Size-Fractionated Filtration Combined with Molecular Methods Reveals the Size and Diversity of Picophytoplankton
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and High-Throughput Sequencing
2.3. Flow-Cytometric Analysis of Picophytoplankton
2.4. Environmental Parameter Measurement
2.5. Data Analysis
3. Results
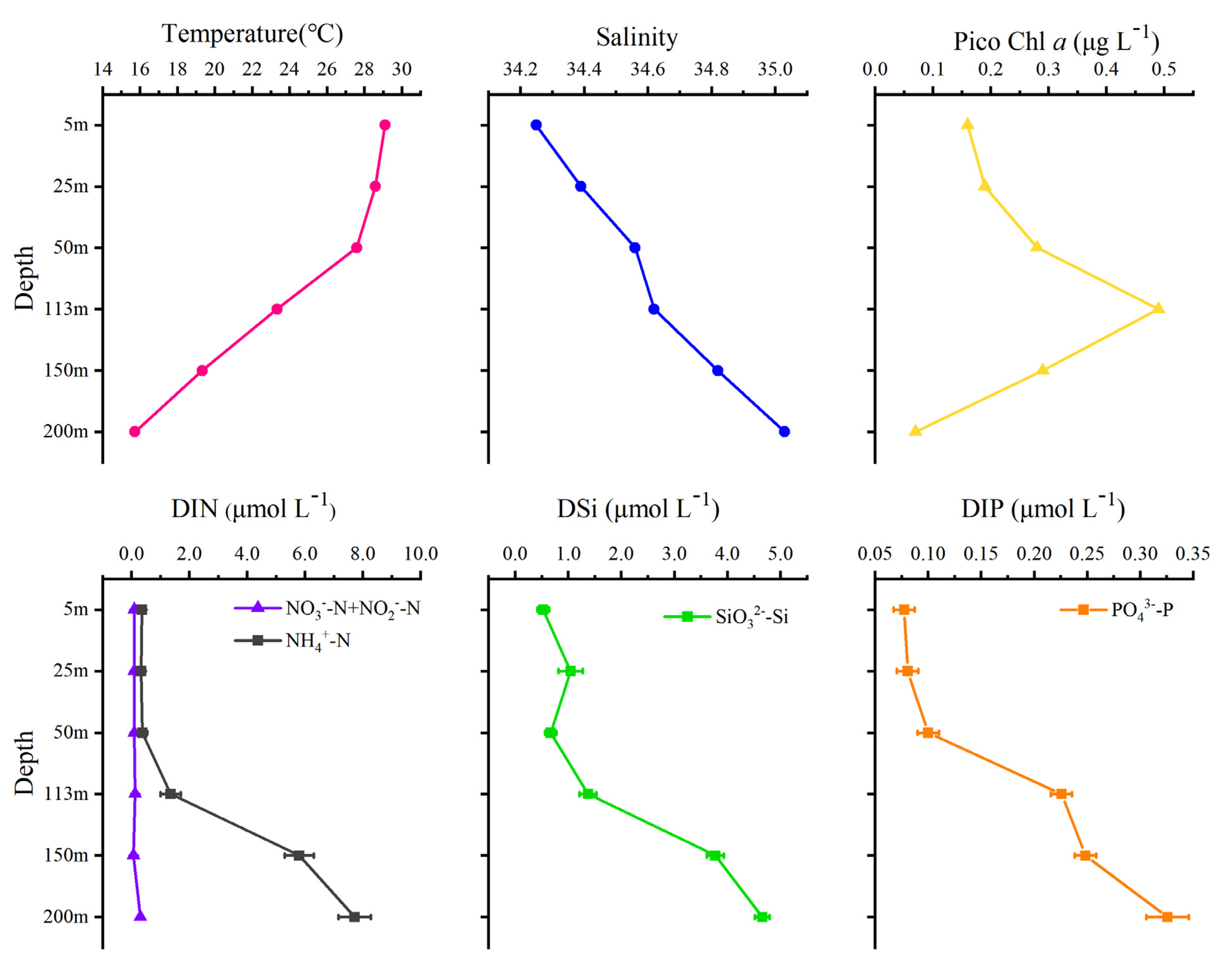
3.1. Environmental Parameters
3.2. Vertical Distribution Patterns and Estimation of the Size of Picophytoplankton Based on Flow Cytometry
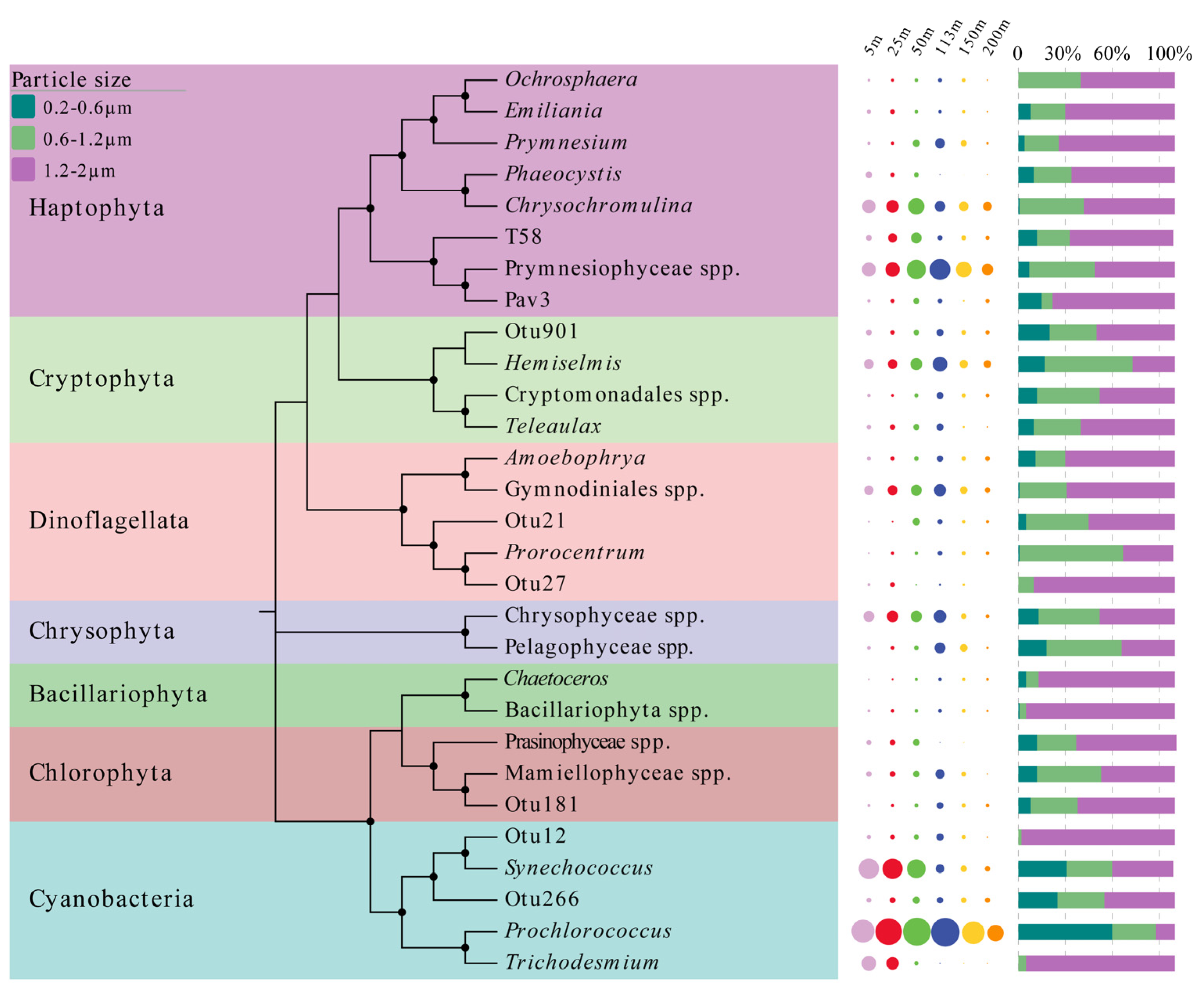
3.3. Vertical Distribution Patterns and Particle Size Composition of Picophytoplankton Based on High-Throughput Sequencing
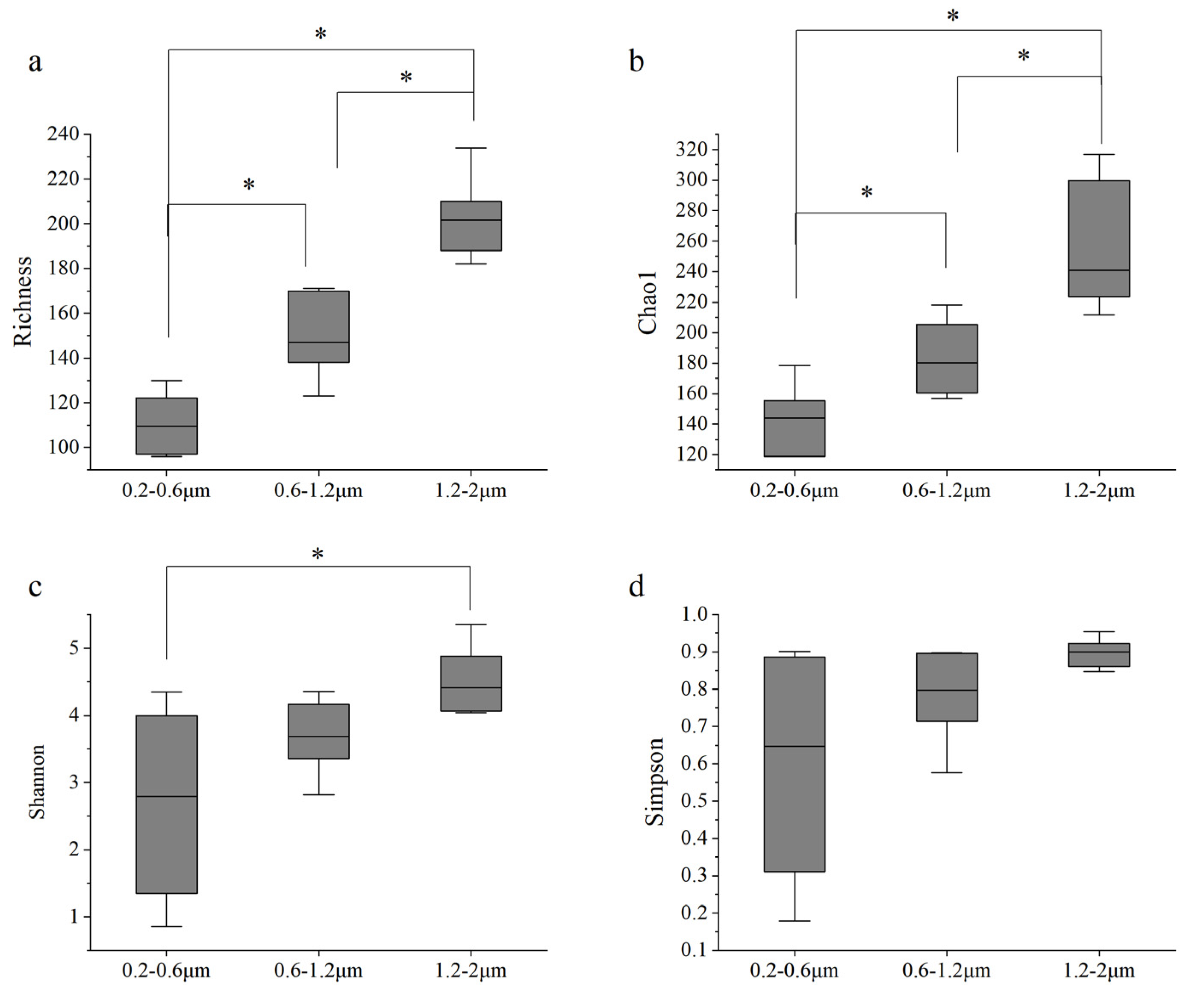
3.4. Diversity and Phylogenetic Relationships of Picophytoplankton
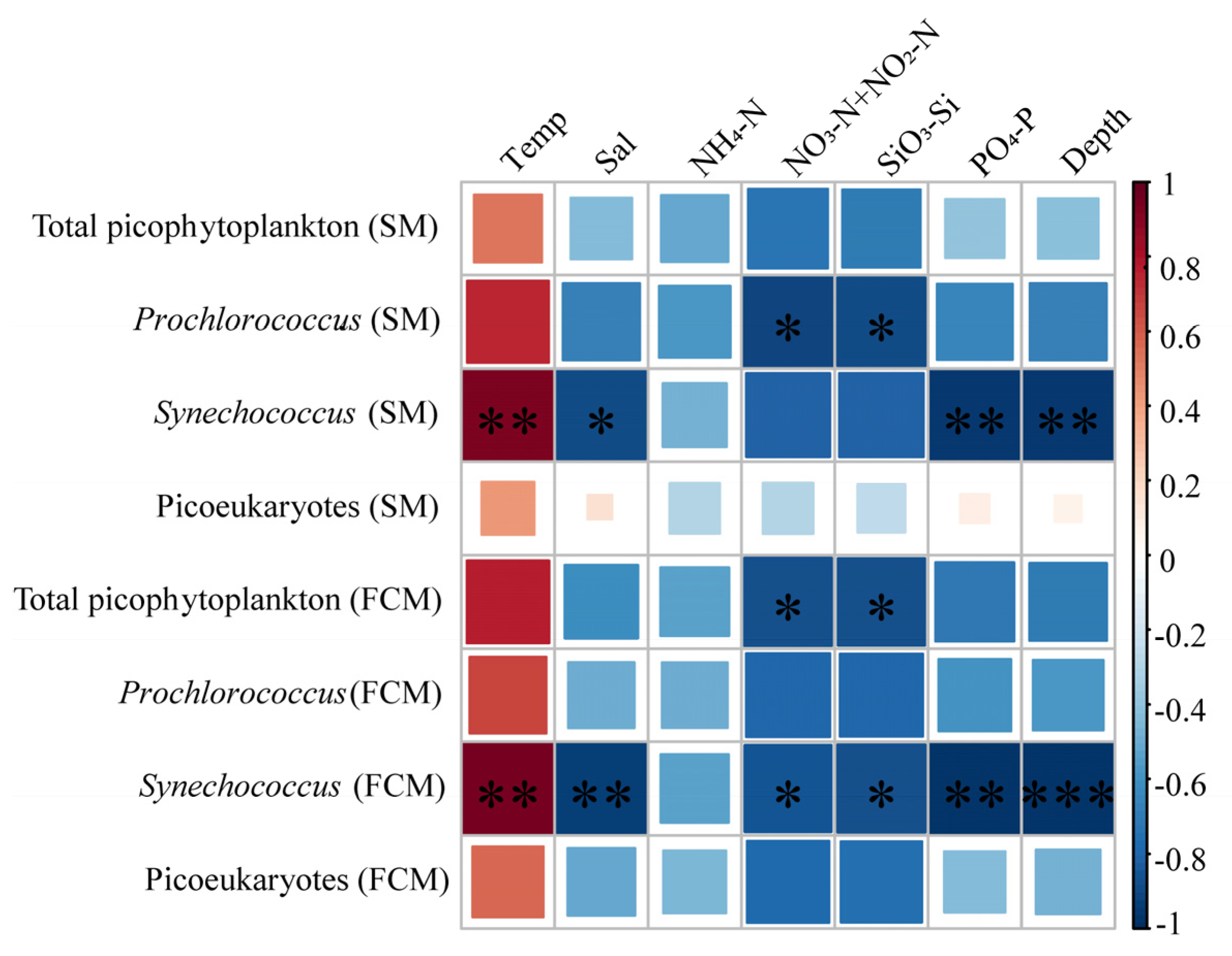
3.5. Comparison of the FCM and SM Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DuRand, M.D.; Olson, R.J.; Chisholm, S.W. Phytoplankton population dynamics at the Bermuda Atlantic Time-series station in the Sargasso Sea. Deep-Sea Res. Part II Top. Stud. Oceanogr. 2001, 48, 1983–2003. [Google Scholar] [CrossRef]
- Guidi, L.; Stemmann, L.; Jackson, G.A.; Ibanez, F.; Claustre, H.; Legendre, L.; Picheral, M.; Gorsky, G. Effects of phytoplankton community on production, size and export of large aggregates: A world-ocean analysis. Limnol. Oceanogr. 2009, 54, 1951–1963. [Google Scholar] [CrossRef] [Green Version]
- Mouw, C.B.; Yoder, J.A.; Doney, S.C. Impact of phytoplankton community size on a linked global ocean optical and ecosystem model. J. Mar. Syst. 2012, 89, 61–75. [Google Scholar] [CrossRef] [Green Version]
- Glover, D.M.; Brewer, P.G. Estimates of wintertime mixed layer nutrient concentrations in the North Atlantic. Deep Sea Res. Part A Oceanogr. Res. Pap. 1998, 35, 1525–1546. [Google Scholar] [CrossRef]
- Goericke, R.; Welschmeyer, N.A. The marine Prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary Production in the Sargasso Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 1993, 40, 2283–2294. [Google Scholar] [CrossRef]
- Poulton, A.J.; Holligan, P.M.; Hickman, A.; Kim, Y.N.; Adey, T.R.; Stinchcombe, M.C.; Holeton, C.; Root, S.; Woodward, E.M.S. Phytoplankton carbon fixation, chlorophyll-biomass and diagnostic pigments in the Atlantic Ocean. Deep-Sea Res. Part II Top. Stud. Oceanogr. 2006, 53, 1593–1610. [Google Scholar] [CrossRef]
- Campbell, L.; Liu, H.; Nolla, H.A.; Vaulot, D. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991–1994 ENSO event. Deep Sea Res. Part I Oceanogr. Res. Pap. 1997, 44, 167–192. [Google Scholar] [CrossRef]
