Going Forward and Back: The Complex Evolutionary History of the GPx
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. GPx Sequence Search
2.2. Sequence Alignments and Evolutionary Analyses
3. Results
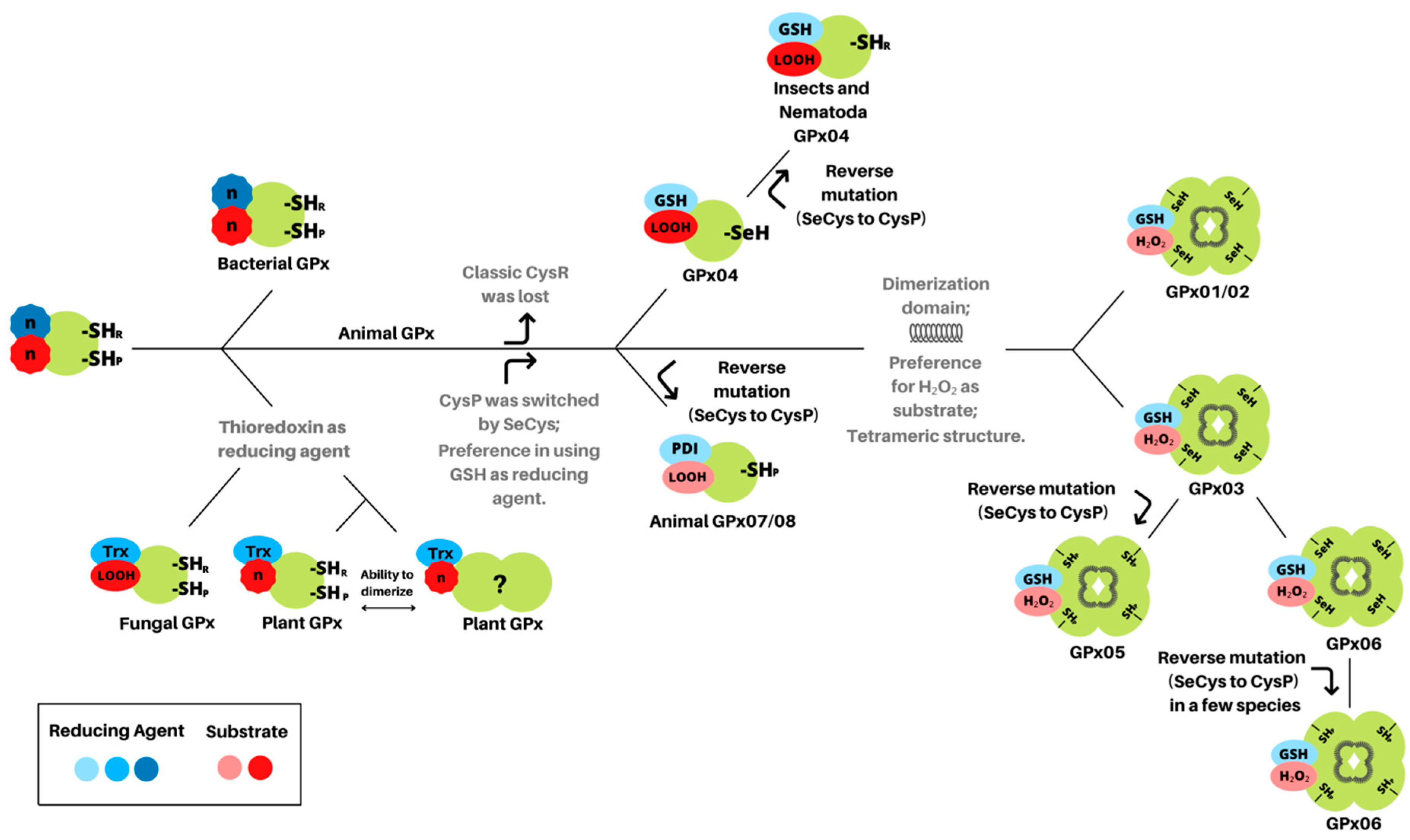
3.1. Glutathione Peroxidases Have a Monomeric Common Ancestor with Peroxidatic Cysteine
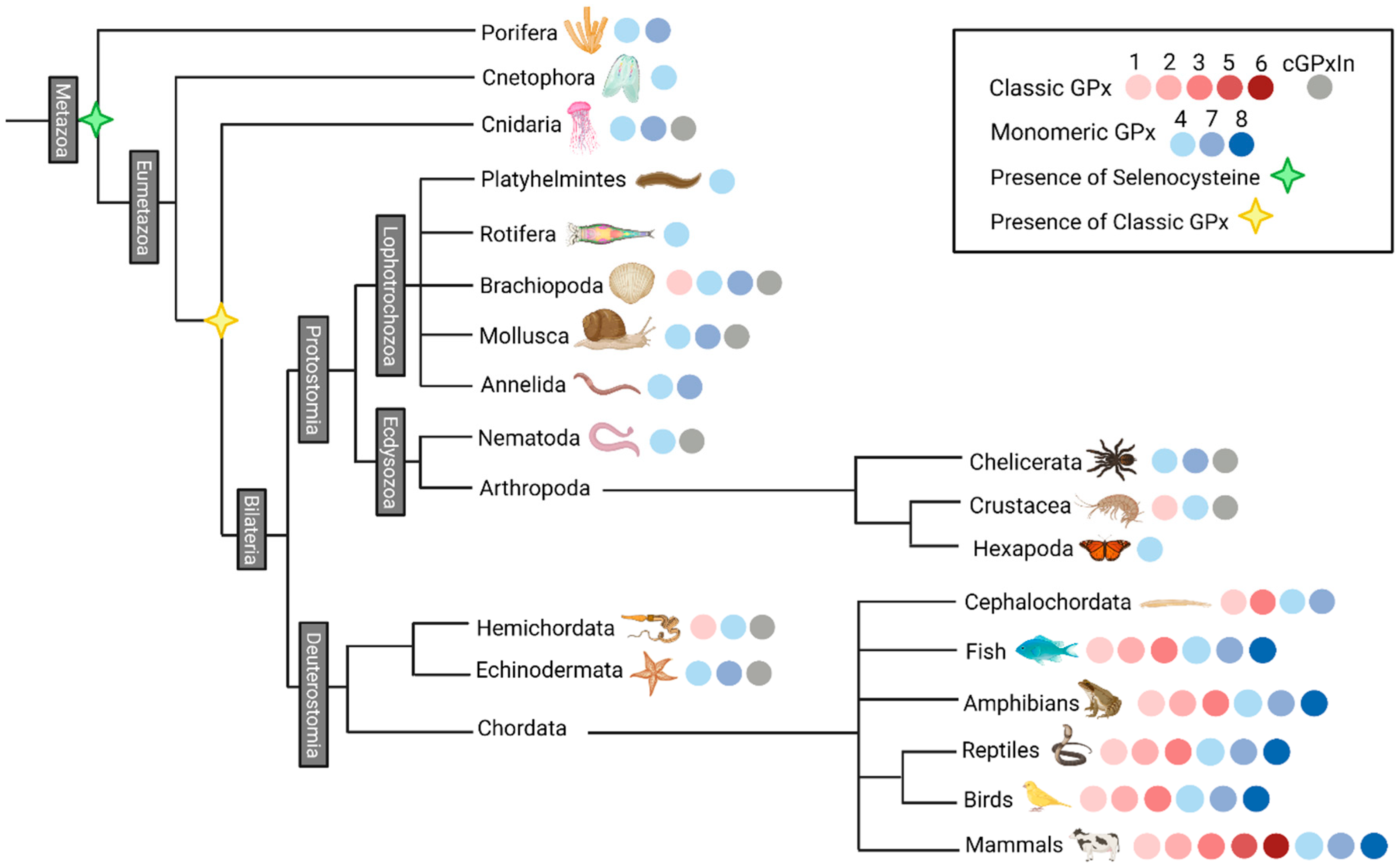
3.2. Selenocysteine Was Introduced into GPx Proteins Right from the Arise of Metazoa Kingdom
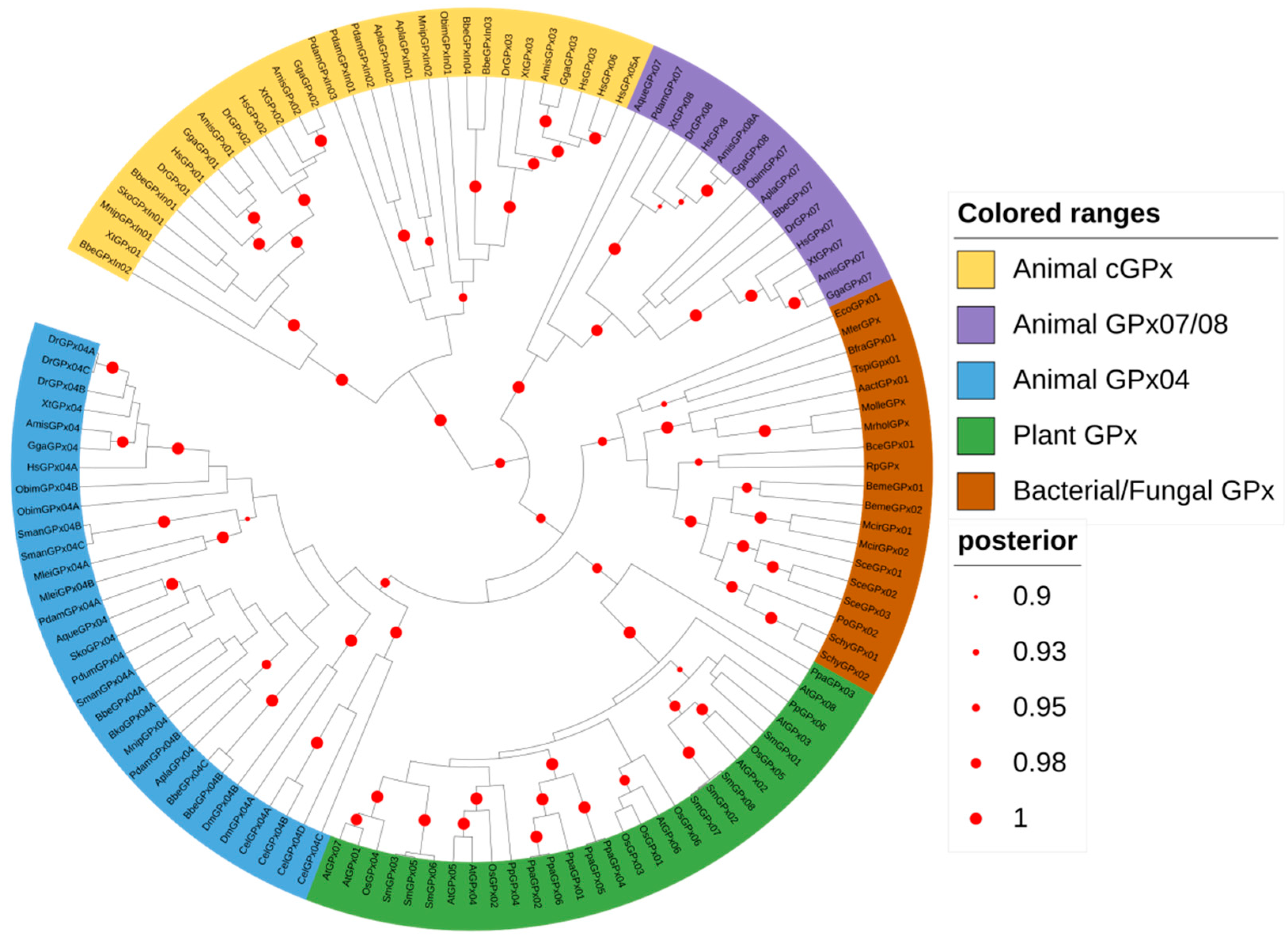
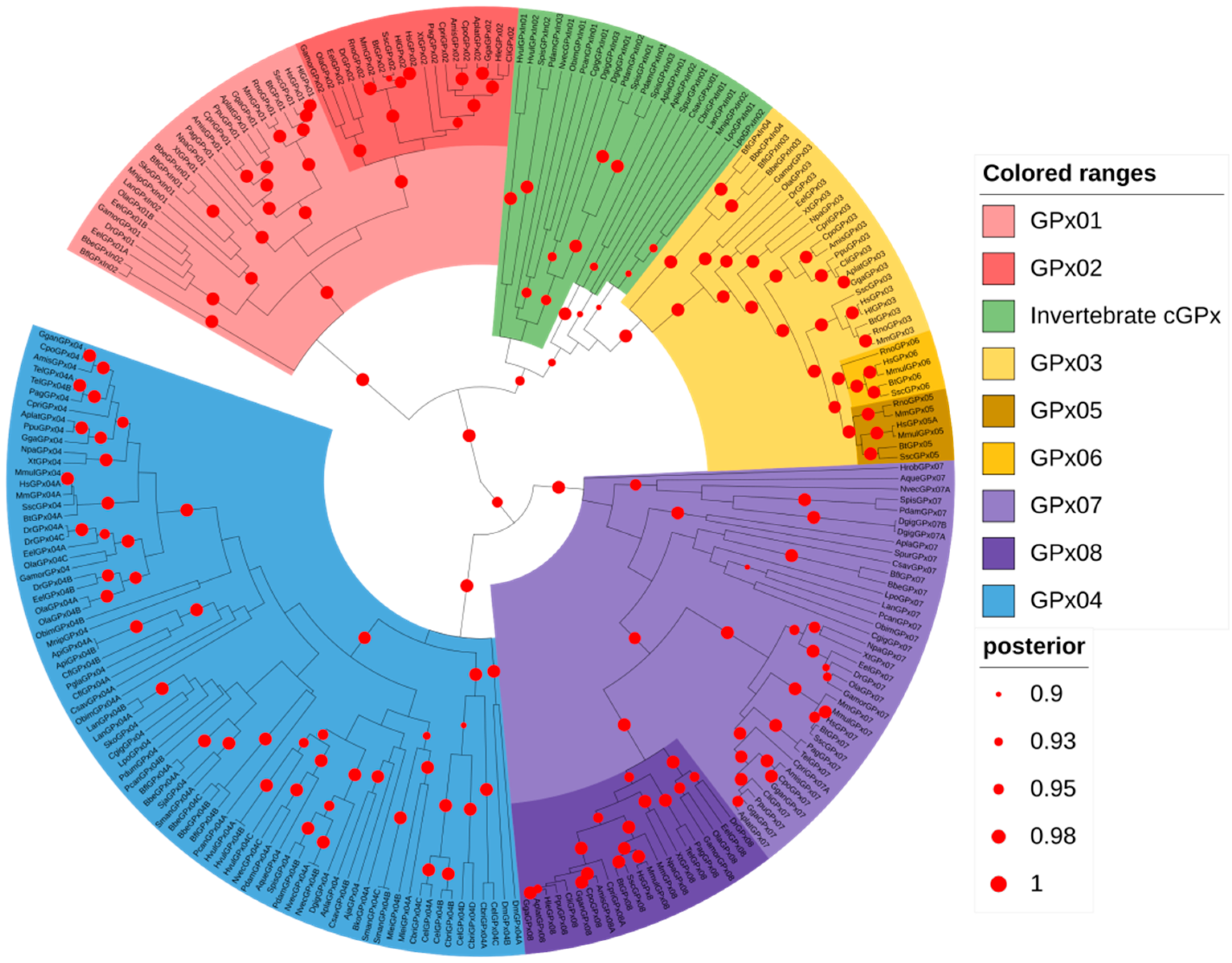
3.3. Animal GPx Proteins Are Evolutionarily Classified into Three Main Groups
3.4. The Radial Spreading of GPx in Animals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reinhard, C.T.; Planavsky, N.J.; Olson, S.L.; Lyons, T.W.; Erwin, D.H. Earth’s oxygen cycle and the evolution of animal life. Proc. Natl. Acad. Sci. USA 2016, 113, 8933–8938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anbar, A.D. Oceans. Elements and evolution. Science 2008, 322, 1481–1483. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta 2013, 1830, 3217–3266. [Google Scholar] [CrossRef] [Green Version]
- Mills, G.C. Hemoglobin catabolism. I. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J. Biol. Chem. 1957, 229, 189–197. [Google Scholar] [CrossRef]
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008, 275, 3959–3970. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta 2013, 1830, 3289–3303. [Google Scholar] [CrossRef]
- Tian, Y.; Fan, M.; Qin, Z.; Lv, H.; Wang, M.; Zhang, Z.; Zhou, W.; Zhao, N.; Li, X.; Han, C.; et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSI-NAZOLE-RESISTANT1 transcription factor. Nat. Commun. 2018, 9, 1063. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-F.; Zheng, J.-J.; Shao, G.-M.; Wang, J.-L.; Liu, X.-S.; Wang, Y.-F. Cloning and functional analysis of glutathione peroxidase gene in red swamp crayfish Procambarus clarkii. Fish Shellfish Immunol. 2013, 34, 1587–1595. [Google Scholar] [CrossRef]
