A Meta-Analysis Indicates Positive Correlation between Genetic Diversity and Species Diversity
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
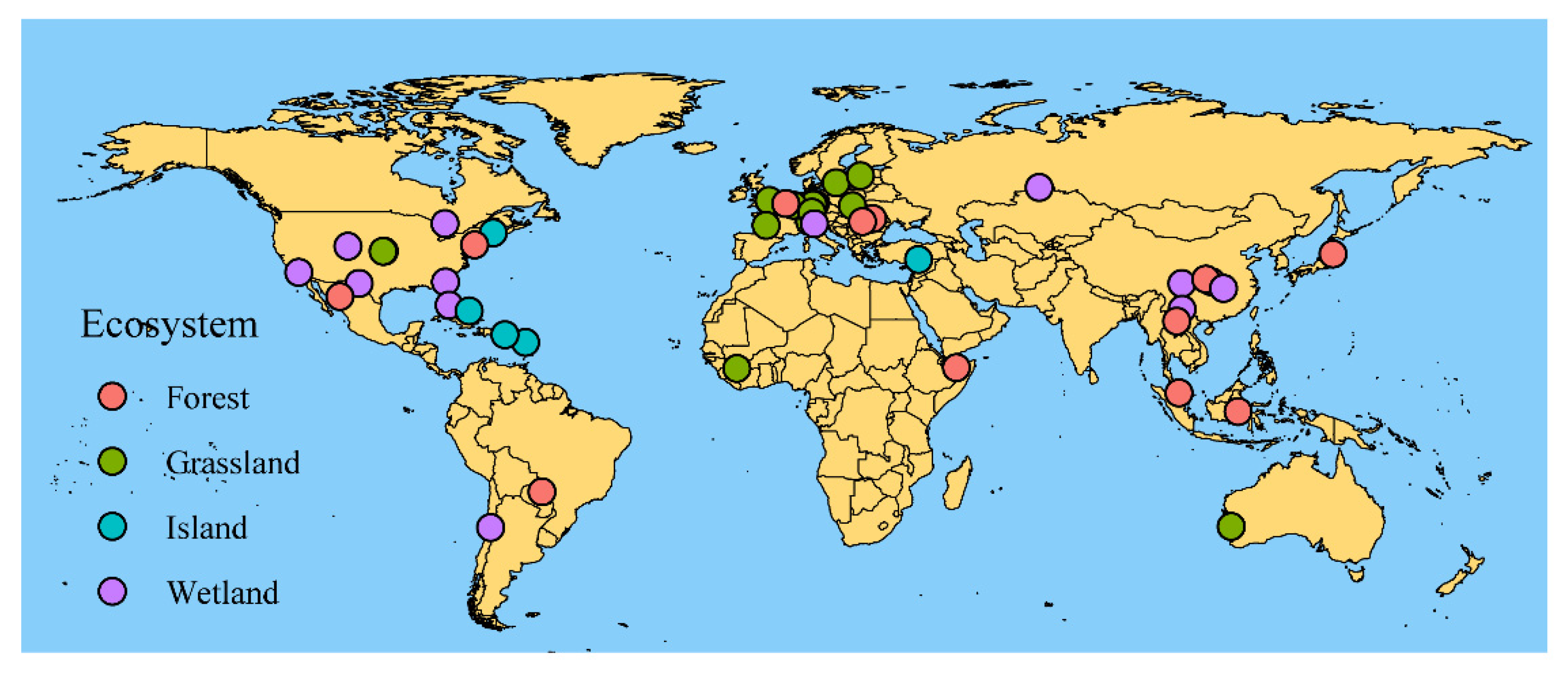
2.1. Data Collection
2.2. Statistical Analyses
3. Results
3.1. Evaluation of Total Heterogeneity
3.2. Factors That Affected SGDC
4. Discussion
4.1. Positive Correlation between SD and GD
4.2. Effects of Species and Experimental Methods on SGDC
4.3. Effects of Climate Change and Numbers of Sampling Units on SGDC
4.4. Implications for Future Experimental Design Regarding SGDC
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, G.K.; Wang, X.; Ma, K.P. Red list of China’s forest ecosystems: A conservation assessment and protected area gap analysis. Biol. Conserv. 2020, 248, 108636. [Google Scholar] [CrossRef]
- Blowes, S.A.; Supp, S.R.; Antao, L.H.; Bates, A.; Bruelheide, H.; Chase, J.M.; Moyes, F.; Magurran, A.; McGill, B.; Myers-Smith, I.H.; et al. The geography of biodiversity change in marine and terrestrial assemblages. Science 2019, 366, 339. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Liao, J.H.; Zou, X.M.; Xu, X.; Yang, J.Y.; Chen, H.Y.H.; Ruan, H.H. Coherent responses of terrestrial C:N stoichiometry to drought across plants, soil, and microorganisms in forests and grasslands. Agric. For. Meteorol. 2020, 292–293, 108104. [Google Scholar] [CrossRef]
- Wang, C.; Sun, Y.; Chen, H.Y.H.; Ruan, H. Effects of elevated CO2 on the C:N stoichiometry of plants, soils, and microorganisms in terrestrial ecosystems. Catena 2021, 201 (Suppl. 1), 105219. [Google Scholar] [CrossRef]
- Vellend, M. Island biogeography of genes and species. Am. Nat. 2003, 162, 358–365. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, J.C.P.; Leiva, F.; Cano-Ortiz, A.; Musarella, C.M.; Quinto-Canas, R.; Pinto-Gomes, C.J.; Cano, E. Impact of Grass Cover Management with Herbicides on Biodiversity, Soil Cover and Humidity in Olive Groves in the Southern Iberian. Agronomy 2021, 11, 412. [Google Scholar] [CrossRef]
- Perrino, E.V.; Calabrese, G. Endangered segetal species in southern Italy: Distribution, conservation status, trends, actions and ethnobotanical notes. Genet. Resour. Crop Evol. 2018, 65, 2107–2134. [Google Scholar] [CrossRef]
- Hammer, K.; Teklu, Y. Plant genetic resources: Selected issues from genetic erosion to genetic engineering. J. Agric. Rural Dev. Trop. 2008, 109, 15–50. [Google Scholar]
- Laroche, F.; Violle, C.; Taudiere, A.; Munoz, F. Analyzing snapshot diversity patterns with the Neutral Theory can show functional groups’ effects on community assembly. Ecology 2020, 101, e02977. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.W.; Fang, Y.M. Landscape Features and Climatic Forces Shape the Genetic Structure and Evolutionary History of an Oak Species (Quercus chenii) in East China. Front. Plant Sci. 2019, 10, 1060. [Google Scholar] [CrossRef]
- Lankau, R.A.; Strauss, S.Y. Mutual feedbacks maintain both genetic and species diversity in a plant community. Science 2007, 317, 1561–1563. [Google Scholar] [CrossRef] [Green Version]
- Wehenkel, C.; Bergmann, F.; Gregorius, H.R. Is there a trade-off between species diversity and genetic diversity in forest tree communities? Plant Ecol. 2006, 185, 151–161. [Google Scholar] [CrossRef]
