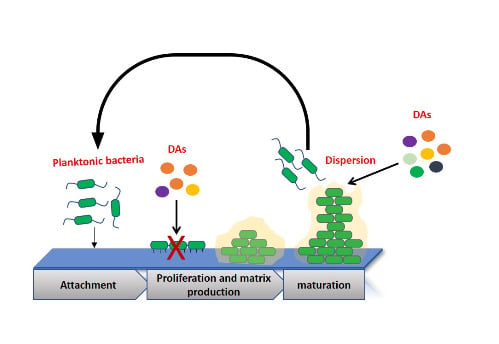
Inhibition of Campylobacter jejuni Biofilm Formation by D-Amino Acids
Abstract
:1. Introduction
2. Results
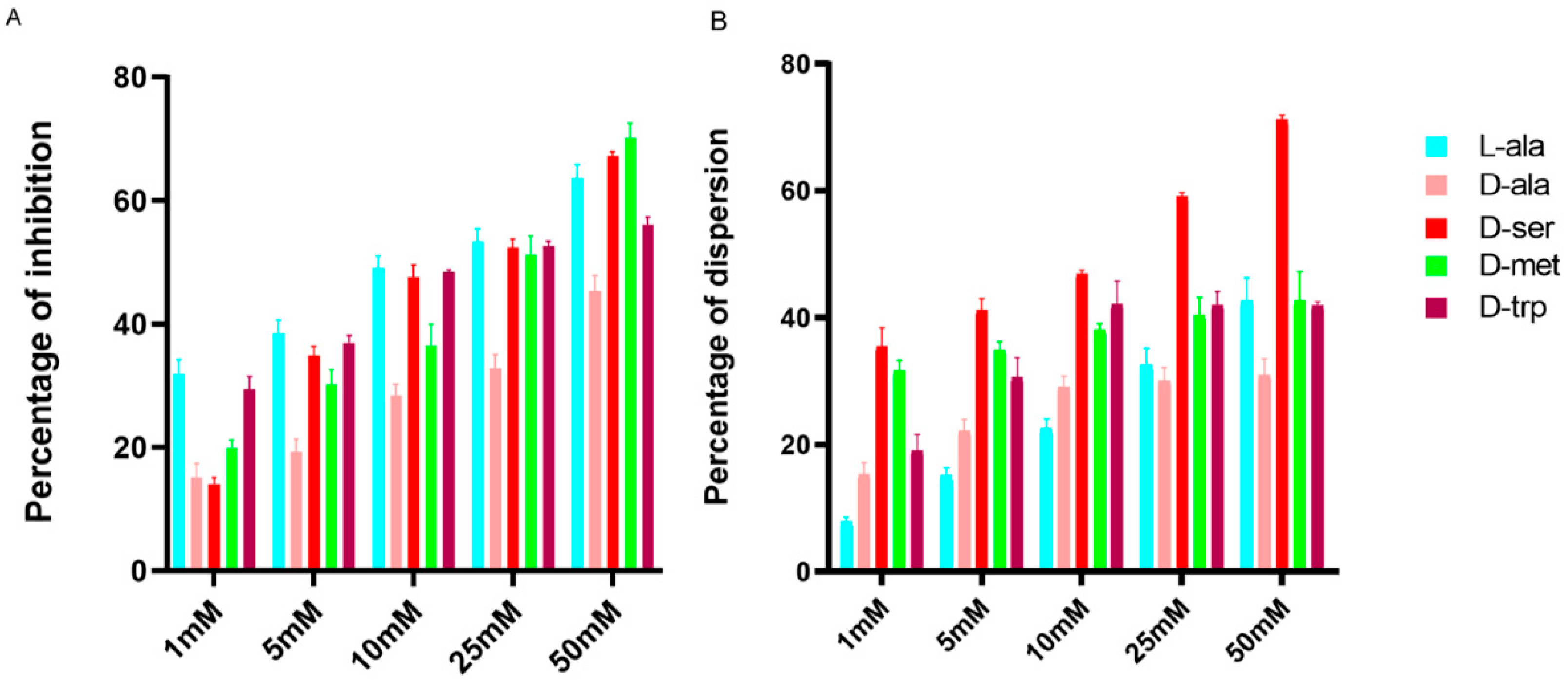
2.1. Effect of LAs and DAs on Biofilm Formation by C. jejuni
2.2. Effect of DAs on Biofilm Formation by Different Campylobacter Strains
2.3. Effect of the Equimolar Mixture of DAs and LAs on C. jejuni 11168-O Biofilm
2.4. Microscopic Characterization of the Dispersion Effect of DAs on Biofilm
2.5. Expression Level of alr and ddlA in the Presence of LAs and DAs
2.6. D-Ala Can Reverse the Inhibitory Effect of DAs and DCS
3. Discussion
4. Materials and Methods
4.1. C. jejuni Strains and Growth Conditions
4.2. Chemical and Reagents Used in this Study
4.3. Biofilm Formation and Dispersion Assays
4.4. RNA Extraction, cDNA Synthesis and RT-qPCR of Alanine Racemase (alr), D-alanine-D-alanine Ligase (ddlA)
4.5. Confocal Laser Scanning Microscopy
4.6. Staining of C. jejuni Cells
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Grantham-McGregor, S.; Cheung, Y.B.; Cueto, S.; Glewwe, P.; Richter, L.; Strupp, B. Developmental potential in the first 5 years for children in developing countries. Lancet 2007, 369, 60–70. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. The Global View of Campylobacteriosis: Report of an Expert Consultation. Utrecht, Netherlands, 9–11 July 2012. Available online: https://apps.who.int/iris/handle/10665/80751 (accessed on 20 November 2020).
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Futur. Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iovine, N.M. Resistance mechanisms in Campylobacter jejuni. Virulence 2013, 4, 230–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engberg, J. Quinolone and Macrolide Resistance in Campylobacter jejuni and Campylobacter coli: Resistance Mechanisms and Trends in Human Isolates. Emerg. Infect. Dis. 2001, 7, 24–34. [Google Scholar] [CrossRef]
