A Layered View on Focal Adhesions
Abstract
:Simple Summary
Abstract
1. Introduction
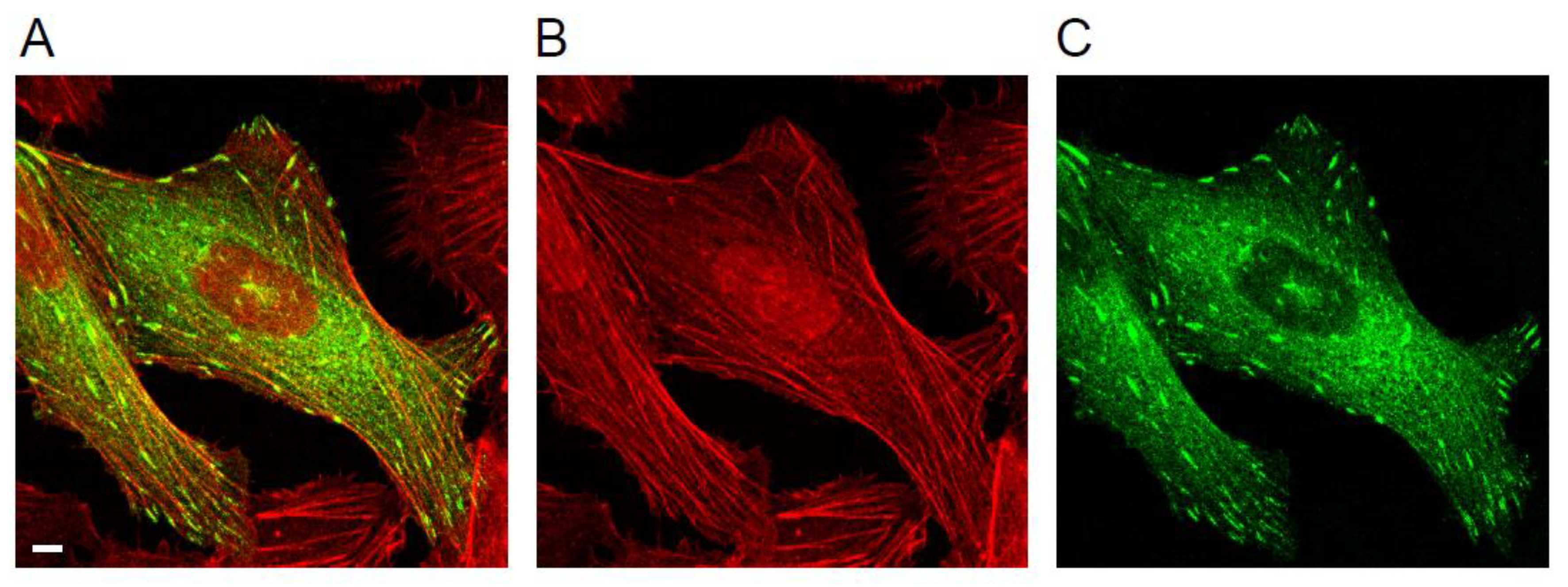
1.1. Focal Adhesions and the Actin Cytoskeleton
1.2. Focal Adhesions in Health and Disease
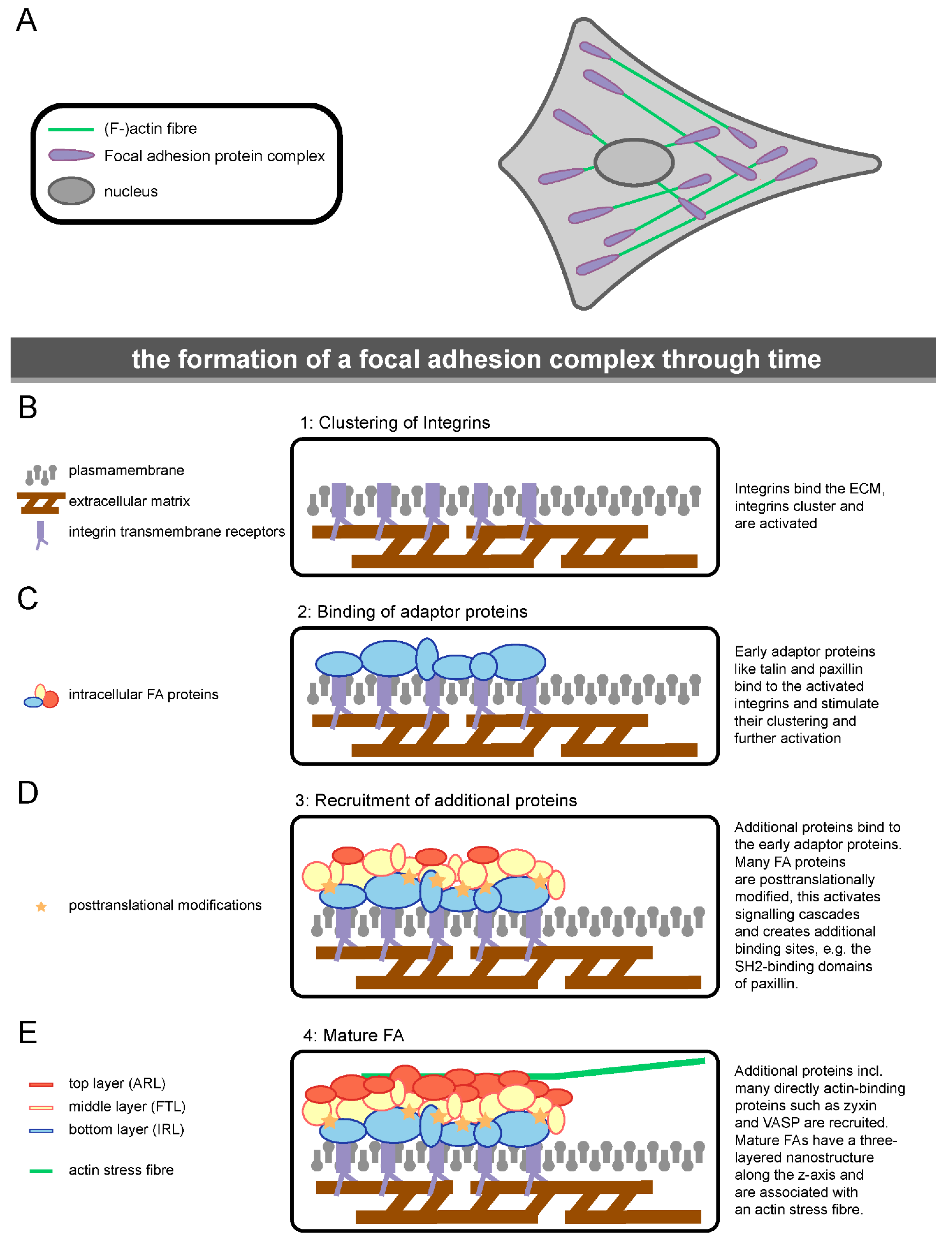
1.3. The Molecular Composition of Focal Adhesions
1.4. Focal Adhesions In Vitro and In Vivo
2. Intracellular Focal Adhesion Proteins
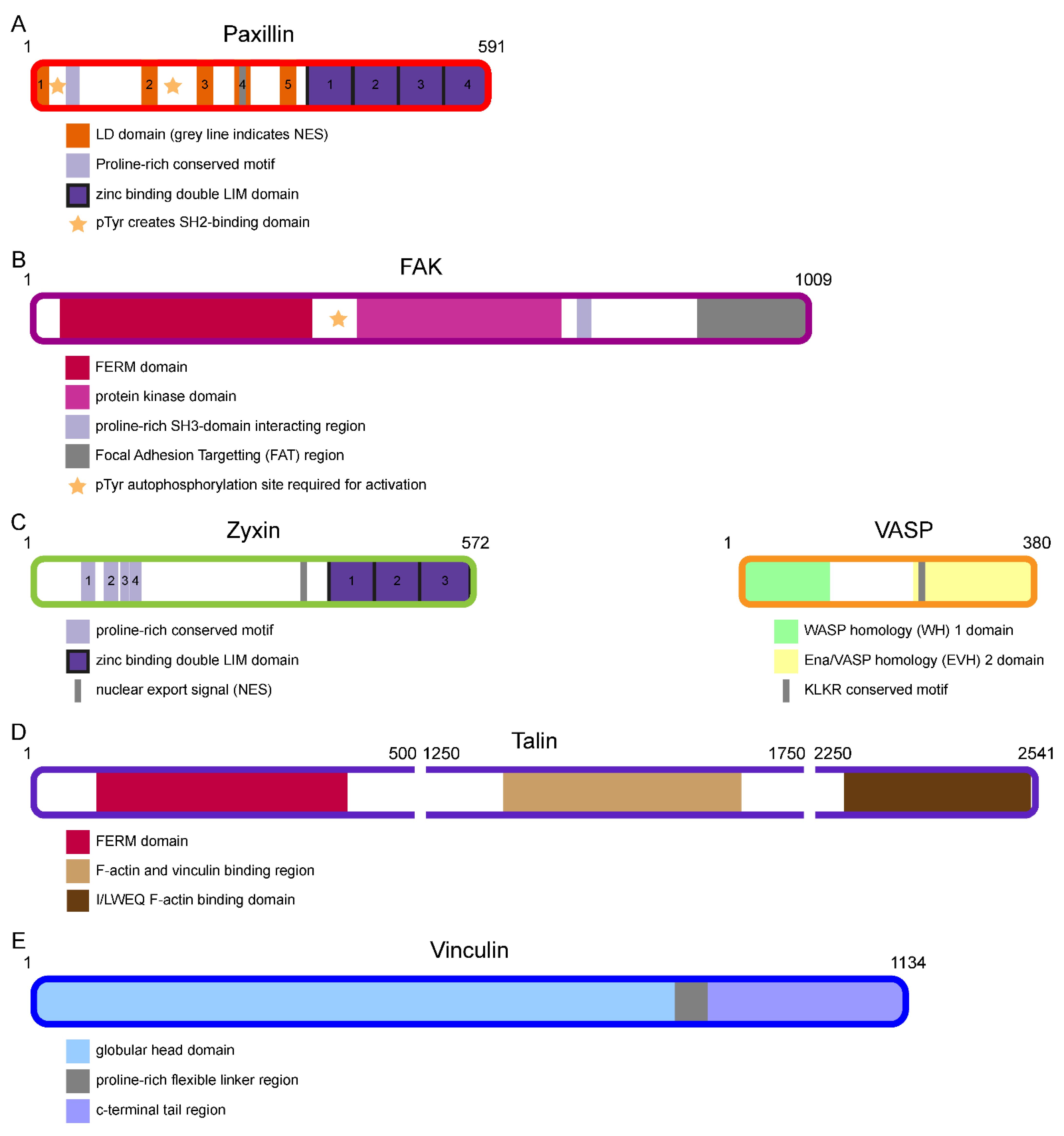
2.1. Bottom Integrin Signalling Layer (ISL) Proteins Paxillin and FAK
2.2. Top Actin Regulatory Layer (ARL) Proteins Zyxin and VASP
2.3. Middle Force Transduction Layer (FTL) Proteins Talin and Vinculin
2.4. The Importance of Individual FA Proteins In Vivo
2.5. FA Proteins and Regulation of Gene Expression
3. Advanced Light Microscopy Techniques
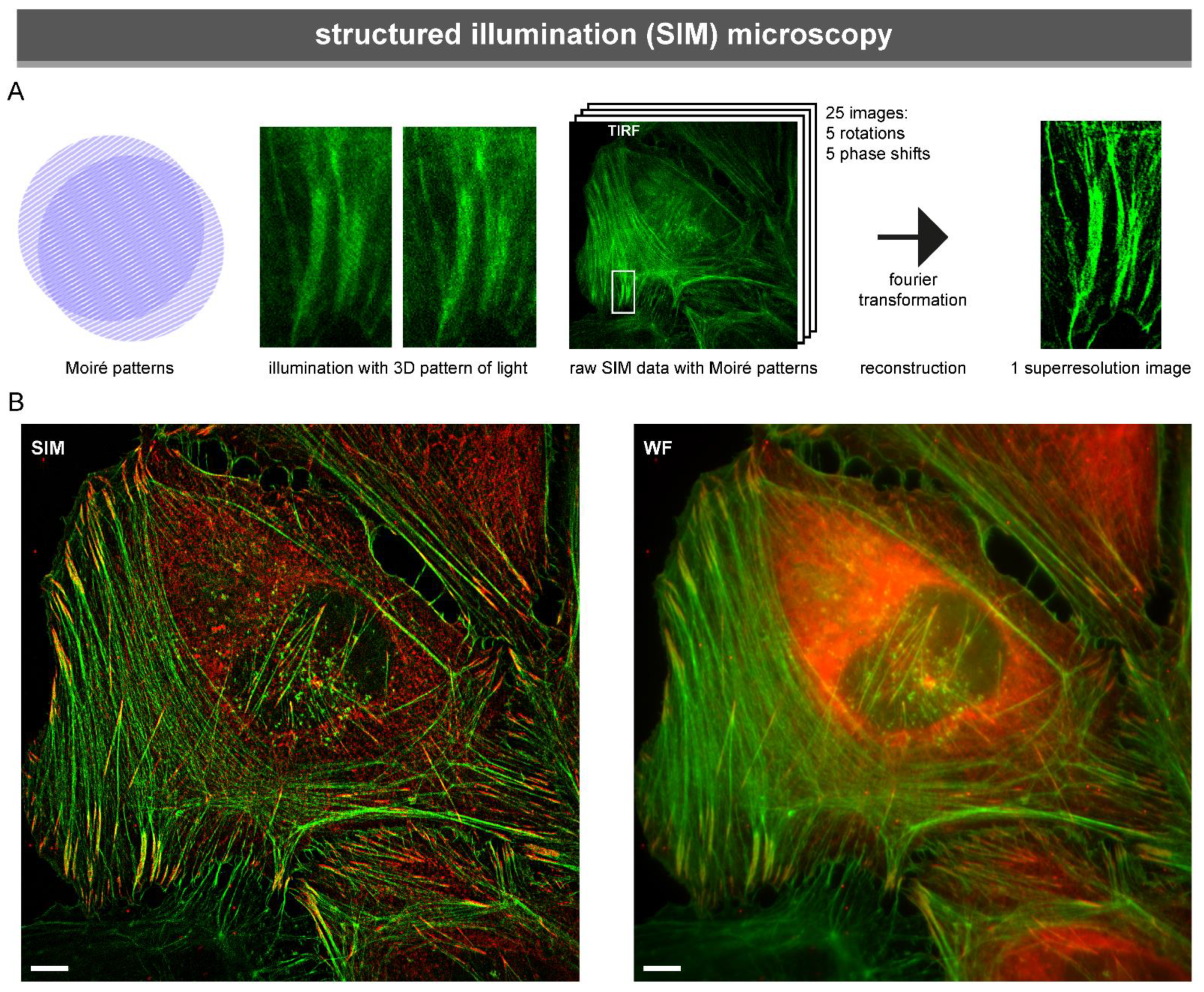
3.1. Advanced Light Microscopy Techniques Circumventing the Resolution Limit
3.1.1. Total Internal Reflection Microscopy
3.1.2. Structured Illumination Microscopy
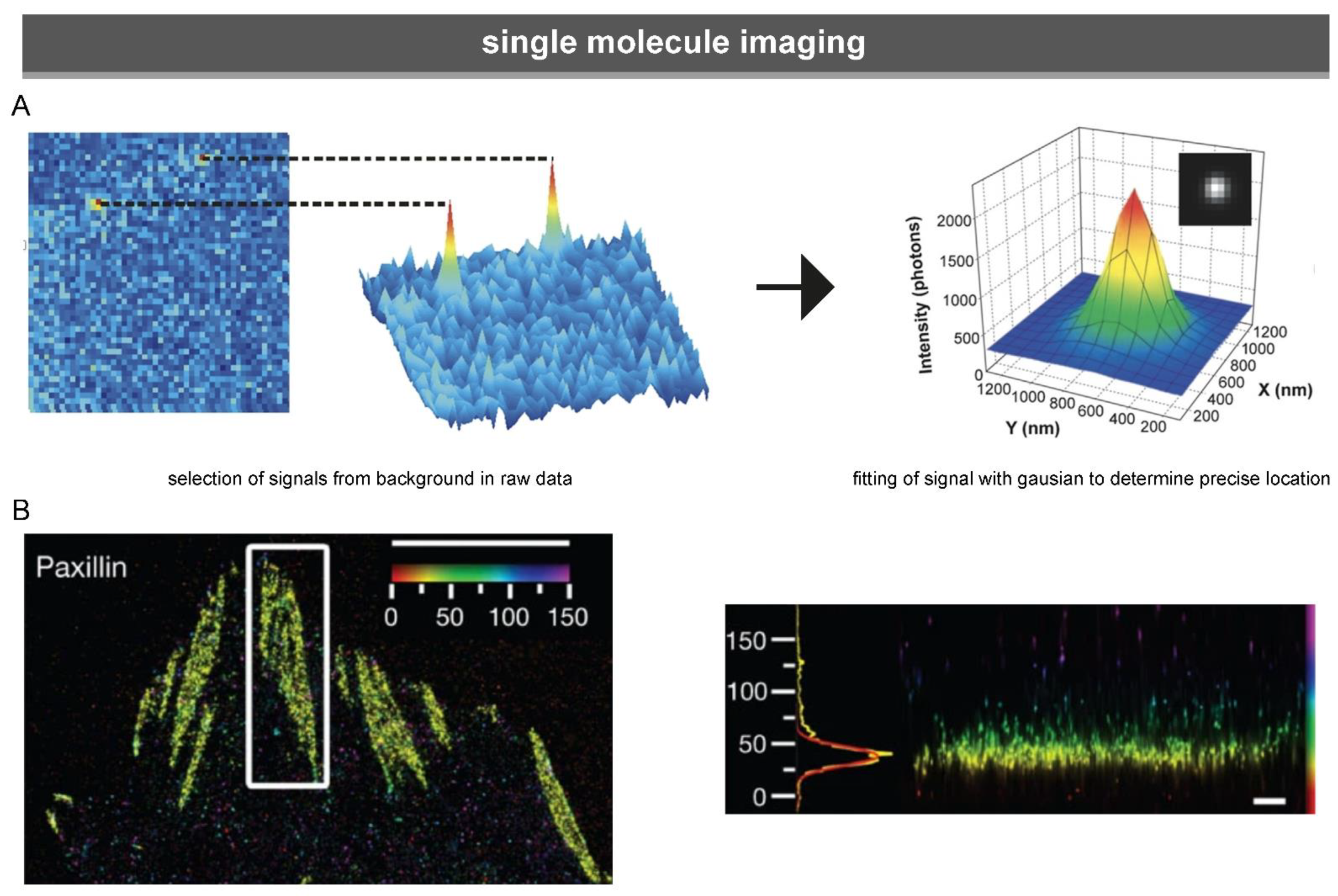
3.1.3. Single-Molecule Microscopy
3.2. Advanced Imaging Assays to Study Protein Dynamics
3.2.1. Fluorescence Recovery after Photobleaching
3.2.2. Photoactivation and Photoswitching
3.3. Advances in Data Analysis
4. Focal Adhesion Protein Dynamics
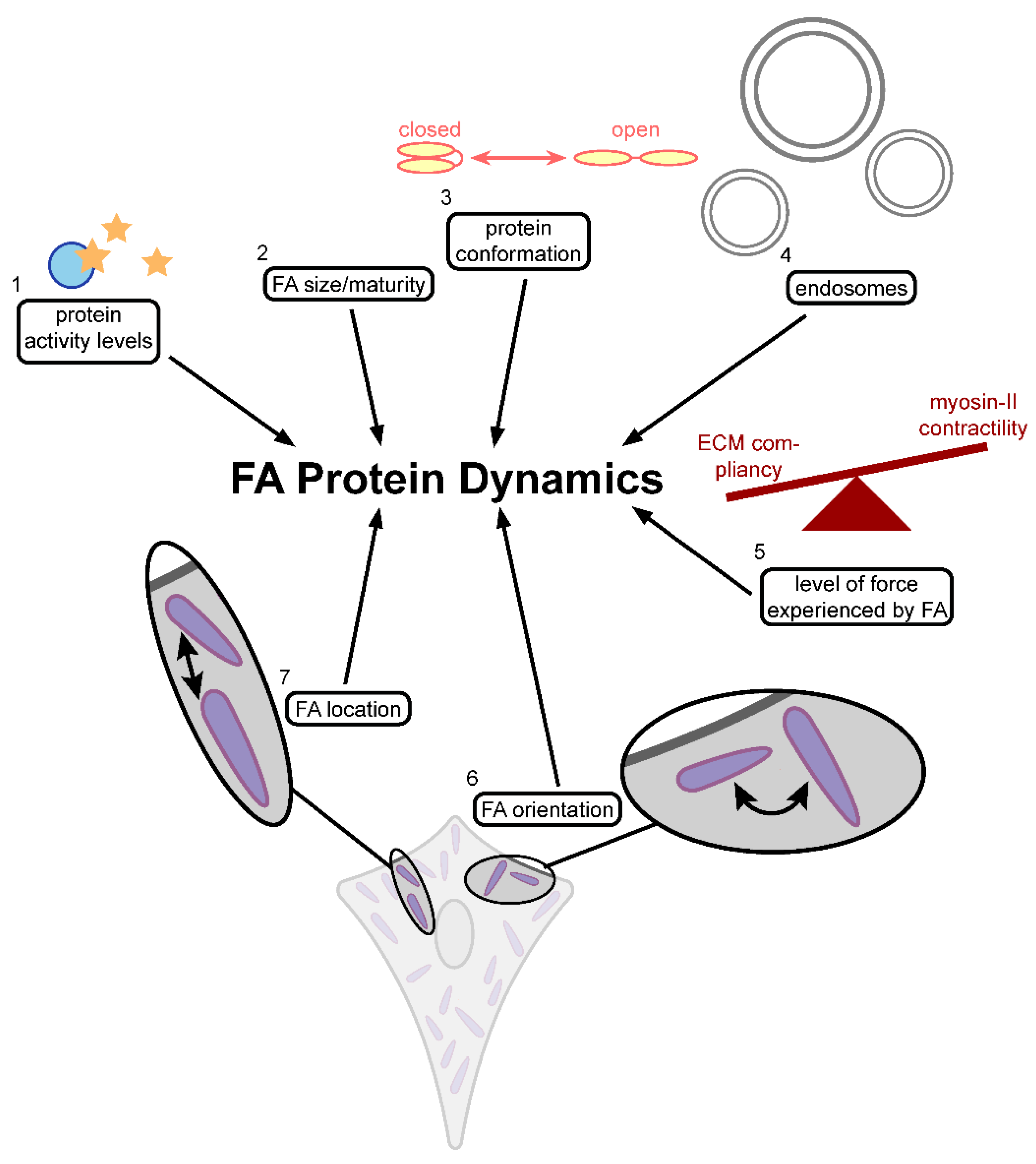
4.1. FAK Activity and the Dynamics of Focal Adhesion Protein
4.2. The Effects of Force
4.3. The Effect of Protein Conformation and FA Size and Maturity
4.4. Endosomes and Paxillin Dynamics
4.5. Relative Dynamics of FA Proteins
4.6. FA Orientation and Location and Its Protein Dynamics
4.7. Paxillin and Vinculin versus Zyxin and VASP—A Recurring Theme
5. Protein Organisation within FAs
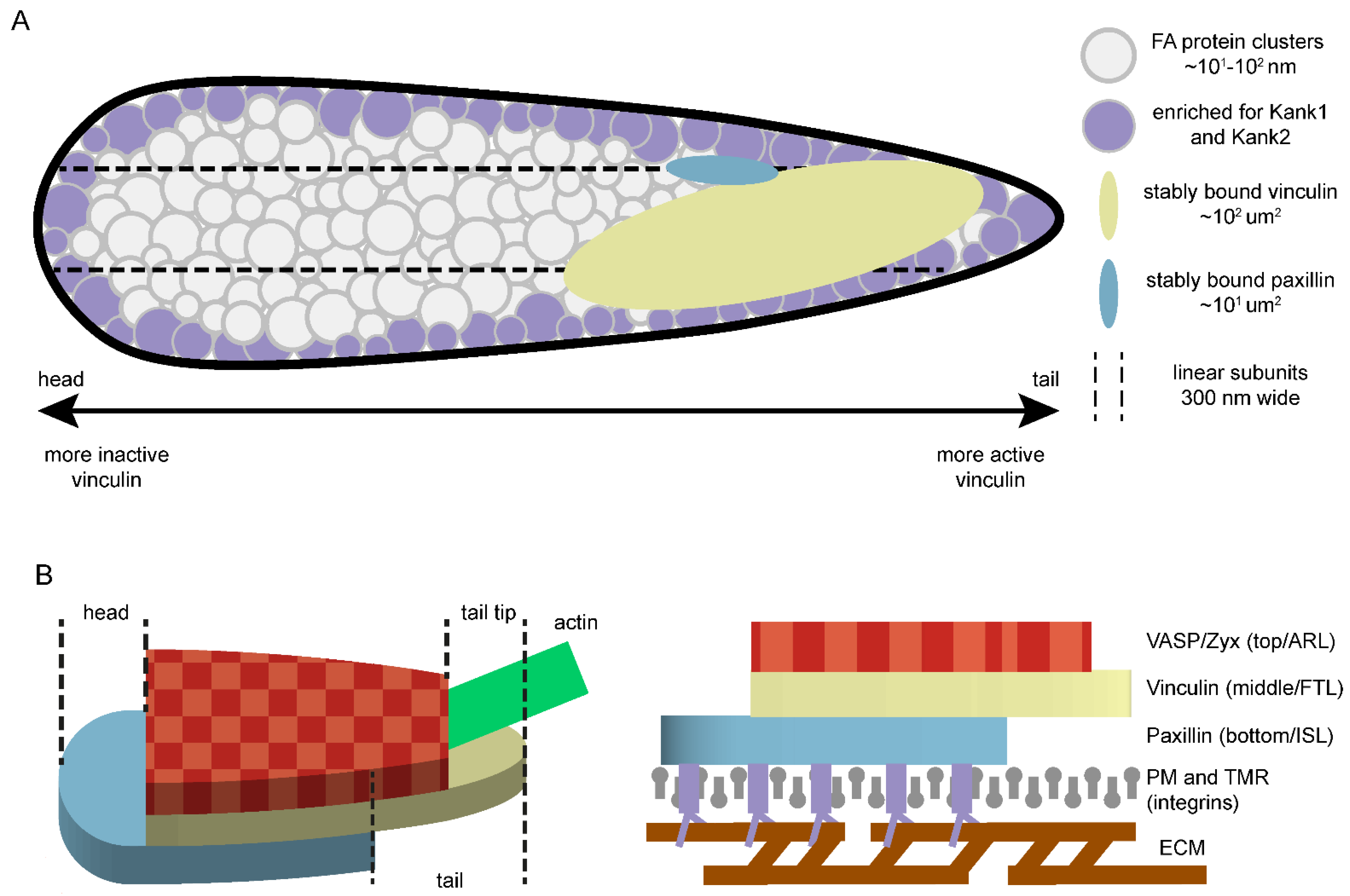
5.1. The Distribution of Focal Adhesion Proteins at the Nanoscale
5.2. Heterogeneous Distribution of Protein Activity or Binding Dynamics within FAs
5.3. FA Proteins Are Not Strictly Separated into Layers
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pollard, T.D. Actin and Actin-Binding Proteins. Cold Spring Harb Perspect. Biol. 2016, 8, a018226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burridge, K.; Guilluy, C. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 2016, 343, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazarides, E.; Burridge, K. Alpha-actinin: Immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell 1975, 6, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Cramer, L.P.; Siebert, M.; Mitchison, T.J. Identification of novel graded polarity actin filament bundles in locomoting heart fibroblasts: Implications for the generation of motile force. J. Cell Biol. 1997, 136, 1287–1305. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.C. Formation of stable microspikes containing actin and the 55 kDa actin bundling protein, fascin, is a consequence of cell adhesion to thrombospondin-1: Implications for the anti-adhesive activities of thrombospondin-1. J. Cell Sci. 1995, 108 Pt 5, 1977–1990. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ash, J.F.; Singer, S.J. Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc. Natl. Acad. Sci. USA 1975, 72, 4483–4486. [Google Scholar] [CrossRef] [Green Version]
- Lehtimäki, J.I.; Rajakylä, E.K.; Tojkander, S.; Lappalainen, P. Generation of stress fibers through myosin-driven reorganization of the actin cortex. eLife 2021, 10, e60710. [Google Scholar] [CrossRef] [PubMed]
- Small, J.V.; Rottner, K.; Kaverina, I.; Anderson, K.I. Assembling an actin cytoskeleton for cell attachment and movement. Biochim. Biophys. Acta 1998, 1404, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Burnette, D.T.; Shao, L.; Ott, C.; Pasapera, A.M.; Fischer, R.S.; Baird, M.A.; Der Loughian, C.; Delanoe-Ayari, H.; Paszek, M.J.; Davidson, M.W.; et al. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J. Cell Biol. 2014, 205, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Balaban, N.Q.; Schwarz, U.S.; Riveline, D.; Goichberg, P.; Tzur, G.; Sabanay, I.; Mahalu, D.; Safran, S.; Bershadsky, A.; Addadi, L.; et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001, 3, 466–472. [Google Scholar] [CrossRef]
- Beningo, K.A.; Dembo, M.; Kaverina, I.; Small, J.V.; Wang, Y.L. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 2001, 153, 881–888. [Google Scholar] [CrossRef] [Green Version]
- Tadokoro, S.; Shattil, S.J.; Eto, K.; Tai, V.; Liddington, R.C.; de Pereda, J.M.; Ginsberg, M.H.; Calderwood, D.A. Talin binding to integrin beta tails: A final common step in integrin activation. Science 2003, 302, 103–106. [Google Scholar] [CrossRef]
- Chrzanowska-Wodnicka, M.; Burridge, K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 1996, 133, 1403–1415. [Google Scholar] [CrossRef] [Green Version]
- Riveline, D.; Zamir, E.; Balaban, N.Q.; Schwarz, U.S.; Ishizaki, T.; Narumiya, S.; Kam, Z.; Geiger, B.; Bershadsky, A.D. Focal contacts as mechanosensors: Externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 2001, 153, 1175–1186. [Google Scholar] [CrossRef]
- Wang, H.B.; Dembo, M.; Hanks, S.K.; Wang, Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc. Natl. Acad. Sci. USA 2001, 98, 11295–11300. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, C.G.; Yamada, K.M.; Sheetz, M.P. The relationship between force and focal complex development. J. Cell Biol. 2002, 159, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Kaverina, I.; Krylyshkina, O.; Beningo, K.; Anderson, K.; Wang, Y.L.; Small, J.V. Tensile stress stimulates microtubule outgrowth in living cells. J. Cell Sci. 2002, 115, 2283–2291. [Google Scholar] [CrossRef] [PubMed]
- Pasapera, A.M.; Schneider, I.C.; Rericha, E.; Schlaepfer, D.D.; Waterman, C.M. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 2010, 188, 877–890. [Google Scholar] [CrossRef] [Green Version]
- Winograd-Katz, S.E.; Fassler, R.; Geiger, B.; Legate, K.R. The integrin adhesome: From genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P. Cell adhesion in development: A complex signaling network. Curr. Opin. Genet. Dev. 2003, 13, 365–371. [Google Scholar] [CrossRef]