- Ren, Q.; Sun, R.R.; Zhao, X.F.; Wang, J.X. A selenium-dependent glutathione peroxidase (Se-GPx) and two gluta-thione S-transferases (GSTs) from Chinese shrimp (Fenneropenaeus chinensis). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 149, 613–623. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Ma, H.-L.; Deng, Y.-Q.; Feng, J.; Chen, X.-L.; Guo, Z.-X. Glutathione peroxidase 3 in the mud crab Scylla paramamosain: Characterization and regulation under nitrite stress. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 229, 108673. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Zou, Z.; Liu, S.; Lin, P.; Wang, Y.; Zhang, Z. Selenium-dependent glutathione peroxidase gene expression during gonad development and its response to LPS and H2O2; challenge in Scylla paramamosain. Fish Shellfish Immunol. 2012, 33, 532–542. [Google Scholar] [CrossRef]
- Bathige, S.; Umasuthan, N.; Godahewa, G.; Thulasitha, W.S.; Whang, I.; Won, S.H.; Kim, C.; Lee, J. Two variants of selenium-dependent glutathione peroxidase from the disk abalone Haliotis discus discus: Molecular characterization and immune responses to bacterial and viral stresses. Fish Shellfish Immunol. 2015, 45, 648–655. [Google Scholar] [CrossRef]
- Qu, C.; Liu, S.; Tang, Z.; Li, J.; Liao, Z.; Qi, P. Response of a novel selenium-dependent glutathione peroxidase from thick shell mussel Mytilus coruscus exposed to lipopolysaccharide, copper and benzo [α] pyrene. Fish Shellfish Immunol. 2019, 89, 595–602. [Google Scholar] [CrossRef]
- Ursini, F.; Maiorino, M.; Gregolin, C. The selenoenzyme phospholipid hydroperoxide glutathione peroxidase. Biochim. Biophys. Acta 1985, 839, 62–70. [Google Scholar] [CrossRef]