- Bae, J.; Oh, E.; Jeon, B. Enhanced transmission of antibiotic resistance in Campylobacter jejuni biofilms by natural transformation. Antimicrob. Agents Chemother. 2014, 58, 7573–7575. [Google Scholar] [CrossRef] [Green Version]
- Ica, T.; Caner, V.; Istanbullu, O.; Nguyen, H.D.; Ahmed, B.; Call, D.R.; Beyenal, H. Characterization of Mono- and Mixed-Culture Campylobacter jejuni biofilms. Appl. Environ. Microbiol. 2011, 78, 1033–1038. [Google Scholar] [CrossRef] [Green Version]
- Zimmer, M.; Barnhart, H.; Idris, U.; Lee, M.D. Detection of Campylobacter jejuni strains in the water lines of a commercial broiler house and their relationship to the strains that colonized the chickens. Avian Dis. 2003, 47, 101–107. [Google Scholar] [CrossRef]
- Joshua, G.W.P.; Guthrie-Irons, C.; Karlyshev, A.V.; Wren, B.W. Biofilm formation in Campylobacter jejuni. Microbiology 2006, 152, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Bronowski, C.; James, C.E.; Winstanley, C. Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol. Lett. 2014, 356, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.E.; Barton, M.D.; Blair, I.S.; Corcoran, D.; Dooley, J.S.; Fanning, S.; Kempf, I.; Lastovica, A.J.; Lowery, C.J.; Matsuda, M.; et al. The epidemiology of antibiotic resistance in Campylobacter. Microbes Infect. 2006, 8, 1955–1966. [Google Scholar] [CrossRef]
- Smith, J.L.; Fratamico, P.M. Fluoroquinolone resistance in Campylobacter. J. Food Prot. 2010, 73, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization, WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 20 November 2020).