- Maartens, A.P.; Brown, N.H. The many faces of cell adhesion during Drosophila muscle development. Dev. Biol. 2015, 401, 62–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahl, S.M.; Feldman, G.M.; McCarthy, J.B. Regulation of leukocyte adhesion and signaling in inflammation and disease. J. Leukoc. Biol. 1996, 59, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.K.; Schlaepfer, D.D. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006, 18, 516–523. [Google Scholar] [CrossRef]
- Bianchi-Smiraglia, A.; Paesante, S.; Bakin, A.V. Integrin beta5 contributes to the tumorigenic potential of breast cancer cells through the Src-FAK and MEK-ERK signaling pathways. Oncogene 2013, 32, 3049–3058. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.K.; Shields, D.J.; Murphy, E.A.; Desgrosellier, J.S.; Anand, S.; Huang, M.; Kato, S.; Lim, S.T.; Weis, S.M.; Stupack, D.G.; et al. EGFR-mediated carcinoma cell metastasis mediated by integrin alphavbeta5 depends on activation of c-Src and cleavage of MUC1. PLoS ONE 2012, 7, e36753. [Google Scholar] [CrossRef]
- Bianconi, D.; Unseld, M.; Prager, G.W. Integrins in the Spotlight of Cancer. Int. J. Mol. Sci. 2016, 17, 2037. [Google Scholar] [CrossRef] [Green Version]
- Ata, R.; Antonescu, C.N. Integrins and Cell Metabolism: An Intimate Relationship Impacting Cancer. Int. J. Mol. Sci. 2017, 18, 189. [Google Scholar] [CrossRef] [Green Version]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Zhang, J.; Hu, G.; Liu, L.; Liang, W. Hypoxia and TGF-β1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation. Cancer Cell Int. 2017, 17, 54. [Google Scholar] [CrossRef]
- Eckert, M.A.; Santiago-Medina, M.; Lwin, T.M.; Kim, J.; Courtneidge, S.A.; Yang, J. ADAM12 induction by Twist1 promotes tumor invasion and metastasis via regulation of invadopodia and focal adhesions. J. Cell Sci. 2017, 130, 2036–2048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wipff, P.-J.; Hinz, B. Integrins and the activation of latent transforming growth factor beta1—An intimate relationship. Eur. J. Cell Biol. 2008, 87, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Bianchi-Smiraglia, A.; Kunnev, D.; Limoge, M.; Lee, A.; Beckerle, M.C.; Bakin, A.V. Integrin-beta5 and zyxin mediate formation of ventral stress fibers in response to transforming growth factor beta. Cell Cycle 2013, 12, 3377–3389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchi, A.; Gervasi, M.E.; Bakin, A. Role of beta5-integrin in epithelial-mesenchymal transition in response to TGF-beta. Cell Cycle 2010, 9, 1647–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.M.; Geiger, B. Molecular interactions in cell adhesion complexes. Curr. Opin. Cell Biol. 1997, 9, 76–85. [Google Scholar] [CrossRef]
- Geiger, B.; Bershadsky, A.; Pankov, R.; Yamada, K.M. Transmembrane crosstalk between the extracellular matrix cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001, 2, 793–805. [Google Scholar] [CrossRef]
- Kanchanawong, P.; Shtengel, G.; Pasapera, A.M.; Ramko, E.B.; Davidson, M.W.; Hess, H.F.; Waterman, C.M. Nanoscale architecture of integrin-based cell adhesions. Nature 2010, 468, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-H.; Wirtz, D. Focal adhesion size uniquely predicts cell migration. FASEB J. 2013, 27, 1351–1361. [Google Scholar] [CrossRef]
- Orré, T.; Rossier, O.; Giannone, G. The inner life of integrin adhesion sites: From single molecules to functional macromolecular complexes. Exp. Cell Res. 2019, 379, 235–244. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Goh, W.I.; Goh, H.; Baird, M.A.; Ruehland, S.; Teo, S.; Bate, N.; Critchley, D.R.; Davidson, M.W.; et al. Talin determines the nanoscale architecture of focal adhesions. Proc. Natl. Acad. Sci. USA 2015, 112, E4864–E4873. [Google Scholar] [CrossRef] [Green Version]
- Paszek, M.J.; DuFort, C.C.; Rubashkin, M.G.; Davidson, M.W.; Thorn, K.S.; Liphardt, J.T.; Weaver, V.M. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nat. Methods 2012, 9, 825–827. [Google Scholar] [CrossRef]
- Stubb, A.; Guzman, C.; Narva, E.; Aaron, J.; Chew, T.L.; Saari, M.; Miihkinen, M.; Jacquemet, G.; Ivaska, J. Superresolution architecture of cornerstone focal adhesions in human pluripotent stem cells. Nat. Commun. 2019, 10, 4756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, L.B.; Baird, M.A.; Shtengel, G.; Campbell, S.L.; Hess, H.F.; Davidson, M.W.; Waterman, C.M. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 2015, 17, 880–892. [Google Scholar] [CrossRef] [Green Version]
- Pedrosa, A.T.; Murphy, K.N.; Nogueira, A.T.; Brinkworth, A.J.; Thwaites, T.R.; Aaron, J.; Chew, T.L.; Carabeo, R.A. A post-invasion role for Chlamydia type III effector TarP in modulating the dynamics and organization of host cell focal adhesions. J. Biol. Chem. 2020, 295, 14763–14779. [Google Scholar] [CrossRef]
- Orré, T.; Joly, A.; Karatas, Z.; Kastberger, B.; Cabriel, C.; Böttcher, R.T.; Lévêque-Fort, S.; Sibarita, J.B.; Fässler, R.; Wehrle-Haller, B.; et al. Molecular motion and tridimensional nanoscale localization of kindlin control integrin activation in focal adhesions. Nat. Commun. 2021, 12, 3104. [Google Scholar] [CrossRef]
- Dong, J.M.; Tay, F.P.; Swa, H.L.; Gunaratne, J.; Leung, T.; Burke, B.; Manser, E. Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Sci. Signal. 2016, 9, rs4. [Google Scholar] [CrossRef] [PubMed]
- Chastney, M.R.; Lawless, C.; Humphries, J.D.; Warwood, S.; Jones, M.C.; Knight, D.; Jorgensen, C.; Humphries, M.J. Topological features of integrin adhesion complexes revealed by multiplexed proximity biotinylation. J. Cell Biol. 2020, 219, e202003038. [Google Scholar] [CrossRef]
- Wu, C.; Keivens, V.M.; O'Toole, T.E.; McDonald, J.A.; Ginsberg, M.H. Integrin activation and cytoskeletal interaction are essential for the assembly of a fibronectin matrix. Cell 1995, 83, 715–724. [Google Scholar] [CrossRef] [Green Version]
- Kechagia, J.Z.; Ivaska, J.; Roca-Cusachs, P. Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol. 2019, 20, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Shattil, S.J.; Kim, C.; Ginsberg, M.H. The final steps of integrin activation: The end game. Nat. Rev. Mol. Cell Biol. 2010, 11, 288–300. [Google Scholar] [CrossRef] [Green Version]
