Abstract
Bioweathering crusts are widely distributed on the limestone sculptures of the Longmen Grottoes, a UNESCO World Heritage Site. However, information on microbial communities in this archaeological site is missing. To fill this gap, we identified bacterial communities involved in the formation of bioweathering crusts. The composition analysis showed that Cyanobacteria, Acidobacteria, Chloroflexi, Actinobacteria, Firmicutes, and Planctomycetota are the major components of the bacterial communities in the crusts. Cyanobacteria might be one of the major contributors to the formation of the crusts. A culture-based analysis indicated the presence of bacterial isolates (e.g., Enterobacter sp. and Bacillus sp.) with a potential role in bioprotection. Moreover, five types of biogenic organic acids were detected in the crusts, implying a consequence of organic acid corrosion. Scanning electron microscopy analysis further confirmed microbial penetration into the stone monuments with a biomechanical attack. Our reports provide a microbiological reference for heritage conservators to treat bioweathering crust.
1. Introduction
The Longmen Grottoes is a famous UNESCO World Heritage Site, which is located aside the Yi River in the south of the ancient capital of Luoyang, Henan province, China (Figure 1). The archeological site comprises more than 2300 caves and niches that are carved into steep limestone cliffs over a 1 km long stretch. Inside these caves and niches are about 110,000 Buddhist stone statues, more than 60 stupas, and 2800 inscriptions, which can be dated from the late Northern Wei Dynasty (the end of the 5th century) and early Tang Dynasty (the mid-8th century). The sculptures, particularly the giant sculptures in the Fengxiansi Cave, are the most representative of the Royal Cave Temples, making great contributions to the development of sculptural art over the world.

Figure 1.
Geographical location of the Longmen Grottoes.
However, long-term exposure to the open environment makes the sculptures in this site suffer from severe weathering crusts widely spreading over the surfaces of the monuments and cliffs (Figure 1). The formation of weathering crusts on historical monuments is a result of both abiotic and biotic attacks responsive to distinct pollutants [1,2]. Epilithic microorganisms have been widely reported to deteriorate the materials of stone monuments and historical buildings [3,4,5], though there are always abiotic impacts [6]. Microorganisms of different domains, including Archaea, Bacteria, and Eukarya, can grow in the form of biofilms on stone monuments. In the presence of atmospheric pollution, they cooperate to drive the biogeochemical cycles of carbon, nitrogen, and sulfur, giving rise to both biochemical and biomechanical damage to the stone materials [7,8,9]. Some recent studies suggest that the microbial population colonizing the limestone monuments might partially be attributed to the deposition of airborne biological particles on the stone surfaces where S-, N-, and P-rich pollutants from the atmosphere provide nutrients and energy for bacteria communities, thus indirectly facilitating biofilm formation, the growth of bioweathering crusts, and biodeterioration of limestone [10]. Recently, it has been found that stress-resistant fungi and dark organotrophic bacteria are the main inhabitants in superficial crusts on the stone monuments in Saint Petersburg, suggesting that these lithobiotic communities are the core microbiome responsible for crust formation [11]. However, the causes of bioweathering crusts in the Longmen Grottoes site have not yet been studied. Information on microbial communities colonizing the sculptures at this site is missing, largely limiting the development of the conservation of these stone monuments.
To figure out the microbial profiles in this archaeological site, we attempted to investigate the typical bioweathering crusts on the limestone sculptures of the site. Both next-genome sequencing and culture-based techniques will be employed to identify the bacterial biodeteriogens that might result in bioweathering crusts. Furthermore, biogenic organic acids will be examined to unveil whether or not bioweathering crusts might be a consequence of the production of organic acids. We expected that our findings would appeal for more attention on microbial attacks to stone cultural heritage, especially for the formation of bioweathering crusts.