- Alfredson, D.A.; Korolik, V. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol. Lett. 2007, 277, 123–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miflin, J.K.; Templeton, J.M.; Blackall, P.J. Antibiotic resistance in Campylobacter jejuni and Campylobacter coli isolated from poultry in the South-East Queensland region. J. Antimicrob. Chemother. 2007, 59, 775–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tambur, Z.; Miljkovic-Selimovic, B.; Bokonjic, D. Determination of sensitivity to antibiotics of Campilobacter jejuni and Campilobacter coli isolated from human feces. Vojn. Pregl. 2009, 66, 49–52. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. 2019, 8, 1–10. [Google Scholar] [CrossRef]
- Kaplan, J. Biofilm Dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [Green Version]
- Sauer, K.; Cullen, M.C.; Rickard, A.H.; Zeef, L.A.H.; Davies, D.G.; Gilbert, P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004, 186, 7312–7326. [Google Scholar] [CrossRef] [Green Version]
- Rumbaugh, K.P.; Ahmad, I. Antibiofilm Agents: From Diagnosis to Treatment and Prevention, 1st ed.; Rumbaugh, K.P., Ahmad, I., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 8. [Google Scholar]
- Nahar, S.; Mizan, F.R.; Ha, A.J.-W.; Ha, S.-D. Advances and future prospects of enzyme-based biofilm prevention approaches in the food industry. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1484–1502. [Google Scholar] [CrossRef] [Green Version]
- Brandenburg, K.S.; Rodriguez, K.J.; McAnulty, J.F.; Murphy, C.J.; Abbott, N.L.; Schurr, M.J.; Czuprynski, C.J. Tryptophan inhibits biofilm formation by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 1921–1925. [Google Scholar] [CrossRef] [Green Version]
- Vlamakis, H.; Chai, Y.; Beauregard, P.B.; Losick, R.; Kolter, R. Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Genet. 2013, 11, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Leiman, S.A.; May, J.M.; Lebar, M.D.; Kahne, D.; Kolter, R.; Losick, R. D-Amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J. Bacteriol. 2013, 195, 5391–5395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Liu, Y. Reduced microbial attachment by d-amino acid-inhibited AI-2 and EPS production. Water Res. 2011, 45, 5796–5804. [Google Scholar] [CrossRef] [PubMed]
- Zilm, P.; Butnejski, V.; Rossi-Fedele, G.; Kidd, S.P.; Edwards, S.; Vasilev, K. D-amino acids reduce Enterococcus faecalis biofilms in vitro and in the presence of antimicrobials used for root canal treatment. PLoS ONE 2017, 12, e0170670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliashkevich, A.; Alvarez, L.; Cava, F. New Insights into the mechanisms and biological roles of D-Amino Acids in complex eco-systems. Front. Microbiol. 2018, 9, 683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azua, I.; Goiriena, I.; Baña, Z.; Iriberri, J.; Unanue, M. Release and consumption of d-amino acids during growth of marine prokaryotes. Microb. Ecol. 2013, 67, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rendueles, O.; Ghigo, J.-M. Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol. Rev. 2012, 36, 972–989. [Google Scholar] [CrossRef]
- Lam, H.; Oh, D.-C.; Cava, F.; Takacs, C.N.; Clardy, J.; de Pedro, M.A.; Waldor, M.K. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 2009, 325, 1552–1555. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic Era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, Z.; Jiao, N. Effects of d-Amino Acids on the EPS production and cell aggregation of Alteromonas macleodii Strain JL2069. Curr. Microbiol. 2014, 68, 751–755. [Google Scholar] [CrossRef]
- Cava, F.; Lam, H.; De Pedro, M.A.; Waldor, M.K. Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cell. Mol. Life Sci. 2011, 68, 817–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramón-Peréz, M.L.; Diaz-Cedillo, F.; Ibarra, J.A.; Torales-Cardeña, A.; Rodríguez-Martínez, S.; Jan-Roblero, J.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. D-Amino acids inhibit biofilm formation in Staphylococcus epidermidis strains from ocular infections. J. Med. Microbiol. 2014, 63, 1369–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochbaum, A.I.; Kolodkin-Gal, I.; Foulston, L.; Kolter, R.; Aizenberg, J.; Losick, R. Inhibitory Effects of D-amino acids on Staphylococcus aureus biofilm development. J. Bacteriol. 2011, 193, 5616–5622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lark, C.; Lark, K.G. Studies on the mechanism by which d-amino acids block cell wall synthesis. Biochim. Biophys. Acta. 1961, 49, 308–322. [Google Scholar] [CrossRef]