- Grob, C.; Hartmann, M.; Zubkov, M.V.; Scanlan, D.J. Invariable biomass-specific primary production of taxonomically discrete picoeukaryote groups across the Atlantic Ocean. Environ. Microbiol. 2011, 13, 3266–3274. [Google Scholar] [CrossRef]
- Buitenhuis, E.T.; Li, W.K.W.; Vaulot, D.; Lomas, M.W.; Landry, M.R.; Partensky, F.; Karl, D.M.; Ulloa, O.; Campbell, L.; Jacquet, S.; et al. Picophytoplankton biomass distribution in the global ocean. Earth Syst. Sci. Data 2012, 4, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Johnson, Z.I.; Zinser, E.R.; Coe, A.; McNulty, N.P.; Woodward, E.M.; Chisholm, S.W. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 2006, 311, 1737–1740. [Google Scholar] [CrossRef] [Green Version]
- Shalapyonok, A.; Olson, R.J.; Shalapyonok, L.S. Arabian Sea phytoplankton during Southwest and Northeast Monsoons 1995: Composition, size structure and biomass from individual cell properties measured by flow cytometry. Deep Sea Res. Part II Top. Stud. Oceanogr. 2001, 48, 1231–1261. [Google Scholar] [CrossRef]
- Flombaum, P.; Gallegos, J.L.; Gordillo, R.A.; Rincon, J.; Zabala, L.L.; Jiao, N.; Karl, D.M.; Li, W.K.; Lomas, M.W.; Veneziano, D.; et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA 2013, 110, 9824–9829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bopp, L.; Aumont, O.; Cadule, P.; Alvain, S.; Gehlen, M. Response of diatoms distribution to global warming and potential implications: A global model study. Geophys. Res. Lett. 2005, 32, 19. [Google Scholar] [CrossRef]
- Bopp, L.; Monfray, P.; Aumont, O.; Dufresne, J.-L.; Le Treut, H.; Madec, G.; Terray, L.; Orr, J.C. Potential impact of climate change on marine export production. Glob. Biogeochem. Cycles 2001, 15, 81–99. [Google Scholar] [CrossRef] [Green Version]
- Beaugrand, G.; Edwards, M.; Legendre, L. Marine biodiversity, ecosystem functioning, and carbon cycles. Proc. Natl. Acad. Sci. USA 2010, 107, 10120–10124. [Google Scholar] [CrossRef] [Green Version]
- MorÁN, X.A.G.; LÓPez-Urrutia, Á.; Calvo-DÍAz, A.; Li, W.K.W. Increasing importance of small phytoplankton in a warmer ocean. Glob. Chang. Biol. 2010, 16, 1137–1144. [Google Scholar] [CrossRef]
- Pauly, D.; Cheung, W.W.L. Sound physiological knowledge and principles in modeling shrinking of fishes under climate change. Glob. Chang. Biol. 2018, 24, e15–e26. [Google Scholar] [CrossRef] [Green Version]
- Vaulot, D.; Eikrem, W.; Viprey, M.; Moreau, H. The diversity of small eukaryotic phytoplankton (<or =3 microm) in marine ecosystems. FEMS Microbiol. Rev. 2008, 32, 795–820. [Google Scholar] [CrossRef] [Green Version]