- Laukaitis, C.M.; Webb, D.J.; Donais, K.; Horwitz, A.F. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 2001, 153, 1427–1440. [Google Scholar] [CrossRef] [Green Version]
- Webb, D.J.; Donais, K.; Whitmore, L.A.; Thomas, S.M.; Turner, C.E.; Parsons, J.T.; Horwitz, A.F. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004, 6, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, P.W.; Brown, C.M.; Webb, D.J.; Hebert, B.; Johnson, N.L.; Squier, J.A.; Ellisman, M.H.; Horwitz, A.F. Spatial mapping of integrin interactions and dynamics during cell migration by image correlation microscopy. J. Cell Sci. 2004, 117, 5521–5534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, C.K.; Vicente-Manzanares, M.; Zareno, J.; Whitmore, L.A.; Mogilner, A.; Horwitz, A.R. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008, 10, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.D.; Kiosses, W.B.; Sieg, D.J.; Otey, C.A.; Schlaepfer, D.D.; Schwartz, M.A. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J. Cell Sci. 2000, 113 Pt 20, 3673–3678. [Google Scholar] [CrossRef]
- Gupton, S.L.; Waterman-Storer, C.M. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 2006, 125, 1361–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaidel-Bar, R.; Milo, R.; Kam, Z.; Geiger, B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 2007, 120, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Legerstee, K.; Geverts, B.; Slotman, J.A.; Houtsmuller, A.B. Dynamics and distribution of paxillin, vinculin, zyxin and VASP depend on focal adhesion location and orientation. Sci. Rep. 2019, 9, 10460. [Google Scholar] [CrossRef] [Green Version]
- Zaidel-Bar, R.; Itzkovitz, S.; Ma'ayan, A.; Iyengar, R.; Geiger, B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 2007, 9, 858–867. [Google Scholar] [CrossRef]
- Berrier, A.L.; Yamada, K.M. Cell-matrix adhesion. J. Cell. Physiol. 2007, 213, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Yamada, K.M. Molecular architecture and function of matrix adhesions. Cold Spring Harb. Perspect. Biol. 2011, 3, a005033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavelin, I.; Wolfenson, H.; Patla, I.; Henis, Y.I.; Medalia, O.; Volberg, T.; Livne, A.; Kam, Z.; Geiger, B. Differential effect of actomyosin relaxation on the dynamic properties of focal adhesion proteins. PLoS ONE 2013, 8, e73549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, J.-C.; Han, X.; Hsiao, C.-T.; Yates, J.R., 3rd; Waterman, C.M. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 2011, 13, 383–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiller, H.B.; Friedel, C.C.; Boulegue, C.; Fässler, R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011, 12, 259–266. [Google Scholar] [CrossRef] [Green Version]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef]
- Adutler-Lieber, S.; Zaretsky, I.; Platzman, I.; Deeg, J.; Friedman, N.; Spatz, J.P.; Geiger, B. Engineering of synthetic cellular microenvironments: Implications for immunity. J. Autoimmun. 2014, 54, 100–111. [Google Scholar] [CrossRef]
- Chautard, E.; Fatoux-Ardore, M.; Ballut, L.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011, 39, D235–D240. [Google Scholar] [CrossRef]
- Doyle, A.D.; Carvajal, N.; Jin, A.; Matsumoto, K.; Yamada, K.M. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat. Commun. 2015, 6, 8720. [Google Scholar] [CrossRef] [Green Version]
- Singer, I.I.; Kawka, D.W.; Kazazis, D.M.; Clark, R.A. In vivo co-distribution of fibronectin and actin fibers in granulation tissue: Immunofluorescence and electron microscope studies of the fibronexus at the myofibroblast surface. J. Cell Biol. 1984, 98, 2091–2106. [Google Scholar] [CrossRef] [Green Version]
- Gunawan, F.; Gentile, A.; Fukuda, R.; Tsedeke, A.T.; Jiménez-Amilburu, V.; Ramadass, R.; Iida, A.; Sehara-Fujisawa, A.; Stainier, D.Y.R. Focal adhesions are essential to drive zebrafish heart valve morphogenesis. J. Cell Biol. 2019, 218, 1039–1054. [Google Scholar] [CrossRef]
- Fischer, R.S.; Lam, P.-Y.; Huttenlocher, A.; Waterman, C.M. Filopodia and focal adhesions: An integrated system driving branching morphogenesis in neuronal pathfinding and angiogenesis. Dev. Biol. 2019, 451, 86–95. [Google Scholar] [CrossRef]
- Yue, J.; Zhang, Y.; Liang, W.G.; Gou, X.; Lee, P.; Liu, H.; Lyu, W.; Tang, W.-J.; Chen, S.-Y.; Yang, F.; et al. In vivo epidermal migration requires focal adhesion targeting of ACF7. Nat. Commun. 2016, 7, 11692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haage, A.; Goodwin, K.; Whitewood, A.; Camp, D.; Bogutz, A.; Turner, C.T.; Granville, D.J.; Lefebvre, L.; Plotnikov, S.; Goult, B.T.; et al. Talin Autoinhibition Regulates Cell-ECM Adhesion Dynamics and Wound Healing In Vivo. Cell Rep. 2018, 25, 2401–2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaller, M.D.; Otey, C.A.; Hildebrand, J.D.; Parsons, J.T. Focal adhesion kinase and paxillin bind to peptides mimicking beta integrin cytoplasmic domains. J. Cell Biol. 1995, 130, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yamaguchi, R.; Sabe, H.; Sekiguchi, K.; Healy, J.M. Paxillin association in vitro with integrin cytoplasmic domain peptides. FEBS Lett. 1996, 399, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Thomas, S.M.; Woodside, D.G.; Rose, D.M.; Kiosses, W.B.; Pfaff, M.; Ginsberg, M.H. Binding of paxillin to alpha4 integrins modifies integrin-dependent biological responses. Nature 1999, 402, 676–681. [Google Scholar] [CrossRef]
- Young, B.A.; Taooka, Y.; Liu, S.; Askins, K.J.; Yokosaki, Y.; Thomas, S.M.; Sheppard, D. The cytoplasmic domain of the integrin alpha9 subunit requires the adaptor protein paxillin to inhibit cell spreading but promotes cell migration in a paxillin-independent manner. Mol. Biol. Cell 2001, 12, 3214–3225. [Google Scholar] [CrossRef]
- Brown, M.C.; Perrotta, J.A.; Turner, C.E. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 1996, 135, 1109–1123. [Google Scholar] [CrossRef] [Green Version]
- Green, H.J.; Brown, N.H. Integrin intracellular machinery in action. Exp. Cell Res. 2019, 378, 226–231. [Google Scholar] [CrossRef]