- Puscas, M.; Taberlet, P.; Choler, P. No positive correlation between species and genetic diversity in European alpine grasslands dominated by Carex curvula. Divers. Distrib. 2008, 14, 852–861. [Google Scholar] [CrossRef]
- Silvertown, J.; Biss, P.M.; Freeland, J. Community genetics: Resource addition has opposing effects on genetic and species diversity in a 150-year experiment. Ecol. Lett. 2009, 12, 165–170. [Google Scholar] [CrossRef]
- Taberlet, P.; Zimmermann, N.E.; Englisch, T.; Tribsch, A.; Holderegger, R.; Alvarez, N.; Niklfeld, H.; Coldea, G.; Mirek, Z.; Moilanen, A.; et al. Genetic diversity in widespread species is not congruent with species richness in alpine plant communities. Ecol. Lett. 2012, 15, 1439–1448. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Ponce-Reyes, R.; Clegg, S.M.; Carvalho, S.B.; McDonald-Madden, E.; Possingham, H.P. Geographical surrogates of genetic variation for selecting island populations for conservation. Divers. Distrib. 2014, 20, 640–651. [Google Scholar] [CrossRef] [Green Version]
- Evanno, G.; Castella, E.; Antoine, C.; Paillat, G.; Goudet, J. Parallel changes in genetic diversity and species diversity following a natural disturbance. Mol. Ecol. 2009, 18, 1137–1144. [Google Scholar] [CrossRef] [Green Version]
- Struebig, M.J.; Kingston, T.; Petit, E.J.; Le Comber, S.C.; Zubaid, A.; Mohd-Adnan, A.; Rossiter, S.J. Parallel declines in species and genetic diversity in tropical forest fragments. Ecol. Lett. 2011, 14, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Vellend, M. Species diversity and genetic diversity: Parallel processes and correlated patterns. Am. Nat. 2005, 166, 199–215. [Google Scholar] [CrossRef] [Green Version]
- Antonovics, J. Toward community genomics? Ecology 2003, 84, 598–601. [Google Scholar] [CrossRef]
- Vellend, M.; Geber, M.A. Connections between species diversity and genetic diversity. Ecol. Lett. 2005, 8, 767–781. [Google Scholar] [CrossRef]
- Lamy, T.; Laroche, F.; David, P.; Massol, F.; Jarne, P. The contribution of species-genetic diversity correlations to the understanding of community assembly rules. Oikos 2017, 126, 759–771. [Google Scholar] [CrossRef]
- Vellend, M. Parallel effects of land-use history on species diversity and genetic diversity of forest herbs. Ecology 2004, 85, 3043–3055. [Google Scholar] [CrossRef] [Green Version]
- Reisch, C.; Schmid, C. Species and genetic diversity are not congruent in fragmented dry grasslands. Ecol. Evol. 2019, 9, 664–671. [Google Scholar] [CrossRef]
- Xu, W.; Liu, L.; He, T.; Cao, M.; Sha, L.; Hu, Y.; Li, Q.; Li, J. Soil properties drive a negative correlation between species diversity and genetic diversity in a tropical seasonal rainforest. Sci. Rep. 2016, 6, 20652. [Google Scholar] [CrossRef]