- Worden, A.Z.; Nolan, J.K.; Palenik, B. Assessing the dynamics and ecology of marine picophytoplankton: The importance of the eukaryotic component. Limnol. Oceanogr. 2004, 49, 168–179. [Google Scholar] [CrossRef]
- Collado-Fabbri, S.; Vaulot, D.; Ulloa, O. Structure and seasonal dynamics of the eukaryotic picophytoplankton community in a wind-driven coastal upwelling ecosystem. Limnol. Oceanogr. 2011, 56, 2334–2346. [Google Scholar] [CrossRef]
- Shi, X.L.; Marie, D.; Jardillier, L.; Scanlan, D.J.; Vaulot, D. Groups without cultured representatives dominate eukaryotic picophytoplankton in the oligotrophic South East Pacific Ocean. PLoS ONE 2009, 4, e7657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellec, L.; Clerissi, C.; Edern, R.; Foulon, E.; Simon, N.; Grimsley, N.; Desdevises, Y. Cophylogenetic interactions between marine viruses and eukaryotic picophytoplankton. BMC Evol. Biol. 2014, 14, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.K.W. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: Measurements from flow cytometric sorting. Limnol. Oceanogr. 1994, 39, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Bec, B.; Collos, Y.; Souchu, P.; Vaquer, A.; Lautier, J.; Fiandrino, A.; Laugier, T. Distribution of picophytoplankton and nanophytoplankton along an anthropogenic eutrophication gradient in French Mediterranean coastal lagoons. Aquat. Microb. Ecol. 2011, 63, 29–45. [Google Scholar] [CrossRef] [Green Version]
- Brunet, C.; Casotti, R.; Vantrepotte, V.; Corato, F.; Conversano, F. Picophytoplankton diversity and photoacclimation in the Strait of Sicily (Mediterranean Sea) in summer. I. Mesoscale variations. Aquat. Microb. Ecol. 2006, 44, 127–141. [Google Scholar] [CrossRef]
- Brunet, C.; Casotti, R.; Vantrepotte, V.; Conversano, F. Vertical variability and diel dynamics of picophytoplankton in the Strait of Sicily, Mediterranean Sea, in summer. Mar. Ecol. Prog. Ser. 2007, 346, 15–26. [Google Scholar] [CrossRef]
- Rii, Y.M.; Karl, D.M.; Church, M.J. Temporal and vertical variability in picophytoplankton primary productivity in the North Pacific Subtropical Gyre. Mar. Ecol. Prog. Ser. 2016, 562, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Mitbavkar, S.; Anil, A.C.; Narale, D.D.; Chitari, R.; Rao, V.T.; Gopalakrishna, V.V. Environmental influence on the picophytoplankton community structure in the central and northern Bay of Bengal. Reg. Stud. Mar. Sci. 2020, 40, 101528. [Google Scholar] [CrossRef]