- Deakin, N.O.; Turner, C.E. Paxillin comes of age. J. Cell Sci. 2008, 121, 2435–2444. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Yi, Q.; Tang, L. The roles of nuclear focal adhesion kinase (FAK) on Cancer: A focused review. J. Exp. Clin. Cancer Res. 2019, 38, 250. [Google Scholar] [CrossRef] [Green Version]
- Schaller, M.D.; Parsons, J.T. pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for Crk. Mol. Cell. Biol. 1995, 15, 2635–2645. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.C.; Appeddu, P.A.; Parsons, J.T.; Hildebrand, J.D.; Schaller, M.D.; Guan, J.L. Interaction of focal adhesion kinase with cytoskeletal protein talin. J. Biol. Chem. 1995, 270, 16995–16999. [Google Scholar] [CrossRef] [Green Version]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef]
- Bellis, S.L.; Miller, J.T.; Turner, C.E. Characterization of tyrosine phosphorylation of paxillin in vitro by focal adhesion kinase. J. Biol. Chem. 1995, 270, 17437–17441. [Google Scholar] [CrossRef] [Green Version]
- Richardson, A.; Malik, R.K.; Hildebrand, J.D.; Parsons, J.T. Inhibition of cell spreading by expression of the C-terminal domain of focal adhesion kinase (FAK) is rescued by coexpression of Src or catalytically inactive FAK: A role for paxillin tyrosine phosphorylation. Mol. Cell. Biol. 1997, 17, 6906–6914. [Google Scholar] [CrossRef] [Green Version]
- Tsubouchi, A.; Sakakura, J.; Yagi, R.; Mazaki, Y.; Schaefer, E.; Yano, H.; Sabe, H. Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J. Cell Biol. 2002, 159, 673–683. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.C.; Turner, C.E. Paxillin: Adapting to change. Physiol. Rev. 2004, 84, 1315–1339. [Google Scholar] [CrossRef]
- Drees, B.; Friederich, E.; Fradelizi, J.; Louvard, D.; Beckerle, M.C.; Golsteyn, R.M. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 2000, 275, 22503–22511. [Google Scholar] [CrossRef] [Green Version]
- Reinhard, M.; Jouvenal, K.; Tripier, D.; Walter, U. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein). Proc. Natl. Acad. Sci. USA 1995, 92, 7956–7960. [Google Scholar] [CrossRef] [Green Version]
- Hirata, H.; Tatsumi, H.; Sokabe, M. Mechanical forces facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J. Cell Sci. 2008, 121, 2795–2804. [Google Scholar] [CrossRef] [Green Version]
- Uemura, A.; Nguyen, T.N.; Steele, A.N.; Yamada, S. The LIM domain of zyxin is sufficient for force-induced accumulation of zyxin during cell migration. Biophys. J. 2011, 101, 1069–1075. [Google Scholar] [CrossRef] [Green Version]
- Drees, B.E.; Andrews, K.M.; Beckerle, M.C. Molecular dissection of zyxin function reveals its involvement in cell motility. J. Cell Biol. 1999, 147, 1549–1560. [Google Scholar] [CrossRef] [Green Version]
- Crawford, A.W.; Michelsen, J.W.; Beckerle, M.C. An interaction between zyxin and alpha-actinin. J. Cell Biol. 1992, 116, 1381–1393. [Google Scholar] [CrossRef]
- Bear, J.E.; Gertler, F.B. Ena/VASP: Towards resolving a pointed controversy at the barbed end. J. Cell Sci. 2009, 122, 1947–1953. [Google Scholar] [CrossRef] [Green Version]
- Yoshigi, M.; Hoffman, L.M.; Jensen, C.C.; Yost, H.J.; Beckerle, M.C. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 2005, 171, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, L.M.; Jensen, C.C.; Kloeker, S.; Wang, C.L.; Yoshigi, M.; Beckerle, M.C. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 2006, 172, 771–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golsteyn, R.M.; Beckerle, M.C.; Koay, T.; Friederich, E. Structural and functional similarities between the human cytoskeletal protein zyxin and the ActA protein of Listeria monocytogenes. J. Cell Sci. 1997, 110 Pt 16, 1893–1906. [Google Scholar] [CrossRef]
- Nix, D.A.; Fradelizi, J.; Bockholt, S.; Menichi, B.; Louvard, D.; Friederich, E.; Beckerle, M.C. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 2001, 276, 34759–34767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fradelizi, J.; Noireaux, V.; Plastino, J.; Menichi, B.; Louvard, D.; Sykes, C.; Golsteyn, R.M.; Friederich, E. ActA and human zyxin harbour Arp2/3-independent actin-polymerization activity. Nat. Cell Biol. 2001, 3, 699–707. [Google Scholar] [CrossRef]
- Brühmann, S.; Ushakov, D.S.; Winterhoff, M.; Dickinson, R.B.; Curth, U.; Faix, J. Distinct VASP tetramers synergize in the processive elongation of individual actin filaments from clustered arrays. Proc. Natl. Acad. Sci. USA 2017, 114, E5815–E5824. [Google Scholar] [CrossRef] [Green Version]
- Rottner, K.; Krause, M.; Gimona, M.; Small, J.V.; Wehland, J. Zyxin is not colocalized with vasodilator-stimulated phosphoprotein (VASP) at lamellipodial tips and exhibits different dynamics to vinculin, paxillin, and VASP in focal adhesions. Mol. Biol. Cell 2001, 12, 3103–3113. [Google Scholar] [CrossRef]
- Furman, C.; Sieminski, A.L.; Kwiatkowski, A.V.; Rubinson, D.A.; Vasile, E.; Bronson, R.T.; Fassler, R.; Gertler, F.B. Ena/VASP is required for endothelial barrier function in vivo. J. Cell Biol. 2007, 179, 761–775. [Google Scholar] [CrossRef]
- Hirata, H.; Tatsumi, H.; Sokabe, M. Zyxin emerges as a key player in the mechanotransduction at cell adhesive structures. Commun. Integr. 2008, 1, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.A.; Blankman, E.; Gardel, M.L.; Luettjohann, L.; Waterman, C.M.; Beckerle, M.C. A zyxin-mediated mechanism for actin stress fiber maintenance and repair. Dev. Cell 2010, 19, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Calderwood, D.A.; Zent, R.; Grant, R.; Rees, D.J.; Hynes, R.O.; Ginsberg, M.H. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 1999, 274, 28071–28074. [Google Scholar] [CrossRef] [Green Version]
- Goldmann, W.H.; Guttenberg, Z.; Kaufmann, S.; Hess, D.; Ezzell, R.M.; Isenberg, G. Examining F-actin interaction with intact talin and talin head and tail fragment using static and dynamic light scattering. Eur. J. Biochem. 1997, 250, 447–450. [Google Scholar] [CrossRef] [Green Version]
- Critchley, D.R. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 2009, 38, 235–254. [Google Scholar] [CrossRef]
- Ciobanasu, C.; Faivre, B.; Le Clainche, C. Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nat. Commun. 2014, 5, 3095. [Google Scholar] [CrossRef] [Green Version]
