Tissue Specificity in Social Context-Dependent lysozyme Expression in Bumblebees
Abstract
:1. Introduction
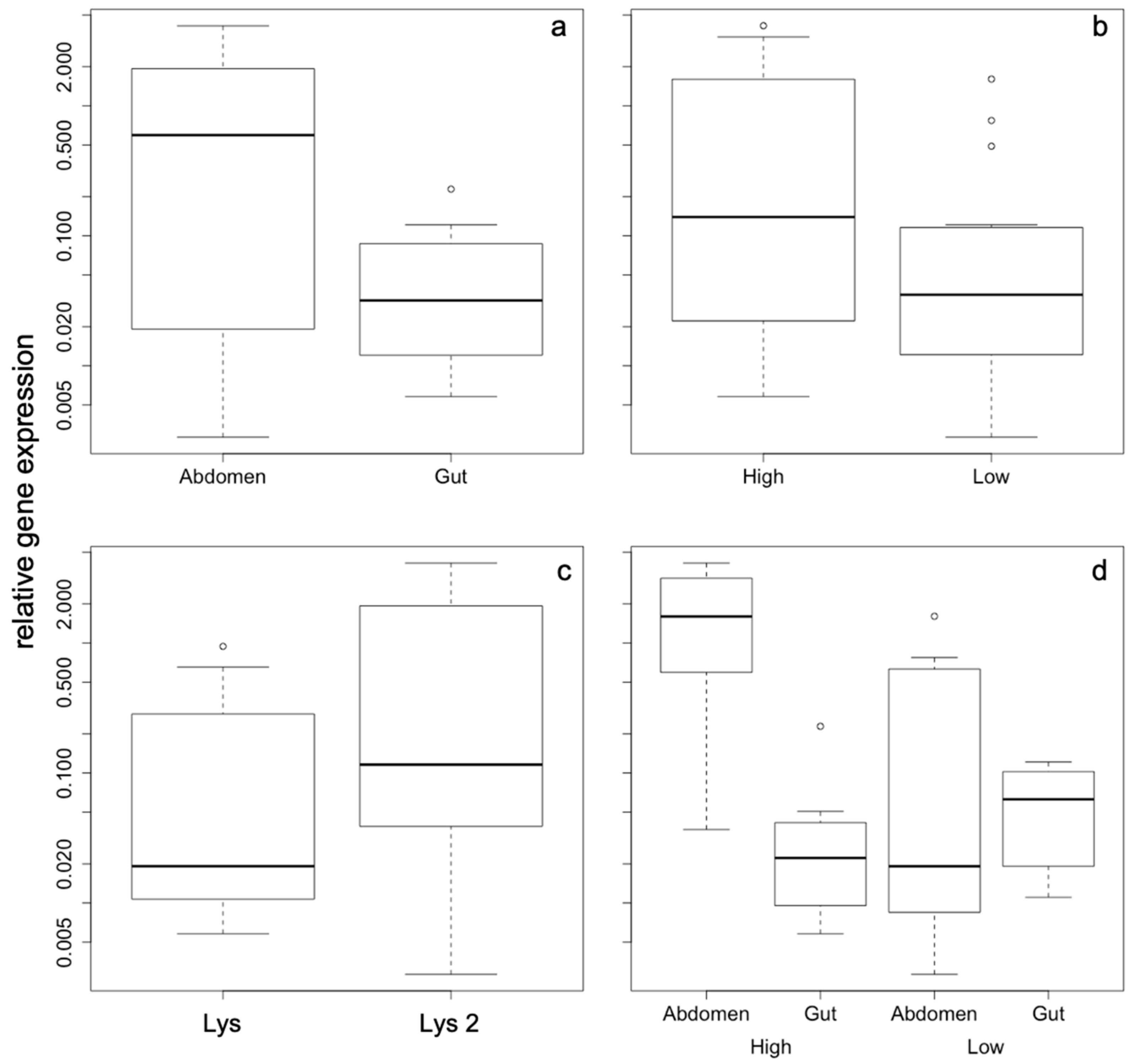
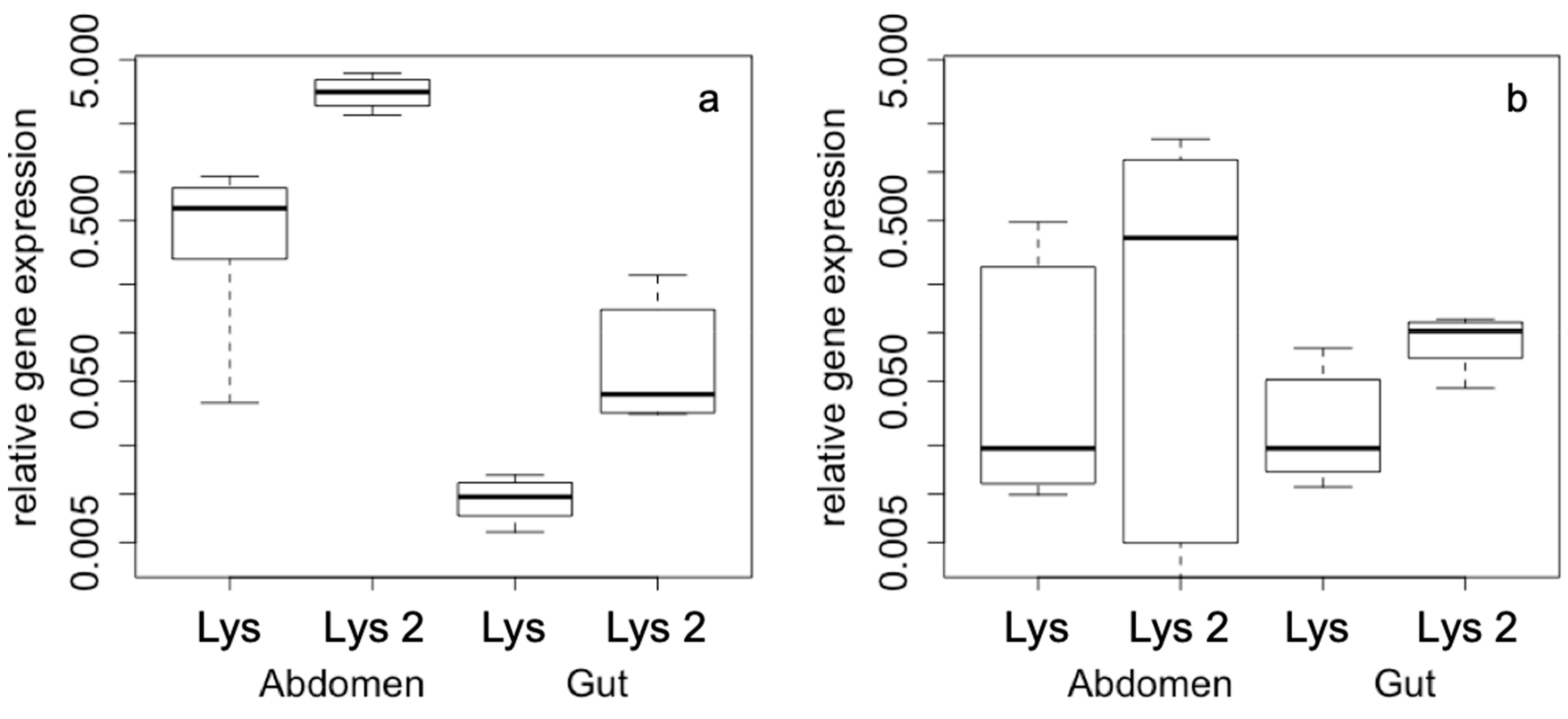
2. Results
3. Discussion
4. Materials and Methods
4.1. Bee Samples and Experimental Manipulation
4.2. RNA Extraction and Gene Expression Analysis
4.3. Data Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boomsma, J.J.; Schmid-Hempel, P.; Hughes, W.O.H. Life histories and parasite pressure across the major groups of social insects. In Insect Evolutionary Ecology; Fellowes, M.D.E., Holloway, G.J., Rolff, J., Eds.; CABI Publishing: Oxon, UK, 2005; pp. 139–175. [Google Scholar]
- Reber, A.; Castella, G.; Christe, P.; Chapuisat, M. Experimentally increased group diversity improves disease resistance in an ant species. Ecol. Lett. 2008, 11, 682–689. [Google Scholar] [CrossRef] [Green Version]
- Schmid-Hempel, P. Parasites in Social Insects; Princeton University Press: Princeton, NJ, USA, 1998. [Google Scholar]
- Cremer, S.; Armitage, S.A.O.; Schmid-Hempel, P. Social immunity. Curr. Biol. 2007, 17, 693–702. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barribeau, S.M.; Sadd, B.M.; Du Plessis, L.; Brown, M.J.F.; Buechel, S.D.; Cappelle, K.; Carolan, J.C.; Christiaens, O.; Colgan, T.J.; Erler, S.; et al. A depauperate immune repertoire precedes evolution of sociality in bees. Genome Biol. 2015, 16, 83. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-González, M.X.; Moret, Y.; Brown, M.J.F. Rapid induction of immune density-dependent prophylaxis in adult social insects. Biol. Lett. 2009, 5, 781–783. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.; Helbing, S.; Erler, S.; Lattorff, H.M.G. Social context dependent immune gene expression in bumblebees (Bombus terrestris). Behav. Ecol. Sociobiol. 2012, 66, 791–796. [Google Scholar] [CrossRef]
- Viljakainen, L.; Jurvansuu, J.; Holmberg, I.; Pamminger, T.; Erler, S.; Cremer, S. Social environment affects the transcriptomic response to bacteria in ant queens. Ecol. Evol. 2018, 8, 11031–11070. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A.; Reichhart, J.M. Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 2002, 3, 121–126. [Google Scholar] [CrossRef]