- Wei, Y.; Zhang, G.; Chen, J.; Wang, J.; Ding, C.; Zhang, X.; Sun, J. Dynamic responses of picophytoplankton to physicochemical variation in the eastern Indian Ocean. Ecol. Evol. 2019, 9, 5003–5017. [Google Scholar] [CrossRef]
- Hopkinson, C.S., Jr.; Sherr, B.; Wiebe, W.J. Size fractionated metabolism of coastal microbial plankton. Mar. Ecol. Prog. Ser. 1989, 51, 155–166. [Google Scholar] [CrossRef]
- Bradford-Grieve, J.M.; Chang, F.H.; Gall, M.; Pickmere, S.; Richards, F. Size-fractionated phytoplankton standing stocks and primary production during austral winter and spring 1993 in the Subtropical Convergence region near New Zealand. N. Z. J. Mar. Freshw. Res. 1997, 31, 201–224. [Google Scholar] [CrossRef] [Green Version]
- Ganesh, S.; Parris, D.J.; DeLong, E.F.; Stewart, F.J. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014, 8, 187–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maki, A.; Salmi, P.; Mikkonen, A.; Kremp, A.; Tiirola, M. Sample Preservation, DNA or RNA Extraction and Data Analysis for High-Throughput Phytoplankton Community Sequencing. Front. Microbiol. 2017, 8, 1848. [Google Scholar] [CrossRef] [Green Version]
- Penna, A.; Casabianca, S.; Guerra, A.F.; Vernesi, C.; Scardi, M. Analysis of phytoplankton assemblage structure in the Mediterranean Sea based on high-throughput sequencing of partial 18S rRNA sequences. Mar. Genom. 2017, 36, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Sogge, H.; Lagesen, K.; Tooming-Klunderud, A.; Jakobsen, K.S.; Rohrlack, T. Use of high throughput sequencing and light microscopy show contrasting results in a study of phytoplankton occurrence in a freshwater environment. PLoS ONE 2014, 9, e106510. [Google Scholar] [CrossRef] [PubMed]
- van den Engh, G.J.; Doggett, J.K.; Thompson, A.W.; Doblin, M.A.; Gimpel, C.N.G.; Karl, D.M. Dynamics of Prochlorococcus and Synechococcus at Station ALOHA Revealed through Flow Cytometry and High-Resolution Vertical Sampling. Front. Mar. Sci. 2017, 4, 359. [Google Scholar] [CrossRef] [Green Version]
- Sommaruga, R.; Hofer, J.S.; Alonso-Saez, L.; Gasol, J.A. Differential sunlight sensitivity of picophytoplankton from surface Mediterranean coastal waters. Appl. Environ. Microbiol. 2005, 71, 2154–2157. [Google Scholar] [CrossRef] [Green Version]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.; Breiner, H.W.; Richards, T.A. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010, 19 (Suppl. 1), 21–31. [Google Scholar] [CrossRef]