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
In August 2020, six samples (S1–S6) of bioweathering crusts with different morphologies were taken on the wall of the different grottoes (G437, G439, G332, G404, G419, and G1280, respectively) in the Longmen Grottoes site. Specifically, about 10 g of crusts that were similar to the crusts on the sculpture were generally collected from the area around the corresponding sculpture (not from the sculpture surface). Samples were then packaged with the ice bag and immediately delivered to the laboratory for analysis. Approximately 3 g of the crusts was fully mixed into 10 mL of sterile water to prepare the suspension for isolation. Bacterial isolation was performed in two different agar media: BG11 for phototrophic Cyanobacteria and M9 for heterotrophic bacteria. Bacterial isolates were identified functionally and biochemically via the GeoChip 5.0 technique [12].
2.2. Total DNA Extraction
To guarantee the homogeneity of samples, about 10 g of the crusts was fully suspended into the distilled water. Then, the suspension was centrifuged at 10,000× g for 15 min and the pellets were completely recovered for total DNA extraction using the PowerSoil® DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA), strictly following the manufacturer’s protocol. After isolation, the quality of total DNA extracts was measured using NanoDrop™ ND-2000 spectrophotometer (Thermo Scientific, Inc., Waltham, MA, USA). The DNA samples of good quality were packed with ice bags and quickly delivered to the third-party company for high throughput sequencing.
2.3. High Throughput Sequencing and Taxonomic Analysis
The universal primers (338F, 5′-ACTCCTACG GGAGGC AGC A-3′; 806R, 5′-GGACTACHV GGG TWTCTA AT-3′) were used to amplify the V3–V4 fragment of the bacterial 16S rRNA genes. The unique molecular barcode was fused to the 5′-end of the forward primer for polymerase chain reaction (PCR)-free libraries, which were sequenced through Illumina HiSeq 2500 (PE250) by MAGIGENE (Guangzhou, China).
Raw data with up to 80,000 reads for each sample were imported into the QIIME 2 software for quality checking following the standard procedure [13]. Specifically, raw sequences were first treated with the software packages of DADA2 [14] and Deblur [15] to denoise the invalid reads. Thereafter, the remaining reads were clustered into operational taxonomic units (OTUs) at the threshold of 97% identity. Taxonomic assignment for each OTU was performed referring to the SILVA Release 138. The OTU table was reused for community composition analyses at the level of both phylum and genus. The sequence data were deposited with the reference BioProject PRJNA917797.
2.4. Scanning Electron Microscopy (SEM)
A piece of 1 mm thick crust was selected as the specimen for SEM analysis. Briefly, the specimen was first dried at 50 °C for 2 h and subsequently fixed onto a two-sided conductive adhesive carbon tape by spray-gold coating for 60 s. The fixed specimen was observed under a scanning electron microscope (JEOL JSM-6610LV, JEOL, Ltd., Tokyo, Japan). The operating conditions were as follows: 15 KV accelerating voltage, 25 mm electro focusing, 1 Na current probe, and 60–300 s spectral collection. To guarantee the best view of the specimen and have an entire observation of the specific parts (e.g., hyphal or filamentous cells) of each sample, we tried to take SEM images with different magnifications.
2.5. High-Performance Liquid Chromatography (HPLC)
In total, 1 g of the crusts of each sample was fully mixed with 5 mL of distilled water before being centrifuged at 10,000× g. Then, the supernatant was filtered through 0.2 μm membrane filters and degassed under vacuum before the HPLC analysis. The analysis of HPLC for organic acids in this study referred to the method of Bevilacqua and Califano (1989) [16]. Briefly, oxalic, lactic, citric, succinic, and fumaric acids of HPLC grade (Sigma Chemical Co., St. Louis, MO, USA) were used as standards. A high-performance liquid chromatograph (Alliance HPLC, Waters Associates Inc., Milford, MA, USA) was equipped with a 2998 PDA detector. The wavelength of the detector was set at 214 nm (for other organic acids except for lactic acid) or 285 nm (for lactic acid). The mobile phase was a solution of 0.5% w/v (NH4)2HPO4 and 0.4% v/v acetonitrile (pH 2.24 with H3PO4). The flow rate was set at 1.5 mL/min. A Thermo ScientificTM Hypersil GOLDTM C8 chromatographic column (Thermo Scientific, Inc., Waltham, MA, USA) was used. All chemicals used in the mobile phase were of HPLC grade.