- Herbette, S.; Roeckel-Drevet, P.; Drevet, J. Seleno-independent glutathione peroxidases: More than simple antioxidant scavengers. FEBS J. 2007, 274, 2163–2180. [Google Scholar] [CrossRef] [PubMed]
- Buday, K.; Conrad, M. Emerging roles for non-selenium containing ER-resident glutathione peroxidases in cell signaling and disease. Biol. Chem. 2020, 402, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Maiorino, M.; Bosello-Travain, V.; Cozza, G.; Miotto, G.; Roveri, A.; Toppo, S.; Zaccarin, M.; Ursini, F. Understanding mammalian glutathione peroxidase 7 in the light of its homologs. Free Radic. Biol. Med. 2015, 83, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, C.; Wang, P.; Wang, S.; Lin, H.; Qiu, L. The response of glutathione peroxidase 1 and glutathione peroxidase 7 under different oxidative stresses in black tiger shrimp, Penaeus monodon. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 217, 1–13. [Google Scholar] [CrossRef]
- Ferro, D.; Franchi, N.; Bakiu, R.; Ballarin, L.; Santovito, G. Molecular characterization and metal induced gene ex-pression of the novel glutathione peroxidase 7 from the chordate invertebrate Ciona robusta. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2018, 205, 1–7. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Song, J.-Y.; Kwon, E.-S.; Roe, J.-H. Gpx1 is a stationary phase-specific thioredoxin peroxidase in fission yeast. Biochem. Biophys. Res. Commun. 2008, 367, 67–71. [Google Scholar] [CrossRef]
- Avery, A.M.; Avery, S.V. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 2001, 276, 33730–33735. [Google Scholar] [CrossRef] [Green Version]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef]