- Fuller, N.J.; Campbell, C.; Allen, D.J.; Pitt, F.D.; Zwirglmaierl, K.; Le Gall, F.; Vaulot, D.; Scanlan, D.J. Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquat. Microb. Ecol. 2006, 43, 79–93. [Google Scholar] [CrossRef] [Green Version]
- Phinney, D.A.; Cucci, T.L. Flow cytometry and phytoplankton. Cytom. J. Int. Soc. Anal. Cytol. 1989, 10, 511–521. [Google Scholar] [CrossRef]
- Wei, Y.; Sun, J.; Zhang, X.; Wang, J.; Huang, K. Picophytoplankton size and biomass around equatorial eastern Indian Ocean. Microbiologyopen 2019, 8, e00629. [Google Scholar] [CrossRef]
- Blanchot, J.; André, J.M.; Navarette, C.; Neveux, J.; Radenac, M.H. Picophytoplankton in the equatorial Pacific: Vertical distributions in the warm pool and in the high nutrient low chlorophyll conditions. Deep Sea Res. Part I Oceanogr. Res. Pap. 2001, 48, 297–314. [Google Scholar] [CrossRef]
- Partensky, F.; Hess, W.R.; Vaulot, D. Prochlorococcus, a marine photoSynthetic Prokaryote of global significance. Microbiol. Mol. Biol. Rev. 1999, 63, 106–127. [Google Scholar] [CrossRef] [Green Version]
- Collos, Y.; Yin, K.; Harrison, J.P. A note of caution on reduction conditions when using the cadmium-copper column for nitrate determinations in aquatic environments of varying salinities. Mar. Chem. 1992, 38, 325–329. [Google Scholar] [CrossRef]
- Brzezinski, M.A.; Nelson, D.M. A solvent extraction method for the colorimetric determination of nanomolar concentrations of silicic acid in seawater. Mar. Chem. 1986, 19, 139–151. [Google Scholar] [CrossRef]
- Karl, D.M.; Tien, G. MAGIC: A sensitive and precise method for measuring dissolved phosphorus in aquatic environments. Limnol. Oceanogr. 1992, 37, 105–116. [Google Scholar] [CrossRef]
- Welschmeyer, N.A. Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol. Oceanogr. 1994, 39, 1985–1992. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.Y.; Liang, Y.T.; Xing, Y.; Wang, L.; Mou, S.L.; Cao, C.J.; Xie, R.Z.; Zhang, C.L.; Tian, J.W.; Zhang, Y.Y. Insight Into the Pico- and Nano-Phytoplankton Communities in the Deepest Biosphere, the Mariana Trench. Front. Microbiol. 2018, 9, 2289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieburth, J.M.; Johnson, P.W.; Hargraves, P.E. ULTRASTRUCTURE AND ECOLOGY OF AUREOCOCCUS ANOPHAGEFERENS GEN. ET SP. NOV.(CHRYSOPHYCEAE): THE DOMINANT PICOPLANKTER DURING A BLOOM IN NARRAGANSETT BAY, RHODE ISLAND, SUMMER 19851. J. Phycol. 1988, 24, 416–425. [Google Scholar] [CrossRef]