3. Results and Discussion
3.1. Bacterial Community Compositions
To find out what bacteria give rise to bioweathering crusts on the limestone sculptures of the Longmen Grottoes, we investigated the bacterial community compositions of the six samples. Generally, the bacterial communities mainly include Cyanobacteria, Acidobacteria, Chloroflexi, Actinobacteria, Firmicutes, and Planctomycetota (Figure 2A). Among them, Cyanobacteria were predominant over the six samples, with a relevant abundance of over 20%. Interestingly, Acidobacteria were detected over the six samples, especially for Samples 2 and 6 with a relative abundance of over 30%. Moreover, similar patterns were observed in Chloroflexi, which accounted for over 30% of the bacterial communities in Samples 3 and 6. Additionally, Actinobacteria were observed to share 10% of the bacterial communities over the samples. At the genus level (Figure 2B), Bacillus (15%) and Pseudomonas (10%) were the most dominant community in the six samples, followed by Acinetobacter (8%). However, there were still 20%–30% of the communities that were unclassified. This might be attributed to the incomplete taxonomy of bacteria, especially for Cyanobacteria [17].

Figure 2.
Composition of bacterial communities in the six samples. (A) Bacterial composition at the level of phylum. The phylum with a relative abundance less than 1% is defined as “Minority”. (B) Bacterial composition of the top 20 abundant genera.
Early in 1991, studies have demenstrated that the biofilm of Cyanobacteria and algae grows on stones, spontaneously detaches, and gives rise to the removal of grains from the stone surface, thus causing mechanical deterioration on the colonized materials [18]. Furthermore, Cyanobacteria have been found to dominate in the black crusts on limestone historic baluartes (forts) and the cathedral in Campeche, Yucatan peninsula, Mexico [19]. These filamentous cells penetrated into readily detached surface layers, leading to the formation of biogenic black crusts by a differential heating and water retention effect. In this study, we observed the predominance of Cyanobacteria in the community of each sample, suggesting their important contributions to the formation of bioweathering crusts. Simultaneously, Cyanobacteria require only light and water to live endolithically and can survive extreme stresses [20], and thus they may become even more important as biodeteriogens of stone in the future [21]. If this is true, research priority should be given to control of the growth of Cyanobacteria on stone monuments.
3.2. Culture-Based Identification
To compare with the high-throughput sequencing data, we tried to isolate bacterial isolates from the six samples. As a result, only ten bacterial isolates were obtained, with four isolates identified on the species level and the other five identified on the genus level (Table 1). Among them, Cyanobacteriales sp., Enterobacter sp., Bacillus cereus, Bacillus wiedmannii, and Bacillus thuringiensis were detected over the six samples. Interestingly, a species of Nitrososphaera was present in all the samples (except S4), indicating the potential nitrification of biogeochemical cycle of N as a result of stone biodeterioration [22,23,24].

Table 1.
Culture-based bacteria isolates in the samples.
Recent studies have demonstrated that some Bacillus spp. isolated from stone monuments can precipitate CaCO3, suggesting a new bio-consolidation protocol for sustainable conservation [25,26]. Here, we isolated three Bacillus spp. in the crusts, implying the biocalcification potential of these bacterial communities to the sculptures. However, further work on their interaction with the limestone of the Longmen Grottos is still required to validate the inference. Importantly, some Enterobacter spp. have been demonstrated to kill microalgae by inhibiting their antioxidase activities [27,28]. Here, Enterobacter sp. was observed over the six samples, indicating a potential role as biocontrol agents for microalgae in the communities.
3.3. Analysis of Biogenic Organic Acids
Biogenic acids have been reported to corrode the minerals of the stone monuments. To unveil the potential patterns of bioweathering, we examined the biogenic acids, though no inorganic acids were detected (data not shown). As a result, five organic acids were found in our samples (Table 2). In the S1, oxalic acid, lactic acid, and fumaric acid were present. Citric acid, succinic acid, and fumaric acid were observed in the S2. Lactic acid, citric acid, and fumaric acid were detected in the S3. Both oxalic acid and lactic acid were examined in the S4, whereas only oxalic acid was found in the S5 and only lactic acid in the S6. Lactic acid was present in four of the six samples, followed by oxalic acids present in three samples. Citric acid and fumaric acid were detected both in S2 and S3. Generally speaking, the macro-environmental conditions and bacterial community compositions might be considered identical, especially when the stone sculptures are exposed to the same opening environment. If possible, the difference in the identified acids for each sample could be attributed to the micro-environment, which might affect the expression of metabolic functions of the entire community [4].