- Kahilainen, A.; Puurtinen, M.; Kotiaho, J.S. Conservation implications of species-genetic diversity correlations. Glob. Ecol. Conserv. 2014, 2, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Whitlock, R. Relationships between adaptive and neutral genetic diversity and ecological structure and functioning: A meta-analysis. J. Ecol. 2014, 102, 857–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vellend, M.; Lajoie, G.; Bourret, A.; Murria, C.; Kembel, S.W.; Garant, D. Drawing ecological inferences from coincident patterns of population- and community-level biodiversity. Mol. Ecol. 2014, 23, 2890–2901. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, C.T.; Chen, H.Y.H.; Ruan, H.H. Responses of C:N stoichiometry in plants, soil, and microorganisms to nitrogen addition. Plant Soil 2020, 456, 277–287. [Google Scholar] [CrossRef]
- Vellend, M. The consequences of genetic diversity in competitive communities. Ecology 2006, 87, 304–311. [Google Scholar] [CrossRef] [Green Version]
- Burda, B.U.; O’Connor, E.A.; Webber, E.M.; Redmond, N.; Perdue, L.A. Estimating data from figures with a Web-based program: Considerations for a systematic review. Res. Synth. Methods 2017, 8, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Viechtbauer, W. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef] [Green Version]
- Vellend, M. The Theory of Ecological Communities; Princeton University Press: Princeton, NJ, USA, 2016. [Google Scholar]
- Laroche, F.; Jarne, P.; Lamy, T.; David, P.; Massol, F. A neutral theory for interpreting correlations between species and genetic diversity in communities. Am. Nat. 2015, 185, 59–69. [Google Scholar] [CrossRef]
- Wei, X.; Bao, D.; Meng, H.; Jiang, M. Pattern and drivers of species-genetic diversity correlation in natural forest tree communities across a biodiversity hotspot. J. Plant Ecol. 2017, 11, 761–770. [Google Scholar] [CrossRef]
- Odat, N.; Jetschke, G.; Hellwig, F.H. Genetic diversity of Ranunculus acris L. (Ranunculaceae) populations in relation to species diversity and habitat type in grassland communities. Mol. Ecol. 2004, 13, 1251–1257. [Google Scholar] [CrossRef]
- Avolio, M.L.; Smith, M.D.; Michalet, R. Correlations between genetic and species diversity: Effects of resource quantity and heterogeneity. J. Veg. Sci. 2013, 24, 1185–1194. [Google Scholar] [CrossRef]
- Bacles, C.F.E.; Jump, A.S. Taking a tree’s perspective on forest fragmentation genetics. Trends Plant Sci. 2011, 16, 13–18. [Google Scholar] [CrossRef]
- Wei, X.; Jiang, M. Contrasting relationships between species diversity and genetic diversity in natural and disturbed forest tree communities. New Phytol. 2012, 193, 779–786. [Google Scholar] [CrossRef]
- Pfeiffer, V.W.; Ford, B.M.; Housset, J.; McCombs, A.; Blanco-Pastor, J.L.; Gouin, N.; Manel, S.; Bertin, A. Partitioning genetic and species diversity refines our understanding of species-genetic diversity relationships. Ecol. Evol. 2018, 8, 12351–12364. [Google Scholar] [CrossRef]
- Watanabe, K.; Monaghan, M.T. Comparative tests of the species-genetic diversity correlation at neutral and nonneutral loci in four species of stream insect. Evolution 2017, 71, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Lamy, T.; Jarne, P.; Laroche, F.; Pointier, J.P.; Huth, G.; Segard, A.; David, P. Variation in habitat connectivity generates positive correlations between species and genetic diversity in a metacommunity. Mol. Ecol. 2013, 22, 4445–4456. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ci, X.; Li, J. Parallel effects of environmental properties on genetic diversity and species diversity. Biodivers. Sci. 2017, 25, 481–489. [Google Scholar]