- Balzano, S.; Marie, D.; Gourvil, P.; Vaulot, D. Composition of the summer photosynthetic pico and nanoplankton communities in the Beaufort Sea assessed by T-RFLP and sequences of the 18S rRNA gene from flow cytometry sorted samples. ISME J. 2012, 6, 1480–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belevich, T.A.; Milyutina, I.A.; Abyzova, G.A.; Troitsky, A.V. The pico-sized Mamiellophyceae and a novel Bathycoccus clade from the summer plankton of Russian Arctic Seas and adjacent waters. FEMS Microbiol. Ecol. 2021, 97, fiaa251. [Google Scholar] [CrossRef]
- Fresnel, J.; Probert, I. The ultrastructure and life cycle of the coastal coccolithophorid Ochrosphaera neapolitana (Prymnesiophyceae). Eur. J. Phycol. 2005, 40, 105–122. [Google Scholar] [CrossRef] [Green Version]
- Young, J.R.; Westbroek, P. Genotypic variation in the coccolithophorid speciesEmiliania huxleyi. Mar. Micropaleontol. 1991, 18, 5–23. [Google Scholar] [CrossRef]
- Olson, R.J.; Chisholm, S.W.; Zettler, E.R.; Altabet, M.A.; Dusenberry, J.A. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep Sea Res. Part A Oceanogr. Res. Pap. 1990, 37, 1033–1051. [Google Scholar] [CrossRef]
- Blanchot, J.; Rodier, M. Picophytoplankton abundance and biomass in the western tropical Pacific Ocean during the 1992 El Niño year: Results from flow cytometry. Deep Sea Res. Part I Oceanogr. Res. Pap. 1996, 43, 877–895. [Google Scholar] [CrossRef]
- Chisholm, S.W.; Olson, R.J.; Zettler, E.R.; Goericke, R.; Waterbury, J.B.; Welschmeyer, N.A. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 1988, 334, 340–343. [Google Scholar] [CrossRef]
- Morel, A.; Ahn, Y.H.; Partensky, F.; Vaulot, D.; Claustre, H. Prochlorococcus and Synechococcus: A comparative study of their optical properties in relation to their size and pigmentation. J. Mar. Res. 1993, 51, 617–649. [Google Scholar] [CrossRef]
- Sieracki, M.E.; Haugen, E.M.; Cucci, T.L. Overestimation of heterotrophic bacteria in the Sargasso Sea: Direct evidence by flow and imaging cytometry. Deep Sea Res. Part I Oceanogr. Res. Pap. 1995, 42, 1399–1409. [Google Scholar] [CrossRef]
- Calvo-Díaz, A.; Morán, X.A.G.; Suárez, L.Á. Seasonality of picophytoplankton chlorophyll a and biomass in the central Cantabrian Sea, southern Bay of Biscay. J. Mar. Syst. 2008, 72, 271–281. [Google Scholar] [CrossRef]
- Cabello, A.M.; Latasa, M.; Forn, I.; Morán, X.A.G.; Massana, R. Vertical distribution of major photosynthetic picoeukaryotic groups in stratified marine waters. Environ. Microbiol. 2016, 18, 1578–1590. [Google Scholar] [CrossRef]
- Eikrem, W.; Medlin, L.K.; Henderiks, J.; Rokitta, S.; Rost, B.; Probert, I.; Edvardsen, B. Haptophyta. In Handbook of the Protists; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1–61. [Google Scholar]