- Del Rio, A.; Perez-Jimenez, R.; Liu, R.; Roca-Cusachs, P.; Fernandez, J.M.; Sheetz, M.P. Stretching single talin rod molecules activates vinculin binding. Science 2009, 323, 638–641. [Google Scholar] [CrossRef]
- Yao, M.; Goult, B.T.; Chen, H.; Cong, P.; Sheetz, M.P.; Yan, J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 2014, 4, 4610. [Google Scholar] [CrossRef] [Green Version]
- Humphries, J.D.; Wang, P.; Streuli, C.; Geiger, B.; Humphries, M.J.; Ballestrem, C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 2007, 179, 1043–1057. [Google Scholar] [CrossRef] [Green Version]
- Bakolitsa, C.; Cohen, D.M.; Bankston, L.A.; Bobkov, A.A.; Cadwell, G.W.; Jennings, L.; Critchley, D.R.; Craig, S.W.; Liddington, R.C. Structural basis for vinculin activation at sites of cell adhesion. Nature 2004, 430, 583–586. [Google Scholar] [CrossRef]
- Jones, P.; Jackson, P.; Price, G.J.; Patel, B.; Ohanion, V.; Lear, A.L.; Critchley, D.R. Identification of a talin binding site in the cytoskeletal protein vinculin. J. Cell Biol. 1989, 109, 2917–2927. [Google Scholar] [CrossRef] [Green Version]
- Hagel, M.; George, E.L.; Kim, A.; Tamimi, R.; Opitz, S.L.; Turner, C.E.; Imamoto, A.; Thomas, S.M. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell. Biol. 2002, 22, 901–915. [Google Scholar] [CrossRef] [Green Version]
- Opazo Saez, A.; Zhang, W.; Wu, Y.; Turner, C.E.; Tang, D.D.; Gunst, S.J. Tension development during contractile stimulation of smooth muscle requires recruitment of paxillin and vinculin to the membrane. Am. J. Physiol. Cell Physiol. 2004, 286, C433–C447. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.M.; Hagel, M.; Turner, C.E. Characterization of a focal adhesion protein, Hic-5, that shares extensive homology with paxillin. J. Cell Sci. 1999, 112 Pt 2, 181–190. [Google Scholar] [CrossRef]
- Gertler, F.B.; Niebuhr, K.; Reinhard, M.; Wehland, J.; Soriano, P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 1996, 87, 227–239. [Google Scholar] [CrossRef] [Green Version]
- Brindle, N.P.; Holt, M.R.; Davies, J.E.; Price, C.J.; Critchley, D.R. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 1996, 318 Pt 3, 753–757. [Google Scholar] [CrossRef] [Green Version]
- Reinhard, M.; Rudiger, M.; Jockusch, B.M.; Walter, U. VASP interaction with vinculin: A recurring theme of interactions with proline-rich motifs. FEBS Lett. 1996, 399, 103–107. [Google Scholar] [CrossRef] [Green Version]
- McGregor, A.; Blanchard, A.D.; Rowe, A.J.; Critchley, D.R. Identification of the vinculin-binding site in the cytoskeletal protein alpha-actinin. Biochem. J. 1994, 301 Pt 1, 225–233. [Google Scholar] [CrossRef]
- Xu, W.; Baribault, H.; Adamson, E.D. Vinculin knockout results in heart and brain defects during embryonic development. Development 1998, 125, 327–337. [Google Scholar] [CrossRef]
- Ilić, D.; Furuta, Y.; Kanazawa, S.; Takeda, N.; Sobue, K.; Nakatsuji, N.; Nomura, S.; Fujimoto, J.; Okada, M.; Yamamoto, T. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 1995, 377, 539–544. [Google Scholar] [CrossRef]
- Monkley, S.J.; Zhou, X.H.; Kinston, S.J.; Giblett, S.M.; Hemmings, L.; Priddle, H.; Brown, J.E.; Pritchard, C.A.; Critchley, D.R.; Fässler, R. Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev. Dyn. 2000, 219, 560–574. [Google Scholar] [CrossRef]
- Hoffman, L.M.; Nix, D.A.; Benson, B.; Boot-Hanford, R.; Gustafsson, E.; Jamora, C.; Menzies, A.S.; Goh, K.L.; Jensen, C.C.; Gertler, F.B.; et al. Targeted disruption of the murine zyxin gene. Mol. Cell. Biol. 2003, 23, 70–79. [Google Scholar] [CrossRef] [Green Version]
- Hauser, W.; Knobeloch, K.P.; Eigenthaler, M.; Gambaryan, S.; Krenn, V.; Geiger, J.; Glazova, M.; Rohde, E.; Horak, I.; Walter, U.; et al. Megakaryocyte hyperplasia and enhanced agonist-induced platelet activation in vasodilator-stimulated phosphoprotein knockout mice. Proc. Natl. Acad. Sci. USA 1999, 96, 8120–8125. [Google Scholar] [CrossRef] [Green Version]
- Shen, T.L.; Park, A.Y.; Alcaraz, A.; Peng, X.; Jang, I.; Koni, P.; Flavell, R.A.; Gu, H.; Guan, J.L. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J. Cell Biol. 2005, 169, 941–952. [Google Scholar] [CrossRef] [Green Version]
- Volberg, T.; Geiger, B.; Kam, Z.; Pankov, R.; Simcha, I.; Sabanay, H.; Coll, J.L.; Adamson, E.; Ben-Ze’ev, A. Focal adhesion formation by F9 embryonal carcinoma cells after vinculin gene disruption. J. Cell Sci. 1995, 108 Pt 6, 2253–2260. [Google Scholar] [CrossRef]
- Xu, W.; Coll, J.L.; Adamson, E.D. Rescue of the mutant phenotype by reexpression of full-length vinculin in null F9 cells; effects on cell locomotion by domain deleted vinculin. J. Cell Sci. 1998, 111 Pt 11, 1535–1544. [Google Scholar] [CrossRef]
- Mierke, C.T.; Kollmannsberger, P.; Zitterbart, D.P.; Diez, G.; Koch, T.M.; Marg, S.; Ziegler, W.H.; Goldmann, W.H.; Fabry, B. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J. Biol. Chem. 2010, 285, 13121–13130. [Google Scholar] [CrossRef] [Green Version]
- Thievessen, I.; Thompson, P.M.; Berlemont, S.; Plevock, K.M.; Plotnikov, S.V.; Zemljic-Harpf, A.; Ross, R.S.; Davidson, M.W.; Danuser, G.; Campbell, S.L.; et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 2013, 202, 163–177. [Google Scholar] [CrossRef] [Green Version]
- Damiano-Guercio, J.; Kurzawa, L.; Mueller, J.; Dimchev, G.; Schaks, M.; Nemethova, M.; Pokrant, T.; Brühmann, S.; Linkner, J.; Blanchoin, L.; et al. Loss of Ena/VASP interferes with lamellipodium architecture, motility and integrin-dependent adhesion. eLife 2020, 9. [Google Scholar] [CrossRef]
- Wang, Y.; Gilmore, T.D. Zyxin and paxillin proteins: Focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta 2003, 1593, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Bubb, M.R.; Spector, I.; Beyer, B.B.; Fosen, K.M. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 2000, 275, 5163–5170. [Google Scholar] [CrossRef] [Green Version]
- Amsellem, V.; Kryszke, M.H.; Hervy, M.; Subra, F.; Athman, R.; Leh, H.; Brachet-Ducos, C.; Auclair, C. The actin cytoskeleton-associated protein zyxin acts as a tumor suppressor in Ewing tumor cells. Exp. Cell Res. 2005, 304, 443–456. [Google Scholar] [CrossRef]