Table 2.
Determination of organic acids in the samples.
Limestone is mainly composed of carbonate masses and thus is more susceptive to biogenic acids [29]. For example, organic acids, oxalic acid in particular, can solubilize stone calcium carbonate, giving rise to the formation of oxalate films as part of the bioweathering crusts [30]. Here, five types of organic acids were detected in the bioweathering crusts, indicating biogenic organic acids as one of the important reasons for the formation of crusts.
3.4. Penetration of Biodeteriogens
To validate the attack of biodeteriogens on the sculptures, we carried out the SEM analysis for the samples under different magnifications (Figure 3). It was obvious that bacterial cells penetrated into the crusts of our samples. For S1 and S3, the crusts were almost dropped from the surface and the SEM analysis showed that hyphae were penetrated in the crusts intensively. This was consistent with the compositions of bacterial communities in S1 and S3, where hyphal or filamentous bacteria (e.g., Cyanobacteria and Actinobacteriota) were very abundant (Figure 2A). Despite this, further identification should be performed to determine whether or not fungal hyphae are widely present in such samples. For S4 and S5, there seemed to be some Cyanobacteria that grew into the crusts. However, the crusts of S2 and S6 almost developed into efflorescence, and they showed tinier particles of the mineral structure, compared with others. Such SEM analyses provided direct evidence for microbial colonization of the crusts, proving the biomechanical attack through the penetration of microbial cells into the stone materials [10].
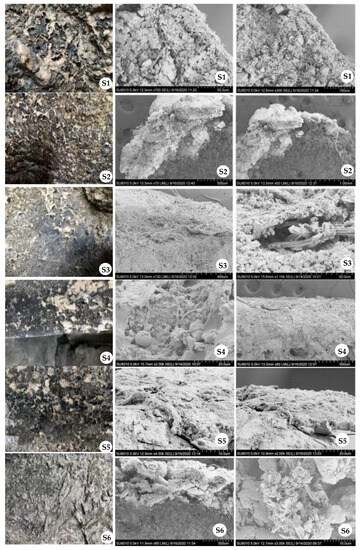
Figure 3.
SEM analysis of the six samples. Images in the first row are the on-site photos where the samples were taken. SEM images for each sample with different magnification times were displayed in the last two rows. The first-row images were made without any magnification.
4. Conclusions
In this study, we revealed that Cyanobacteria, Acidobacteria, Chloroflexi, Actinobacteria, Firmicutes, and Planctomycetota are the major colonizers in bioweathering crusts on the limestone sculptures in the Longmen Grottoes site. Furthermore, the culture-based analysis indicated the presence of bacteria with some of them having a potential role of bioprotection or biodeterioration. The formation of the crusts might be a consequence of the corrosion of biogenic organic acids or attributed to the biomechanical attack through microbial penetration into the stone monuments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/coatings13091506/s1, Figure S1: Liquid chromatogram of the standard substances of oxalic acid (a), lactic acid (b), citric acid (c), succinic acid (d), fumaric acid (e), and (f) of mixture of the four standard substances; Figure S2: Liquid chromatogram of the solution of Sample 1 where oxalic acid, lactic acid and succinic acid were detected; Figure S3: Liquid chromatogram of the solution of Sample 2 where citric acid, succinic acid and fumaric acid were detected; Figure S4: Liquid chromatogram of the solution of Sample 3 where lactic acid, citric acid and fumaric acid were detected; Figure S5: Liquid chromatogram of the solution of Sample 4 where oxalic acid and lactic acid were detected; Figure S6: Liquid chromatogram of the solution of Sample 5 where oxalic acid was detected; Figure S7: Liquid chromatogram of the solution of Sample 6 where citric acid was detected.