- Wink, D.A.; Hines, H.B.; Cheng, R.Y.; Switzer, C.H.; Flores-Santana, W.; Vitek, M.P.; Ridnour, L.A.; Colton, C.A. Nitric oxide and redox mechanisms in the immune response. J. Leukoc. Biol. 2011, 89, 873–891. [Google Scholar] [CrossRef] [Green Version]
- Tzou, P.; Ohresser, S.; Ferrandon, D.; Capovilla, M.; Reichhart, J.M.; Lemaitre, B.; Hoffmann, J.A.; Imler, J.L. Tissue-Specific Inducible Expression of Antimicrobial Peptide Genes in Drosophila Surface Epithelia. Immunity 2000, 13, 737–748. [Google Scholar] [CrossRef] [Green Version]
- Fujita, A. Lysozymes in insects: What role do they play in nitrogen metabolism? Physiol. Entomol. 2004, 29, 305–310. [Google Scholar] [CrossRef]
- Lattorff, H.M.G. Increased stress levels in caged honey bee (Apis mellifera) (Hymenoptera: Apidae) workers. J. Apic. Res. 2020. under review. [Google Scholar]
- Stow, A.; Briscoe, D.; Gillings, M.; Holley, M.; Smith, S.; Leys, R.; Silberbauer, T.; Turnbull, C.; Beattie, A. Antimicrobial defences increase with sociality in bees. Biol. Lett. 2007, 3, 422–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbull, C.; Hoggard, S.; Gillings, M.; Palmer, C.; Stow, A.; Beattie, D.; Briscoe, D.; Smith, S.; Wilson, P.; Beattie, A. Antimicrobial strength increases with group size: Implications for social evolution. Biol. Lett. 2011, 7, 249–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viljakainen, L.; Evans, J.D.; Hasselmann, M.; Rueppell, O.; Tingek, S.; Pamilo, P. Rapid evolution of immune proteins in social insects. Mol. Biol. Evol. 2009, 26, 1791–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erler, S.; Lhomme, P.; Rasmont, P.; Lattorff, H.M.G. Rapid evolution of antimicrobial peptide genes in an insect host-social parasite system. Infect. Genet. Evol. 2014, 23, 129–137. [Google Scholar] [CrossRef]
- Fouks, B.; Lattorff, H.M.G. Contrasting evolutionary rates between social and parasitic bumblebees for three social effect genes. Front. Ecol. Evol. 2016, 4, 64. [Google Scholar] [CrossRef] [Green Version]