- Avery, A.M.; Willetts, S.A.; Avery, S. Genetic Dissection of the Phospholipid Hydroperoxidase Activity of Yeast Gpx3 Reveals Its Functional Importance. J. Biol. Chem. 2004, 279, 46652–46658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attacha, S.; Solbach, D.; Bela, K.; Moseler, A.; Wagner, S.; Schwarzländer, M.; Aller, I.; Müller, S.J.; Meyer, A.J. Glutathione peroxidase-like enzymes cover five distinct cell compartments and membrane surfaces in Arabidopsis thaliana. Plant Cell Environ. 2017, 40, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Yabuta, Y.; Takeda, T.; Nakano, Y.; Shigeoka, S. Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J. 2006, 273, 5589–5597. [Google Scholar] [CrossRef]
- Hazebrouck, S.; Camoin, L.; Faltin, Z.; Strosberg, A.D.; Eshdat, Y. Substituting Selenocysteine for Catalytic Cysteine 41 Enhances Enzymatic Activity of Plant Phospholipid Hydroperoxide Glutathione Peroxidase Expressed in Escherichia coli. J. Biol. Chem. 2000, 275, 28715–28721. [Google Scholar] [CrossRef] [Green Version]
- Arenas, F.A.; Díaz, W.A.; Leal, C.A.; Pérez-Donoso, J.M.; Imlay, J.A.; Vásquez, C.C. The Escherichia coli btuE gene, encodes a glutathione peroxidase that is induced under oxidative stress conditions. Biochem. Biophys. Res. Commun. 2010, 398, 690–694. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Han, H.; Cui, B.; Hou, Y.; Wang, Y.; Wang, Q. A glutathione peroxidase from Antarctic psychrotrophic bacterium Pseudoalteromonas sp. ANT506: Cloning and heterologous expression of the gene and characterization of recombinant enzyme. Bioengineered 2017, 8, 742–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.M.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019, 26, 101247. [Google Scholar] [CrossRef]
- Churin, Y.; Schilling, S.; Börner, T. A gene family encoding glutathione peroxidase homologues in Hordeum vulgare(barley). FEBS Lett. 1999, 459, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Hlaing, S.M.M.; Lou, J.; Cheng, J.; Xun, X.; Li, M.; Lu, W.; Hu, X.; Bao, Z. Tissue-Biased and Species-Specific Regulation of Glutathione Peroxidase. Toxins 2020, 13, 21. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hoehna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Ayres, D.L.; Darling, A.; Zwickl, D.J.; Beerli, P.; Holder, M.; Lewis, P.O.; Huelsenbeck, J.P.; Ronquist, F.; Swofford, D.L.; Cummings, M.P.; et al. BEAGLE: An Application Programming Interface and High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 2011, 61, 170–173. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE 2010), New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Rambaut, A.; Drummond, A.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Geng, Y.; Yang, Y.; Seim, I.; Yang, G. Oxidative stress drives divergent evolution of the glutathione peroxidase (GPX) gene family in mammals. Integr. Zoσl. 2021, 16, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Bersweiler, A.; D’Autréaux, B.; Mazon, H.; Kriznik, A.; Belli, G.; Delaunay-Moisan, A.; Toledano, M.B.; Rahuel-Clermont, S. A scaffold protein that chaperones a cysteine-sulfenic acid in H2O2 signaling. Nat. Chem. Biol. 2017, 13, 909–915. [Google Scholar] [CrossRef] [Green Version]
- Passaia, G.; Margis-Pinheiro, M. Glutathione peroxidases as redox sensor proteins in plant cells. Plant Sci. 2015, 234, 22–26. [Google Scholar] [CrossRef]
- Delaunay, A.; Pflieger, D.; Barrault, M.B.; Vinh, J.; Toledano, M.B. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 2002, 111, 471–481. [Google Scholar] [CrossRef] [Green Version]
- Bae, Y.-A.; Cai, G.-B.; Kim, S.-H.; Zo, Y.-G.; Kong, Y. Modular evolution of glutathione peroxidase genes in association with different biochemical properties of their encoded proteins in invertebrate animals. BMC Evol. Biol. 2009, 9, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxidative Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.-P.; Rouhier, N. Plant Glutathione Peroxidases Are Functional Peroxiredoxins Distributed in Several Subcellular Compartments and Regulated during Biotic and Abiotic Stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [Green Version]