Author Contributions
Conceptualization, C.M., Z.F., X.L. (Xinjian Li) and X.L. (Xiaobo Liu); writing—original draft preparation, C.M. and X.L. (Xiaobo Liu); writing—review and editing, C.M., Z.F. and X.L. (Xiaobo Liu). All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Nos. 32370105 and 32100101) and the Fundamental Research Funds for the Central Universities (Grant Nos. 30922010305 and 1225011021289).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Maravelaki-Kalaitzaki, P.; Biscontin, G. Origin, characteristics and morphology of weathering crusts on Istria stone in Venice. Atmos. Environ. 1999, 33, 1699–1709. [Google Scholar] [CrossRef]
- Wilhelm, K.; Longman, J.; Orr, S.A.; Viles, H. Stone-built heritage as a proxy archive for long-term historical air quality: A study of weathering crusts on three generations of stone sculptures on Broad Street, Oxford. Sci. Total Environ. 2021, 759, 143916. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Koestler, R.J.; Warscheid, T.; Katayama, Y.; Gu, J.-D. Microbial deterioration and sustainable conservation of stone monuments and buildings. Nat. Sustain. 2020, 3, 991–1004. [Google Scholar] [CrossRef]
- Meng, S.; Qian, Y.; Liu, X.; Wang, Y.; Wu, F.; Wang, W.; Gu, J.-D. Community structures and biodeterioration processes of epilithic biofilms imply the significance of micro-environments. Sci. Total Environ. 2023, 876, 162665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Gong, C.; Gu, J.; Katayama, Y.; Someya, T.; Gu, J.-D. Biochemical reactions and mechanisms involved in the biodeterioration of stone world cultural heritage under the tropical climate conditions. Int. Biodeterior. Biodegrad. 2019, 143, 104723. [Google Scholar] [CrossRef]
- Liu, X.; Qian, Y.; Wu, F.; Wang, Y.; Wang, W.; Gu, J.-D. Biofilms on stone monuments: Biodeterioration or bioprotection? Trends Microbiol. 2022, 30, 816–819. [Google Scholar] [CrossRef]
- Warscheid, T.; Braams, J. Biodeterioration of stone: A review. Int. Biodeterior. Biodegrad. 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Negi, A.; Sarethy, I.P. Microbial biodeterioration of cultural heritage: Events, colonization, and analyses. Microb. Ecol. 2019, 78, 1014–1029. [Google Scholar] [CrossRef]
- Gaylarde, C.; Little, B. Biodeterioration of stone and metal—Fundamental microbial cycling processes with spatial and temporal scale differences. Sci. Total Environ. 2022, 823, 153193. [Google Scholar] [CrossRef]
- Ding, Y.; Salvador, C.S.C.; Caldeira, A.T.; Angelini, E.; Schiavon, N. Biodegradation and microbial contamination of limestone surfaces: An experimental study from Batalha Monastery, Portugal. Corros. Mater. Degrad. 2021, 2, 31–45. [Google Scholar] [CrossRef]
- Sazanova, K.V.; Zelenskaya, M.S.; Vlasov, A.D.; Bobir, S.Y.; Yakkonen, K.L.; Vlasov, D.Y. Microorganisms in Superficial Deposits on the Stone Monuments in Saint Petersburg. Microorganisms 2022, 10, 316. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Gentry, T.J.; Schadt, C.W.; Wu, L.; Liebich, J.; Chong, S.C.; Huang, Z.; Wu, W.; Gu, B.; Jardine, P.; et al. GeoChip: A comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 2007, 1, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Xu, Z.Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2017, 2, e00116–e00191. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, A.E.; Califano, A.N. Determination of organic acids in dairy products by High Performance Liquid Chromatography. J. Food Sci. 1989, 54, 1076. [Google Scholar] [CrossRef]
