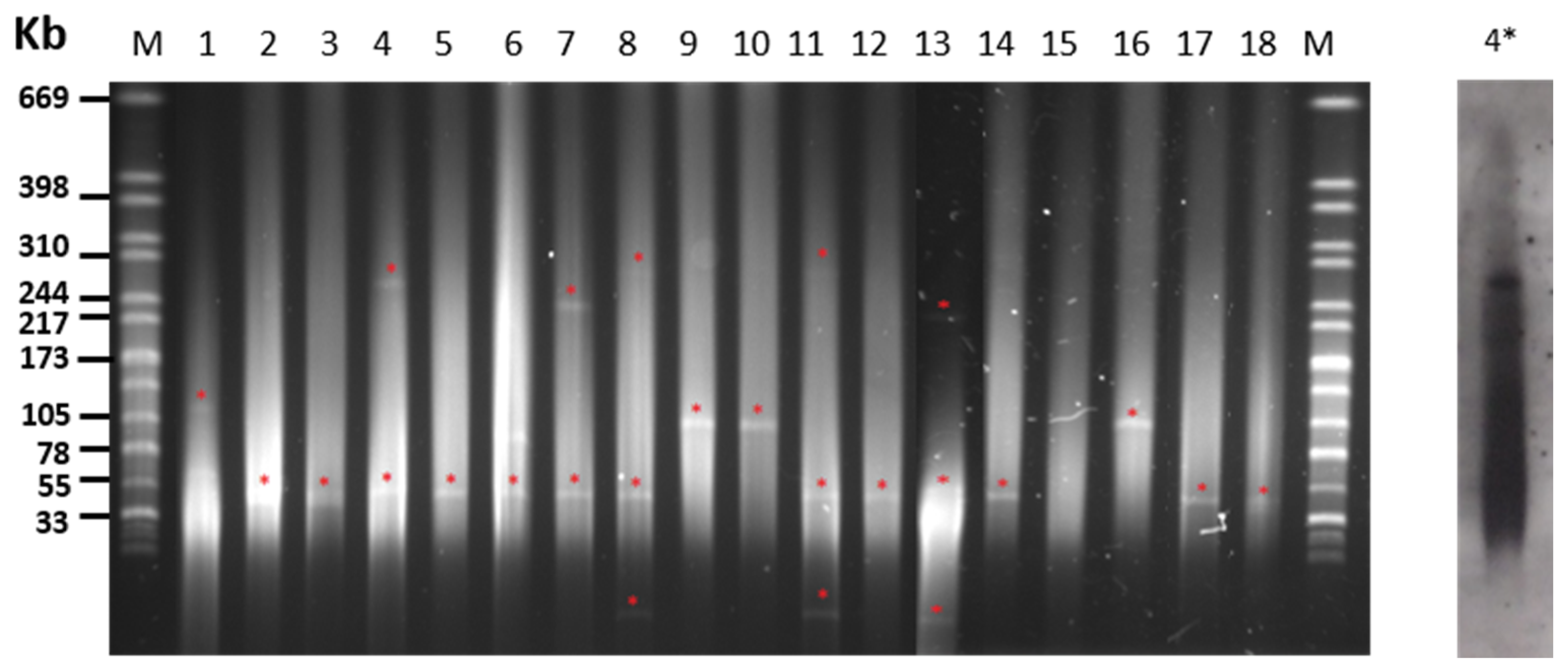

| PT 1 | Isolate | Origin | Resistance Phenotype | Resistance Genotype | Plasmid Size (kb) 2 |
|---|---|---|---|---|---|
| 1 | R145 | WB | AMP–STR–TET–TRS–SUL | blaTEM–aadA1–sul1–sul3–tetA | >105 |
| 5662 | IP | AMP–DOX–TRS–SUL–CHL | blaTEM–aadA1–sul3–Int1 | 55 | |
| 6011 | IP | AMP–STR–TRS–SUL | blaTEM | 55 | |
| 6012 | IP | AMP–STR | blaTEM | ND | |
| 5661 | IP | AMP–DOX | blaTEM | 55 | |
| 5663 | IP | AMP–STR–TRS–SUL | blaTEM | ND | |
| 330 | IP | AMP–GEN–NEO–STR–TET–DOX–TRS–SUL–COL | strA–strB–sul1–mcr–1 | 55 +> 244 3 | |
| 2 | I 82 | WB | AMP–NEO–TET–DOX | blaTEM | 55 |
| 3 | M1452 | IP | AMP–NEO–STR–TET–DOX–TRS–SUL–CHL | blaTEM–tetA–Int1(aadA1)4 | 55 |
| I 144 | WB | - | - | 55 | |
| I 160 | WB | NEO | - | 55 | |
| I 163 | WB | NEO | - | 55 | |
| 4 | I 329 | IP | AMP–STR–TET–DOX–TRS–SUL | blaTEM–strA–strB–sul1–Int1–(blaPSE1)4 | 55 + 244 |
| 5 | R40 | WB | STR–TET–DOX–SUL | aadA1–strA–strB–sul1–tetA | <33 + 55 + 310 |
| R160 | WB | STR–TET–SUL | strA–strB–tetA | <33 + 55 + 240 | |
| I 203 | WB | STR–TRS–SUL | strA–strB–tetA | <33 + 55 + 310 | |
| 6 | 5649 | IP | AMP–TRS–SUL–CHL | blaTEM–aadA1–sul3–Int1 | 105 |
| 5650 | IP | AMP–TRS–SUL–CHL | blaTEM–aadA1–sul3–Int1 | 105 | |
| 5655 | IP | AMP–TRS–SUL–CHL | strA–sul3 | 105 | |
| 7 | I 36 | IP | AMP–NEO–STR–TET–DOX–SUL | aadA1–strA–strB–sul2–tetB | - |
| Antimicrobials | IP | WB | |||
|---|---|---|---|---|---|
| N 1 | Genes 2 | N 1 | Genes 2 | ||
| Sulfonamides | Sulfadiazine | 10 | sul1 (2), sul2 (1), sul3 (4) | 4 | sul1 (2), sul3 (1) |
| Cotrimoxazol | 9 | 2 | |||
| β-lactams | Ampicillin | 12 | blaTEM (9), blaPSE (1) | 2 | blaTEM (2) |
| Aminoglycosides | Gentamycin | 1 | - | 0 | - |
| Neomycin | 3 | - | 3 | - | |