- Moulder, J.W.; Novosel, D.L.; Officer, J.E. Inhibition of the growth of agents of the psittacosis group by d-cycloserine and its specific reversal by d-alanine. J. Bacteriol. 1963, 85, 707–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awasthy, D.; Bharath, S.; Subbulakshmi, V.; Sharma, U. Alanine racemase mutants of Mycobacterium tuberculosis require d-alanine for growth and are defective for survival in macrophages and mice. Microbiology 2012, 158, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Samie, A.; Ramalivhana, J.; Igumbor, E.; Obi, C. Prevalence, haemolytic and haemagglutination activities and antibiotic susceptibility profiles of Campylobacter spp. isolated from human diarrhoeal stools in vhembe district, South Africa. J. Health Popul. Nutr. 2007, 25, 406–413. [Google Scholar]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. D-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef] [Green Version]
- Van Der Hooft, J.J.J.; Alghefari, W.; Watson, E.; Everest, P.; Morton, F.R.; Burgess, K.E.V.; Smith, D.G.E. Unexpected differential metabolic responses of Campylobacter jejuni to the abundant presence of glutamate and fucose. Metabolomics 2018, 14, 144. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, C.J., Jr.; Akers, K.S.; Romano, D.R.; Woodbury, R.L.; Hardy, S.K.; Murray, C.K.; Wenke, J.C. D-amino acids enhance the activity of antimicrobials against biofilms of clinical wound isolates of Staphylococcus aureus and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 4353–4361. [Google Scholar] [CrossRef] [Green Version]
- Wijsman, H.J.W. The characterization of an alanine racemase mutant of Escherichia coli. Genet. Res. 1972, 20, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Sidiq, K.R.; Chow, M.W.; Zhao, Z.; Daniel, R.A. Alanine metabolism in Bacillus subtilis. Mol. Microbiol. 2020, 562850. [Google Scholar] [CrossRef] [PubMed]
- Turonova, H.; Neu, T.R.; Ulbrich, P.; Pazlarova, J.; Tresse, O. The biofilm matrix of Campylobacter jejuni determined by fluorescence lectin-binding analysis. Biofouling 2016, 32, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Cava, F.; de Pedro, M.A.; Lam, H.; Davis, B.M.; Waldor, M.K. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011, 30, 3442–3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, R.K.; Jiang, T.; Kwon, Y.M. Essential genome of Campylobacter jejuni. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Lara-Tejero, M.; Lefebre, M.; Goodman, A.L.; Galán, J.E. Novel components of the flagellar system in epsilonproteobacteria. mBio 2014, 5, e01349-14. [Google Scholar] [CrossRef] [Green Version]
- Chacon, O.; Bermudez, L.E.; Zinniel, D.K.; Chahal, H.K.; Fenton, R.J.; Feng, Z.; Hanford, K.; Adams, L.G.; Barletta, R.G. Impairment of d-alanine biosynthesis in Mycobacterium smegmatis determines decreased intracellular survival in human macrophages. Microbiology 2009, 155, 1440–1450. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Zheng, X.; Wei, Y.; Zhou, X.; Zhang, K.; Wang, S.; Cheng, L.; Li, Y.; Ren, B.; Xu, X.; et al. D-Alanine metabolism is essential for growth and biofilm formation of Streptococcus mutans. Mol. Oral Microbiol. 2016, 31, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Qiu, W.; Zhou, X.-D.; Zheng, X.; Zhang, K.-K.; Wang, S.-D.; Li, Y.-Q.; Cheng, L.; Li, J.-Y.; Xu, X.; et al. Alanine racemase is essential for the growth and interspecies competitiveness of Streptococcus mutans. Int. J. Oral Sci. 2016, 8, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Batson, S.; Rea, D.; Fülöp, V.; Roper, D.I. Crystallization and preliminary X-ray analysis of a D-alanyl-D-alanine ligase (EcDdlB) from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 405–408. [Google Scholar] [CrossRef] [Green Version]