- Koh, C.S.; Didierjean, C.; Navrot, N.; Panjikar, S.; Mulliert, G.; Rouhier, N.; Jacquot, J.-P.; Aubry, A.; Shawkataly, O.; Corbier, C. Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes. J. Mol. Biol. 2007, 370, 512–529. [Google Scholar] [CrossRef]
- Navrot, N.; Skjoldager, N.; Bunkenborg, J.; Svensson, B.; Hägglund, P. A redox-dependent dimerization switch regulates activity and tolerance for reactive oxygen species of barley seed glutathione peroxidase. Plant Physiol. Biochem. 2015, 90, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Selles, B.; Hugo, M.; Trujillo, M.; Srivastava, V.; Wingsle, G.; Jacquot, J.P.; Radi, R.; Rouhier, N. Hydroperoxide and peroxynitrite reductase activity of poplar thioredoxin-dependent glutathione peroxidase 5: Kinetics, catalytic mechanism and oxi-dative inactivation. Biochem. J. 2012, 442, 369–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moosmayer, D.; Hilpmann, A.; Hoffmann, J.; Schnirch, L.; Zimmermann, K.; Badock, V.; Furst, L.; Eaton, J.K.; Viswanathan, V.S.; Schreiber, S.L.; et al. Crystal structures of the selenoprotein glutathione peroxidase 4 in its apo form and in complex with the covalently bound inhibitor ML162. Acta Crystallogr. Sect. D Struct. Biol. 2021, 77, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Flohé, L.; Toppo, S.; Cozza, G.; Ursini, F. A Comparison of Thiol Peroxidase Mechanisms. Antioxid. Redox Signal. 2011, 15, 763–780. [Google Scholar] [CrossRef] [PubMed]