- Blanchard, G.; Birnbaum, P.; Munoz, F. Extinction-immigration dynamics lag behind environmental filtering in shaping the composition of tropical dry forests within a changing landscape. Ecography 2020, 43, 869–881. [Google Scholar] [CrossRef] [Green Version]
- Laliberte, E.; Zemunik, G.; Turner, B.L. Environmental filtering explains variation in plant diversity along resource gradients. Science 2014, 345, 1602–1605. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.C.; Hill, J.K.; Ohlemuller, R.; Roy, D.B.; Thomas, C.D. Rapid Range Shifts of Species Associated with High Levels of Climate Warming. Science 2011, 333, 1024–1026. [Google Scholar] [CrossRef]
- Li, L.; Chen, J. Influence of climate change on wild plants and the conservation strategies. Chin. Biodivers. Sci. 2014, 22, 549–563. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Anastasiou, I.; Spagopoulou, F.; Stalimerou, M.; Terzopoulou, S.; Legakis, A.; Vogler, A.P. Testing the species--genetic diversity correlation in the Aegean archipelago: Toward a haplotype-based macroecology? Am. Nat. 2011, 178, 241–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, C.Z.; Michalski, S.G.; Fischer, M.; Durka, W. Genetic diversity and differentiation follow secondary succession in a multi-species study on woody plants from subtropical China. J. Plant Ecol. 2017, 10, 213–221. [Google Scholar] [CrossRef]
- Frey, D.; Arrigo, N.; Granereau, G.; Sarr, A.; Felber, F.; Kozlowski, G. Parallel declines in species and genetic diversity driven by anthropogenic disturbance: A multispecies approach in a French Atlantic dune system. Evol. Appl. 2016, 9, 479–488. [Google Scholar] [CrossRef] [Green Version]



| Types | Attribute | Estimate | SE | p-Value |
|---|---|---|---|---|
| Sampling methods | Discrete | 0.25 | 0.05 | <0.01 |
| Continuous | 0.05 | 0.05 | 0.39 | |
| Ecosystems | Forest | 0.08 | 0.05 | 0.12 |
| Grassland | 0.13 | 0.09 | 0.12 | |
| Island | 0.45 | 0.17 | <0.01 | |
| Wetland | 0.28 | 0.08 | <0.01 | |
| Species pools | Animal | 0.21 | 0.07 | <0.01 |
| Plant | 0.13 | 0.04 | <0.01 | |
| Molecular markers | AFLP | 0.10 | 0.04 | 0.03 |
| Allozyme | 0.11 | 0.12 | 0.33 | |
| Microsatellite | 0.26 | 0.08 | <0.01 | |
| mtDNA | 0.33 | 0.10 | <0.01 | |
| RAPD | 0.28 | 0.26 | 0.28 | |
| SNP | 0.07 | 0.28 | 0.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, L.; Yang, Y.; Li, Y.; Chen, S.; Feng, Y.; Wang, N.; Lv, T.; Ding, H.; Wang, L.; Fang, Y. A Meta-Analysis Indicates Positive Correlation between Genetic Diversity and Species Diversity. Biology 2021, 10, 1089. https://doi.org/10.3390/biology10111089
Xie L, Yang Y, Li Y, Chen S, Feng Y, Wang N, Lv T, Ding H, Wang L, Fang Y. A Meta-Analysis Indicates Positive Correlation between Genetic Diversity and Species Diversity. Biology. 2021; 10(11):1089. https://doi.org/10.3390/biology10111089
Chicago/Turabian StyleXie, Lei, Yuan Yang, Yao Li, Shuifei Chen, Yueyao Feng, Ningjie Wang, Ting Lv, Hui Ding, Lu Wang, and Yanming Fang. 2021. "A Meta-Analysis Indicates Positive Correlation between Genetic Diversity and Species Diversity" Biology 10, no. 11: 1089. https://doi.org/10.3390/biology10111089