- Mishra, D.; Saraf, A.; Kumar, N.; Pal, S.; Singh, P. Issues in cyanobacterial taxonomy: Comprehensive case study of unbranched, false branched and true branched heterocytous cyanobacteria. FEMS Microbiol. Lett. 2021, 368, fnab005. [Google Scholar] [CrossRef]
- Ortega-Calvo, J.J.; Hernandez-Marine, M.; Saiz-Jimenez, C. Biodeterioration of building materials by cyanobacteria and algae. Int. Biodeterior. Biodegrad. 1991, 28, 165–185. [Google Scholar] [CrossRef]
- Gaylarde, C.C.; Ortega-Morales, B.O.; Bartolo-Pérez, P. Biogenic black crusts on buildings in unpolluted environments. Curr. Microbiol. 2007, 54, 162–166. [Google Scholar] [CrossRef]
- Mondal, A.; Mandal, S.; Rath, J. Seasonal diversity of cyanobacteria and new report of Brasilonema sp. colonizing the monuments of Santiniketan and Bishnupur (India). Int. Biodeterior. Biodegrad. 2022, 167, 105350. [Google Scholar] [CrossRef]
- Gaylarde, C.C. Influence of environment on microbial colonization of historic stone buildings with emphasis on Cyanobacteria. Heritage 2020, 3, 1469–1482. [Google Scholar] [CrossRef]
- Tourna, M.; Stieglmeier, M.; Spang, A.; Könneke, M.; Schintlmeister, A.; Urich, T.; Engel, M.; Schloter, M.; Wagner, M.; Richter, A.; et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. USA 2011, 108, 8420–8425. [Google Scholar] [CrossRef]
- Ding, X.; Lan, W.; Li, Y.; Yan, A.; Katayama, Y.; Koba, K.; Makabe, A.; Fukushima, K.; Yano, M.; Onishi, Y.; et al. An internal recycling mechanism between ammonia/ammonium and nitrate driven by ammonia-oxidizing archaea and bacteria (AOA, AOB, and Comammox) and DNRA on Angkor sandstone monuments. Int. Biodeterior. Biodegrad. 2021, 165, 105328. [Google Scholar] [CrossRef]
- Meng, H.; Luo, L.; Chan, H.W.; Katayama, Y.; Gu, J.-D. Higher diversity and abundance of ammonia-oxidizing archaea than bacteria detected at the Bayon Temple of Angkor Thom in Cambodia. Int. Biodeterior. Biodegrad. 2016, 115, 234–243. [Google Scholar] [CrossRef]
- Andreolli, M.; Lampis, S.; Bernardi, P.; Calò, S.; Vallini, G. Bacteria from black crusts on stone monuments can precipitate CaCO3 allowing the development of a new bio-consolidation protocol for ornamental stone. Int. Biodeterior. Biodegrad. 2020, 153, 105031. [Google Scholar] [CrossRef]
- Omoregie, A.I.; Ong, D.E.L.; Nissom, P.M. Assessing ureolytic bacteria with calcifying abilities isolated from limestone caves for biocalcification. Lett. Appl. Microbiol. 2019, 68, 173–181. [Google Scholar] [CrossRef]
- Liao, C.; Liu, X.; Liu, R.; Shan, L. Two novel algicidal isolates kill Chlorella pyrenoidosa by inhibiting their host antioxidase activities. Appl. Biochem. Biotechnol. 2015, 177, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Niu, X.; Zhang, D.; Ma, J.; Zheng, X.; Xiao, H.; Huang, X.; Lin, Z.; Hu, H. The algicidal efficacy and the mechanism of Enterobacter sp. EA-1 on Oscillatoria dominating in aquaculture system. Environ. Res. 2021, 197, 111105. [Google Scholar] [CrossRef] [PubMed]
- ElBaghdady, K.Z.; Tolba, S.T.; Houssien, S.S. Biogenic deterioration of Egyptian limestone monuments: Treatment and conservation. J. Cult. Herit. 2019, 38, 118–125. [Google Scholar] [CrossRef]
- Di Bonaventura, M.P.; Del Gallo, M.; Cacchio, P.; Ercole, C.; Lepidi, A. Microbial formation of oxalate films on monument surfaces: Bioprotection or biodeterioration? Geomicrobiol. J. 1999, 16, 55–64. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).