- Gornik, S.G.; Hu, I.; Lassadi, I.; Waller, R.E. The Biochemistry and Evolution of the Dinoflagellate Nucleus. Microorganisms 2019, 7, 245. [Google Scholar] [CrossRef] [Green Version]
- Le Bescot, N.; Mahe, F.; Audic, S.; Dimier, C.; Garet, M.J.; Poulain, J.; Wincker, P.; de Vargas, C.; Siano, R. Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding. Environ. Microbiol. 2016, 18, 609–626. [Google Scholar] [CrossRef]
- Didymus, J.M.; Young, J.R.; Mann, S. Construction and morphogenesis of the chiral ultrastructure of coccoliths from the marine alga Emiliania huxleyi. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1994, 258, 237–245. [Google Scholar] [CrossRef]
- Hickman, A.E.; Dutkiewicz, S.; Williams, R.G.; Follows, M.J. Modelling the effects of chromatic adaptation on phytoplankton community structure in the oligotrophic ocean. Mar. Ecol. Prog. Ser. 2010, 406, 1–17. [Google Scholar] [CrossRef]
- Hickman, A.E.; Holligan, P.M.; Moore, C.M.; Sharples, J.; Krivtsov, V.; Palmer, M.R. Distribution and chromatic adaptation of phytoplankton within a shelf sea thermocline. Limnol. Oceanogr. 2009, 54, 525–536. [Google Scholar] [CrossRef]
- Edvardsen, B.; Paasche, E. Bloom Dynamics and Physiology of Prymnesium and Chrysochromulina. Nato Asi Ser. G Ecol. Sci. 1998, 41, 193–208. [Google Scholar] [CrossRef]
- Dimier, C.; Brunet, C.; Geider, R.; Raven, J. Growth and photoregulation dynamics of the picoeukaryote Pelagomonas calceolata in fluctuating light. Limnol. Oceanogr. 2009, 54, 823–836. [Google Scholar] [CrossRef]
- Marty, J.C.; Garcia, N.; Rairnbault, P. Phytoplankton dynamics and primary production under late summer conditions in the NW Mediterranean Sea. Deep-Sea Res. Part I 2008, 55, 1131–1149. [Google Scholar] [CrossRef]
- Raven, J.A. The twelfth Tansley lecture. Small is beautiful-The picophytoplankton. Funct. Ecol. 1998, 12, 503–513. [Google Scholar] [CrossRef]
- Kulk, G.; de Vries, P.; van de Poll, W.H.; Visser, J.V.; Buma, A.G. Temperature-dependent growth and photophysiology of prokaryotic and eukaryotic oceanic picophytoplankton. Mar. Ecol. Prog. Ser. 2012, 466, 43–55. [Google Scholar] [CrossRef]
- Moore, L.R.; Goericke, R.; Chisholm, S.W. Comparative physiology of Synechococcus and Prochlorococcus: Influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 1995, 259–275. Available online: https://www.jstor.org/stable/44635011 (accessed on 18 October 2021). [CrossRef]