- Sanchez-Carbayo, M.; Socci, N.D.; Charytonowicz, E.; Lu, M.; Prystowsky, M.; Childs, G.; Cordon-Cardo, C. Molecular profiling of bladder cancer using cDNA microarrays: Defining histogenesis and biological phenotypes. Cancer Res. 2002, 62, 6973–6980. [Google Scholar]
- Wang, W.; Huper, G.; Guo, Y.; Murphy, S.K.; Olson, J.A., Jr.; Marks, J.R. Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene 2005, 24, 2705–2714. [Google Scholar] [CrossRef] [Green Version]
- Nix, D.A.; Beckerle, M.C. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: A potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 1997, 138, 1139–1147. [Google Scholar] [CrossRef]
- Sen, A.; De Castro, I.; DeFranco, D.B.; Deng, F.-M.; Melamed, J.; Kapur, P.; Raj, G.V.; Rossi, R.; Hammes, S.R. Paxillin mediates extranuclear and intranuclear signaling in prostate cancer proliferation. J. Clin. Investig. 2012, 122, 2469–2481. [Google Scholar] [CrossRef]
- Cattaruzza, M.; Lattrich, C.; Hecker, M. Focal adhesion protein zyxin is a mechanosensitive modulator of gene expression in vascular smooth muscle cells. Hypertension 2004, 43, 726–730. [Google Scholar] [CrossRef] [Green Version]
- Lele, T.P.; Pendse, J.; Kumar, S.; Salanga, M.; Karavitis, J.; Ingber, D.E. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J. Cell. Physiol. 2006, 207, 187–194. [Google Scholar] [CrossRef]
- Woods, A.J.; Roberts, M.S.; Choudhary, J.; Barry, S.T.; Mazaki, Y.; Sabe, H.; Morley, S.J.; Critchley, D.R.; Norman, J.C. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J. Biol. Chem. 2002, 277, 6428–6437. [Google Scholar] [CrossRef] [Green Version]
- Woods, A.J.; Kantidakis, T.; Sabe, H.; Critchley, D.R.; Norman, J.C. Interaction of paxillin with poly(A)-binding protein 1 and its role in focal adhesion turnover and cell migration. Mol. Cell. Biol. 2005, 25, 3763–3773. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [Green Version]
- Sathe, A.R.; Shivashankar, G.V.; Sheetz, M.P. Nuclear transport of paxillin depends on focal adhesion dynamics and FAT domains. J. Cell Sci. 2016, 129, 1981–1988. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.T.; Chen, X.L.; Lim, Y.; Hanson, D.A.; Vo, T.T.; Howerton, K.; Larocque, N.; Fisher, S.J.; Schlaepfer, D.D.; Ilic, D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol. Cell 2008, 29, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Ceccarelli, D.F.; Song, H.K.; Poy, F.; Schaller, M.D.; Eck, M.J. Crystal structure of the FERM domain of focal adhesion kinase. J. Biol. Chem. 2006, 281, 252–259. [Google Scholar] [CrossRef] [Green Version]
- Ossovskaya, V.; Lim, S.T.; Ota, N.; Schlaepfer, D.D.; Ilic, D. FAK nuclear export signal sequences. FEBS Lett. 2008, 582, 2402–2406. [Google Scholar] [CrossRef] [Green Version]
- Axelrod, D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 1981, 89, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Stout, A.L.; Axelrod, D. Evanescent field excitation of fluorescence by epi-illumination microscopy. Appl. Opt. 1989, 28, 5237–5242. [Google Scholar] [CrossRef]
- Gustafsson, M.G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000, 198, 82–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schermelleh, L.; Heintzmann, R.; Leonhardt, H. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 2010, 190, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Walde, M.; Monypenny, J.; Heintzmann, R.; Jones, G.E.; Cox, S. Vinculin binding angle in podosomes revealed by high resolution microscopy. PLoS ONE 2014, 9, e88251. [Google Scholar] [CrossRef] [Green Version]
- Mortensen, K.I.; Churchman, L.S.; Spudich, J.A.; Flyvbjerg, H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods 2010, 7, 377–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deschout, H.; Cella Zanacchi, F.; Mlodzianoski, M.; Diaspro, A.; Bewersdorf, J.; Hess, S.T.; Braeckmans, K. Precisely and accurately localizing single emitters in fluorescence microscopy. Nat. Methods 2014, 11, 253–266. [Google Scholar] [CrossRef]
- Van Royen, M.E.; van Cappellen, W.A.; Geverts, B.; Schmidt, T.; Houtsmuller, A.B.; Schaaf, M.J. Androgen receptor complexes probe DNA for recognition sequences by short random interactions. J. Cell Sci. 2014, 127, 1406–1416. [Google Scholar] [CrossRef] [Green Version]
- Geverts, B.; van Royen, M.E.; Houtsmuller, A.B. Analysis of biomolecular dynamics by FRAP and computer simulation. Methods Mol. Biol. 2015, 1251, 109–133. [Google Scholar] [CrossRef]
- Axelrod, D.; Koppel, D.E.; Schlessinger, J.; Elson, E.; Webb, W.W. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976, 16, 1055–1069. [Google Scholar] [CrossRef] [Green Version]
- Carrero, G.; McDonald, D.; Crawford, E.; de Vries, G.; Hendzel, M.J. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods 2003, 29, 14–28. [Google Scholar] [CrossRef] [Green Version]
- Shcherbakova, D.M.; Verkhusha, V.V. Chromophore chemistry of fluorescent proteins controlled by light. Curr. Opin. Chem. Biol. 2014, 20, 60–68. [Google Scholar] [CrossRef] [Green Version]
- Lukyanov, K.A.; Chudakov, D.M.; Lukyanov, S.; Verkhusha, V.V. Innovation: Photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 2005, 6, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Patterson, G.H.; Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 2002, 297, 1873–1877. [Google Scholar] [CrossRef]
- Nienhaus, K.; Nienhaus, G.U. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 2014, 43, 1088–1106. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [Green Version]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–795. [Google Scholar] [CrossRef] [Green Version]
- Hess, S.T.; Girirajan, T.P.; Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006, 91, 4258–4272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdaasdonk, J.S.; Stephens, A.D.; Haase, J.; Bloom, K. Bending the rules: Widefield microscopy and the Abbe limit of resolution. J. Cell. Physiol. 2014, 229, 132–138. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Arena, E.T.; Rueden, C.T.; Hiner, M.C.; Wang, S.; Yuan, M.; Eliceiri, K.W. Quantitating the cell: Turning images into numbers with ImageJ. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e260. [Google Scholar] [CrossRef]