- Helbing, S.; Lattorff, H.M.G. Patterns of molecular evolution of RNAi genes in social and socially parasitic bumblebees. Infect. Genet. Evol. 2016, 42, 53–59. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Erler, S.; Popp, M.; Lattorff, H.M.G. Dynamics of Immune System Gene Expression upon Bacterial Challenge and Wounding in a Social Insect (Bombus terrestris). PLoS ONE 2011, 6, e18126. [Google Scholar] [CrossRef]
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Bartoń, K. MuMIn: Multi-Model Inference. R Package Version 1.43.6. 2019. Available online: https://CRAN.R-project.org/package=MuMIn (accessed on 22 December 2019).
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
| Factor | Estimate | S.E. | Adj. S.E. | z Value | Pr (>|z|) |
|---|---|---|---|---|---|
| (intercept) | 0 | 0 | 0 | NA | NA |
| Density | −0.696 | 0.233 | 0.244 | 2.850 | 0.0044 |
| Gene | 0.440 | 0.227 | 0.237 | 1.858 | 0.0632 |
| Tissue | −0.905 | 0.210 | 0.221 | 4.093 | 4.2 × 10−5 |
| Density × Tissue | 0.755 | 0.211 | 0.221 | 3.412 | 0.0006 |
| Group size | 0.162 | 0.165 | 0.173 | 0.937 | 0.3488 |
| Density × Gene | −0.214 | 0.210 | 0.221 | 0.969 | 0.3327 |
| Gene × Group size | −0.231 | 0.392 | 0.412 | 0.560 | 0.5752 |
| Gene × Tissue | 0.025 | 0.212 | 0.223 | 0.110 | 0.9124 |
| Density × Group size | 0.076 | 0.438 | 0.460 | 0.166 | 0.8685 |
| Group size × Tissue | 0.068 | 0.401 | 0.422 | 0.162 | 0.8716 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lattorff, H.M.G. Tissue Specificity in Social Context-Dependent lysozyme Expression in Bumblebees. Antibiotics 2020, 9, 130. https://doi.org/10.3390/antibiotics9030130
Lattorff HMG. Tissue Specificity in Social Context-Dependent lysozyme Expression in Bumblebees. Antibiotics. 2020; 9(3):130. https://doi.org/10.3390/antibiotics9030130
Chicago/Turabian StyleLattorff, H. Michael G. 2020. "Tissue Specificity in Social Context-Dependent lysozyme Expression in Bumblebees" Antibiotics 9, no. 3: 130. https://doi.org/10.3390/antibiotics9030130
APA StyleLattorff, H. M. G. (2020). Tissue Specificity in Social Context-Dependent lysozyme Expression in Bumblebees. Antibiotics, 9(3), 130. https://doi.org/10.3390/antibiotics9030130