- Rocap, G.; Larimer, F.W.; Lamerdin, J.; Malfatti, S.; Chain, P.; Ahlgren, N.A.; Chisholm, S.W. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 2003, 424, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Zinser, E.R.; Johnson, Z.I.; Coe, A.; Karaca, E.; Veneziano, D.; Chisholm, S.W. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol. Oceanogr. 2007, 52, 2205–2220. [Google Scholar] [CrossRef]
- Dufresne, A.; Salanoubat, M.; Partensky, F.; Artiguenave, F.; Axmann, I.M.; Barbe, V.; Duprat, S.; Galperin, M.Y.; Koonin, E.V.; Le Gall, F.; et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 2003, 100, 10020–10025. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.R.; Post, A.F.; Rocap, G.; Chisholm, S.W. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 2002, 47, 989–996. [Google Scholar] [CrossRef]
- Stramski, D.; Sciandra, A.; Claustre, H. Effects of temperature, nitrogen, and light limitation on the optical properties of the marine diatom Thalassiosira pseudonana. Limnol. Oceanogr. 2002, 47, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Kirkham, A.R.; Lepère, C.; Jardillier, L.E.; Not, F.; Bouman, H.; Mead, A.; Scanlan, D.J. A global perspective on marine photosynthetic picoeukaryote community structure. ISME J. 2013, 7, 922–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Not, F.; Gausling, R.; Azam, F.; Heidelberg, J.F.; Worden, A.Z. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ. Microbiol. 2007, 9, 1233–1252. [Google Scholar] [CrossRef]




| 0.2–0.6 μm | 0.6–1.2 μm | 1.2–2 μm | |
|---|---|---|---|
| Prochlorococcus | 59.86% | 27.93% | 12.21% |
| Synechococcus | 31.46% | 29.32% | 39.22% |
| PEuks | 14.71% | 32.41% | 52.88% |
| Prochlorococcus | Synechococcus | Picoeukaryotes | |
|---|---|---|---|
| FCM | 0.60 ± 0.22 μm | 0.83 ± 0.10 μm | 1.20 ± 0.47 μm |
| SM | ~0.69 μm | ~1.02 μm | ~1.21 μm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shuwang, X.; Sun, J.; Wei, Y.; Guo, C. Size-Fractionated Filtration Combined with Molecular Methods Reveals the Size and Diversity of Picophytoplankton. Biology 2021, 10, 1280. https://doi.org/10.3390/biology10121280
Shuwang X, Sun J, Wei Y, Guo C. Size-Fractionated Filtration Combined with Molecular Methods Reveals the Size and Diversity of Picophytoplankton. Biology. 2021; 10(12):1280. https://doi.org/10.3390/biology10121280
Chicago/Turabian StyleShuwang, Xinze, Jun Sun, Yuqiu Wei, and Congcong Guo. 2021. "Size-Fractionated Filtration Combined with Molecular Methods Reveals the Size and Diversity of Picophytoplankton" Biology 10, no. 12: 1280. https://doi.org/10.3390/biology10121280