- Dworzański, J.; Strycharz-Dudziak, M.; Kliszczewska, E.; Kiełczykowska, M.; Dworzańska, A.; Drop, B.; Polz-Dacewicz, M. Glutathione peroxidase (GPx) and superoxide dismutase (SOD) activity in patients with diabetes mellitus type 2 infected with Epstein-Barr virus. PLoS ONE 2020, 15, e0230374. [Google Scholar] [CrossRef]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione Peroxidase-1 in Health and Disease: From Molecular Mechanisms to Therapeutic Opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef] [Green Version]
- Ghneim, H.K.; Al-Sheikh, Y.A. Effect of Selenium Supplementation on Glutathione Peroxidase and Catalase Activities in Senescent Cultured Human Fibroblasts. Ann. Nutr. Metab. 2011, 59, 127–138. [Google Scholar] [CrossRef]
- Richie, J.P.; Nichenametla, S.; Neidig, W.; Calcagnotto, A.; Haley, J.S.; Schell, T.D.; Muscat, J.E. Randomized con-trolled trial of oral glutathione supplementation on body stores of glutathione. Eur. J. Nutr. 2015, 54, 251–263. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trenz, T.S.; Delaix, C.L.; Turchetto-Zolet, A.C.; Zamocky, M.; Lazzarotto, F.; Margis-Pinheiro, M. Going Forward and Back: The Complex Evolutionary History of the GPx. Biology 2021, 10, 1165. https://doi.org/10.3390/biology10111165
Trenz TS, Delaix CL, Turchetto-Zolet AC, Zamocky M, Lazzarotto F, Margis-Pinheiro M. Going Forward and Back: The Complex Evolutionary History of the GPx. Biology. 2021; 10(11):1165. https://doi.org/10.3390/biology10111165
Chicago/Turabian StyleTrenz, Thomaz Stumpf, Camila Luiza Delaix, Andreia Carina Turchetto-Zolet, Marcel Zamocky, Fernanda Lazzarotto, and Márcia Margis-Pinheiro. 2021. "Going Forward and Back: The Complex Evolutionary History of the GPx" Biology 10, no. 11: 1165. https://doi.org/10.3390/biology10111165
APA StyleTrenz, T. S., Delaix, C. L., Turchetto-Zolet, A. C., Zamocky, M., Lazzarotto, F., & Margis-Pinheiro, M. (2021). Going Forward and Back: The Complex Evolutionary History of the GPx. Biology, 10(11), 1165. https://doi.org/10.3390/biology10111165