- Von Wichert, G.; Haimovich, B.; Feng, G.S.; Sheetz, M.P. Force-dependent integrin-cytoskeleton linkage formation requires downregulation of focal complex dynamics by Shp2. EMBO J. 2003, 22, 5023–5035. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekar, I.; Stradal, T.E.; Holt, M.R.; Entschladen, F.; Jockusch, B.M.; Ziegler, W.H. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 2005, 118, 1461–1472. [Google Scholar] [CrossRef] [Green Version]
- Cohen, D.M.; Kutscher, B.; Chen, H.; Murphy, D.B.; Craig, S.W. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 2006, 281, 16006–16015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lele, T.P.; Thodeti, C.K.; Pendse, J.; Ingber, D.E. Investigating complexity of protein-protein interactions in focal adhesions. Biochem. Biophys. Res. Commun. 2008, 369, 929–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohl, C.; Kirchgessner, N.; Schafer, C.; Kupper, K.; Born, S.; Diez, G.; Goldmann, W.H.; Merkel, R.; Hoffmann, B. Becoming stable and strong: The interplay between vinculin exchange dynamics and adhesion strength during adhesion site maturation. Cell Motil. Cytoskelet. 2009, 66, 350–364. [Google Scholar] [CrossRef]
- Wolfenson, H.; Lubelski, A.; Regev, T.; Klafter, J.; Henis, Y.I.; Geiger, B. A role for the juxtamembrane cytoplasm in the molecular dynamics of focal adhesions. PLoS ONE 2009, 4, e4304. [Google Scholar] [CrossRef]
- Horton, E.R.; Humphries, J.D.; Stutchbury, B.; Jacquemet, G.; Ballestrem, C.; Barry, S.T.; Humphries, M.J. Modulation of FAK and Src adhesion signaling occurs independently of adhesion complex composition. J. Cell Biol. 2016, 212, 349–364. [Google Scholar] [CrossRef] [Green Version]
- Wolfenson, H.; Bershadsky, A.; Henis, Y.I.; Geiger, B. Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J. Cell Sci. 2011, 124, 1425–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiefermeier, N.; Scheffler, J.M.; de Araujo, M.E.; Stasyk, T.; Yordanov, T.; Ebner, H.L.; Offterdinger, M.; Munck, S.; Hess, M.W.; Wickstrom, S.A.; et al. The late endosomal p14-MP1 (LAMTOR2/3) complex regulates focal adhesion dynamics during cell migration. J. Cell Biol. 2014, 205, 525–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feutlinske, F.; Browarski, M.; Ku, M.C.; Trnka, P.; Waiczies, S.; Niendorf, T.; Stallcup, W.B.; Glass, R.; Krause, E.; Maritzen, T. Stonin1 mediates endocytosis of the proteoglycan NG2 and regulates focal adhesion dynamics and cell motility. Nat. Commun. 2015, 6, 8535. [Google Scholar] [CrossRef] [Green Version]
- Le Devedec, S.E.; Geverts, B.; de Bont, H.; Yan, K.; Verbeek, F.J.; Houtsmuller, A.B.; van de Water, B. The residence time of focal adhesion kinase (FAK) and paxillin at focal adhesions in renal epithelial cells is determined by adhesion size, strength and life cycle status. J. Cell Sci. 2012, 125, 4498–4506. [Google Scholar] [CrossRef] [Green Version]
- Smilenov, L.B.; Mikhailov, A.; Pelham, R.J.; Marcantonio, E.E.; Gundersen, G.G. Focal adhesion motility revealed in stationary fibroblasts. Science 1999, 286, 1172–1174. [Google Scholar] [CrossRef] [PubMed]
- Zamir, E.; Katz, M.; Posen, Y.; Erez, N.; Yamada, K.M.; Katz, B.Z.; Lin, S.; Lin, D.C.; Bershadsky, A.; Kam, Z.; et al. Dynamics and segregation of cell-matrix adhesions in cultured fibroblasts. Nat. Cell Biol. 2000, 2, 191–196. [Google Scholar] [CrossRef]
- Linklater, E.S.; Duncan, E.D.; Han, K.J.; Kaupinis, A.; Valius, M.; Lyons, T.R.; Prekeris, R. Rab40-Cullin5 complex regulates EPLIN and actin cytoskeleton dynamics during cell migration. J. Cell Biol. 2021, 220, e202008060. [Google Scholar] [CrossRef]
- Schneeberger, P.E.; von Elsner, L.; Barker, E.L.; Meinecke, P.; Marquardt, I.; Alawi, M.; Steindl, K.; Joset, P.; Rauch, A.; Zwijnenburg, P.J.G.; et al. Bi-allelic Pathogenic Variants in HS2ST1 Cause a Syndrome Characterized by Developmental Delay and Corpus Callosum, Skeletal, and Renal Abnormalities. Am. J. Hum. Genet. 2020, 107, 1044–1061. [Google Scholar] [CrossRef]
- Barsony, J.; Marx, S.J. Immunocytology on microwave-fixed cells reveals rapid and agonist-specific changes in subcellular accumulation patterns for cAMP or cGMP. Proc. Natl. Acad. Sci. USA 1990, 87, 1188–1192. [Google Scholar] [CrossRef] [Green Version]
- Neher, E.; Augustine, G.J. Calcium gradients and buffers in bovine chromaffin cells. J. Physiol. 1992, 450, 273–301. [Google Scholar] [CrossRef] [PubMed]
- Ponti, A.; Machacek, M.; Gupton, S.L.; Waterman-Storer, C.M.; Danuser, G. Two distinct actin networks drive the protrusion of migrating cells. Science 2004, 305, 1782–1786. [Google Scholar] [CrossRef] [Green Version]
- Nikolaev, V.O.; Bunemann, M.; Schmitteckert, E.; Lohse, M.J.; Engelhardt, S. Cyclic AMP imaging in adult cardiac myocytes reveals far-reaching beta1-adrenergic but locally confined beta2-adrenergic receptor-mediated signaling. Circ. Res. 2006, 99, 1084–1091. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.J.; Baillie, G.S.; Houslay, M.D. cAMP-specific phosphodiesterase-4D5 (PDE4D5) provides a paradigm for understanding the unique non-redundant roles that PDE4 isoforms play in shaping compartmentalized cAMP cell signalling. Biochem. Soc. Trans. 2007, 35, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Dixit, N.; Simon, S.I. Chemokines, selectins and intracellular calcium flux: Temporal and spatial cues for leukocyte arrest. Front. Immunol. 2012, 3, 188. [Google Scholar] [CrossRef] [Green Version]
- Mehta, S.; Aye-Han, N.N.; Ganesan, A.; Oldach, L.; Gorshkov, K.; Zhang, J. Calmodulin-controlled spatial decoding of oscillatory Ca2+ signals by calcineurin. eLife 2014, 3, e03765. [Google Scholar] [CrossRef] [PubMed]
- Van Unen, J.; Reinhard, N.R.; Yin, T.; Wu, Y.I.; Postma, M.; Gadella, T.W.; Goedhart, J. Plasma membrane restricted RhoGEF activity is sufficient for RhoA-mediated actin polymerization. Sci. Rep. 2015, 5, 14693. [Google Scholar] [CrossRef] [Green Version]
- Meddens, M.B.; van den Dries, K.; Cambi, A. Podosomes revealed by advanced bioimaging: What did we learn? Eur. J. Cell Biol. 2014, 93, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Joosten, B.; Willemse, M.; Fransen, J.; Cambi, A.; van den Dries, K. Super-Resolution Correlative Light and Electron Microscopy (SR-CLEM) Reveals Novel Ultrastructural Insights Into Dendritic Cell Podosomes. Front. Immunol. 2018, 9, 1908. [Google Scholar] [CrossRef] [PubMed]