- Halouska, S.; Fenton, R.J.; Zinniel, D.K.; Marshall, D.D.; Barletta, R.G.; Powers, R. Metabolomics Analysis Identifiesd-Alanine-d-Alanine Ligase as the Primary Lethal Target of d-Cycloserine in Mycobacteria. J. Proteome Res. 2014, 13, 1065–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Aart, L. Lizah T.; Lemmens, N.; Van Wamel, W.J.; Van Wezel, G.P. Substrate inhibition of VanA by d-Alanine reduces vancomycin resistance in a VanX-dependent manner. Antimicrob. Agents Chemother. 2016, 60, 4930–4939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Healy, V.L.; Lessard, I.A.; Roper, D.I.; Knox, J.R.; Walsh, C.T. Vancomycin resistance in enterococci: Reprogramming of the d-Ala–d-Ala ligases in bacterial peptidoglycan biosynthesis. Chem. Biol. 2000, 7, R109–R119. [Google Scholar] [CrossRef] [Green Version]
- A Lessard, I.; Healy, V.L.; Park, I.S.; Walsh, C.T. Determinants for differential effects on D-Ala-D-lactate vs D-Ala-D-Ala formation by the VanA ligase from vancomycin-resistant enterococci. Biochemistry 1999, 38, 14006–14022. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, F.; Depardieu, F.; Bourdon, N.; Fines-Guyon, M.; Berger, P.; Camiade, S.; Leclercq, R.; Courvalin, P.; Cattoir, V. D-Ala-d-Ser VanN-Type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 2011, 55, 4606–4612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tram, G.; Korolik, V.; Day, C.J. MBDS Solvent: An Improved method for assessment of biofilms. Adv. Microbiol. 2013, 3, 200–204. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Patil, A.; Prabhune, A.; Goel, G. Inhibition of quorum-sensing-mediated biofilm formation in Cronobacter sakazakii strains. Microbiology 2016, 162, 1708–1714. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Wu, Y.; Elena, G.; Zhong, K.; Gao, H. Insight into the effect of quinic acid on biofilm formed by Staphylococcus aureus. RSC Adv. 2019, 9, 3938–3945. [Google Scholar] [CrossRef] [Green Version]
- Day, C.J.; E Hartley, L.; Shewell, L.K.; King, R.M.; Tram, G.; Day, S.K.; Semchenko, E.A.; Korolik, V. Variation of chemosensory receptor content of Campylobacter jejuni strains and modulation of receptor gene expression under different in vivo and in vitro growth conditions. BMC Microbiol. 2012, 12, 128. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Schefe, J.H.; Lehmann, K.E.; Buschmann, I.R.; Unger, T.; Funke-Kaiser, H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “gene expression’s C T difference” formula. J. Mol. Med. 2006, 84, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khurana, R.; Coleman, C.; Ionescu-Zanetti, C.; Carter, S.A.; Krishna, V.; Grover, R.K.; Roy, R.; Singh, S. Mechanism of thioflavin-T binding to amyloid fibrils. J. Struct. Biol. 2005, 151, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Biancalana, M.; Koide, S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Fold Change | |||||||
|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | ||||||
| Gene Name | D-ala | D-ser | D-met | L-ala | D-ser | D-trp | DCS |
| alr | 38 ± 7 | - | - | 4.18 ± 0.3 | 2.92 ± 0.2 | 1.65 ± 0.3 | 2.85 ± 0.2 |
| ddlA | 10 ± 2 | 2.58 ± 0.6 | - | 1.25 ± 0.1 | - | 3.42 ± 0.4 | 7.15 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elgamoudi, B.A.; Taha, T.; Korolik, V. Inhibition of Campylobacter jejuni Biofilm Formation by D-Amino Acids. Antibiotics 2020, 9, 836. https://doi.org/10.3390/antibiotics9110836
Elgamoudi BA, Taha T, Korolik V. Inhibition of Campylobacter jejuni Biofilm Formation by D-Amino Acids. Antibiotics. 2020; 9(11):836. https://doi.org/10.3390/antibiotics9110836
Chicago/Turabian StyleElgamoudi, Bassam A., Taha Taha, and Victoria Korolik. 2020. "Inhibition of Campylobacter jejuni Biofilm Formation by D-Amino Acids" Antibiotics 9, no. 11: 836. https://doi.org/10.3390/antibiotics9110836
APA StyleElgamoudi, B. A., Taha, T., & Korolik, V. (2020). Inhibition of Campylobacter jejuni Biofilm Formation by D-Amino Acids. Antibiotics, 9(11), 836. https://doi.org/10.3390/antibiotics9110836