| Streptomycin | 7 | aadA (5), strA (3), strB (3) | 4 | aadA (2), strA (3), strB (3) | |
| Tetracyclines | Tetracycline | 4 | tetA (1), tetB (1) | 4 | tetA (4) |
| Doxycycline | 6 | 2 | |||
| Phenicols | Chloramphenicol | 5 | 0 | - | |
| Polymixins | Colistin | 1 | mcr-1 (1) | 0 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil Molino, M.; García, A.; Zurita, S.G.; Martín-Cano, F.E.; García-Jiménez, W.; Risco, D.; Rey, J.; Fernández-Llario, P.; Quesada, A. Spread of Antimicrobial Resistance by Salmonella enterica Serovar Choleraesuis between Close Domestic and Wild Environments. Antibiotics 2020, 9, 750. https://doi.org/10.3390/antibiotics9110750
Gil Molino M, García A, Zurita SG, Martín-Cano FE, García-Jiménez W, Risco D, Rey J, Fernández-Llario P, Quesada A. Spread of Antimicrobial Resistance by Salmonella enterica Serovar Choleraesuis between Close Domestic and Wild Environments. Antibiotics. 2020; 9(11):750. https://doi.org/10.3390/antibiotics9110750
Chicago/Turabian StyleGil Molino, María, Alfredo García, Sofía Gabriela Zurita, Francisco Eduardo Martín-Cano, Waldo García-Jiménez, David Risco, Joaquín Rey, Pedro Fernández-Llario, and Alberto Quesada. 2020. "Spread of Antimicrobial Resistance by Salmonella enterica Serovar Choleraesuis between Close Domestic and Wild Environments" Antibiotics 9, no. 11: 750. https://doi.org/10.3390/antibiotics9110750
APA StyleGil Molino, M., García, A., Zurita, S. G., Martín-Cano, F. E., García-Jiménez, W., Risco, D., Rey, J., Fernández-Llario, P., & Quesada, A. (2020). Spread of Antimicrobial Resistance by Salmonella enterica Serovar Choleraesuis between Close Domestic and Wild Environments. Antibiotics, 9(11), 750. https://doi.org/10.3390/antibiotics9110750