- Shroff, H.; Galbraith, C.G.; Galbraith, J.A.; White, H.; Gillette, J.; Olenych, S.; Davidson, M.W.; Betzig, E. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc. Natl. Acad. Sci. USA 2007, 104, 20308–20313. [Google Scholar] [CrossRef] [Green Version]
- Shroff, H.; Galbraith, C.G.; Galbraith, J.A.; Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 2008, 5, 417–423. [Google Scholar] [CrossRef] [Green Version]
- Chien, F.C.; Kuo, C.W.; Yang, Z.H.; Chueh, D.Y.; Chen, P. Exploring the formation of focal adhesions on patterned surfaces using super-resolution imaging. Small 2011, 7, 2906–2913. [Google Scholar] [CrossRef]
- Xu, L.; Braun, L.J.; Ronnlund, D.; Widengren, J.; Aspenstrom, P.; Gad, A.K.B. Nanoscale localization of proteins within focal adhesions indicates discrete functional assemblies with selective force-dependence. FEBS J. 2018, 285, 1635–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patla, I.; Volberg, T.; Elad, N.; Hirschfeld-Warneken, V.; Grashoff, C.; Fässler, R.; Spatz, J.P.; Geiger, B.; Medalia, O. Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat. Cell Biol. 2010, 12, 909–915. [Google Scholar] [CrossRef]
- Deschout, H.; Platzman, I.; Sage, D.; Feletti, L.; Spatz, J.P.; Radenovic, A. Investigating Focal Adhesion Substructures by Localization Microscopy. Biophys. J. 2017, 113, 2508–2518. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Tee, Y.H.; Kabla, A.; Zaidel-Bar, R.; Bershadsky, A.; Hersen, P. Structured illumination microscopy reveals focal adhesions are composed of linear subunits. Cytoskeleton 2015, 72, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Spiess, M.; Hernandez-Varas, P.; Oddone, A.; Olofsson, H.; Blom, H.; Waithe, D.; Lock, J.G.; Lakadamyali, M.; Stromblad, S. Active and inactive beta1 integrins segregate into distinct nanoclusters in focal adhesions. J. Cell Biol. 2018, 217, 1929–1940. [Google Scholar] [CrossRef] [Green Version]
- Young, L.E.; Higgs, H.N. Focal Adhesions Undergo Longitudinal Splitting into Fixed-Width Units. Curr. Biol. 2018, 28, 2033–2045.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Tseng, H.Y.; Tan, S.; Senger, F.; Kurzawa, L.; Dedden, D.; Mizuno, N.; Wasik, A.A.; Thery, M.; Dunn, A.R.; et al. Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation. Nat. Cell Biol. 2016, 18, 941–953. [Google Scholar] [CrossRef]
- Bouchet, B.P.; Gough, R.E.; Ammon, Y.C.; van de Willige, D.; Post, H.; Jacquemet, G.; Altelaar, A.M.; Heck, A.J.; Goult, B.T.; Akhmanova, A. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife 2016, 5, e18124. [Google Scholar] [CrossRef]
- Legerstee, K.; Abraham, T.E.; van Cappellen, W.A.; Nigg, A.L.; Slotman, J.A.; Houtsmuller, A.B. Growth factor dependent changes in nanoscale architecture of focal adhesions. Sci. Rep. 2021, 11, 2315. [Google Scholar] [CrossRef] [PubMed]
- Coltella, N.; Manara, M.C.; Cerisano, V.; Trusolino, L.; Di Renzo, M.F.; Scotlandi, K.; Ferracini, R. Role of the MET/HGF receptor in proliferation and invasive behavior of osteosarcoma. FASEB J. 2003, 17, 1162–1164. [Google Scholar] [CrossRef] [Green Version]
- Patane, S.; Avnet, S.; Coltella, N.; Costa, B.; Sponza, S.; Olivero, M.; Vigna, E.; Naldini, L.; Baldini, N.; Ferracini, R.; et al. MET overexpression turns human primary osteoblasts into osteosarcomas. Cancer Res. 2006, 66, 4750–4757. [Google Scholar] [CrossRef] [Green Version]
- Patane, S.; Pietrancosta, N.; Hassani, H.; Leroux, V.; Maigret, B.; Kraus, J.L.; Dono, R.; Maina, F. A new Met inhibitory-scaffold identified by a focused forward chemical biological screen. Biochem. Biophys. Res. Commun. 2008, 375, 184–189. [Google Scholar] [CrossRef]
- Pennacchietti, S.; Michieli, P.; Galluzzo, M.; Mazzone, M.; Giordano, S.; Comoglio, P.M. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 2003, 3, 347–361. [Google Scholar] [CrossRef] [Green Version]
- Webb, D.J.; Brown, C.M.; Horwitz, A.F. Illuminating adhesion complexes in migrating cells: Moving toward a bright future. Curr. Opin. Cell Biol. 2003, 15, 614–620. [Google Scholar] [CrossRef]
- Digman, M.A.; Brown, C.M.; Horwitz, A.R.; Mantulin, W.W.; Gratton, E. Paxillin dynamics measured during adhesion assembly and disassembly by correlation spectroscopy. Biophys. J. 2008, 94, 2819–2831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Cohen, D.M.; Choudhury, D.M.; Kioka, N.; Craig, S.W. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 2005, 169, 459–470. [Google Scholar] [CrossRef] [Green Version]
- Rubashkin, M.G.; Cassereau, L.; Bainer, R.; DuFort, C.C.; Yui, Y.; Ou, G.; Paszek, M.J.; Davidson, M.W.; Chen, Y.Y.; Weaver, V.M. Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 2014, 74, 4597–4611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montanez, E.; Ussar, S.; Schifferer, M.; Bösl, M.; Zent, R.; Moser, M.; Fässler, R. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008, 22, 1325–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.Q.; Qin, J.; Wu, C.; Plow, E.F. Kindlin-2 (Mig-2): A co-activator of beta3 integrins. J. Cell Biol. 2008, 181, 439–446. [Google Scholar] [CrossRef] [Green Version]
- Theodosiou, M.; Widmaier, M.; Böttcher, R.T.; Rognoni, E.; Veelders, M.; Bharadwaj, M.; Lambacher, A.; Austen, K.; Müller, D.J.; Zent, R.; et al. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife 2016, 5, e1f0130. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legerstee, K.; Houtsmuller, A.B. A Layered View on Focal Adhesions. Biology 2021, 10, 1189. https://doi.org/10.3390/biology10111189
Legerstee K, Houtsmuller AB. A Layered View on Focal Adhesions. Biology. 2021; 10(11):1189. https://doi.org/10.3390/biology10111189
Chicago/Turabian StyleLegerstee, Karin, and Adriaan B. Houtsmuller. 2021. "A Layered View on Focal Adhesions" Biology 10, no. 11: 1189. https://doi.org/10.3390/biology10111189
APA StyleLegerstee, K., & Houtsmuller, A. B. (2021). A Layered View on Focal Adhesions. Biology, 10(11), 1189. https://doi.org/10.3390/biology10111189




