Comparison of Myosepta Development and Transcriptome Profiling between Blunt Snout Bream with and Tilapia without Intermuscular Bones
Simple Summary
Abstract
1. Introduction
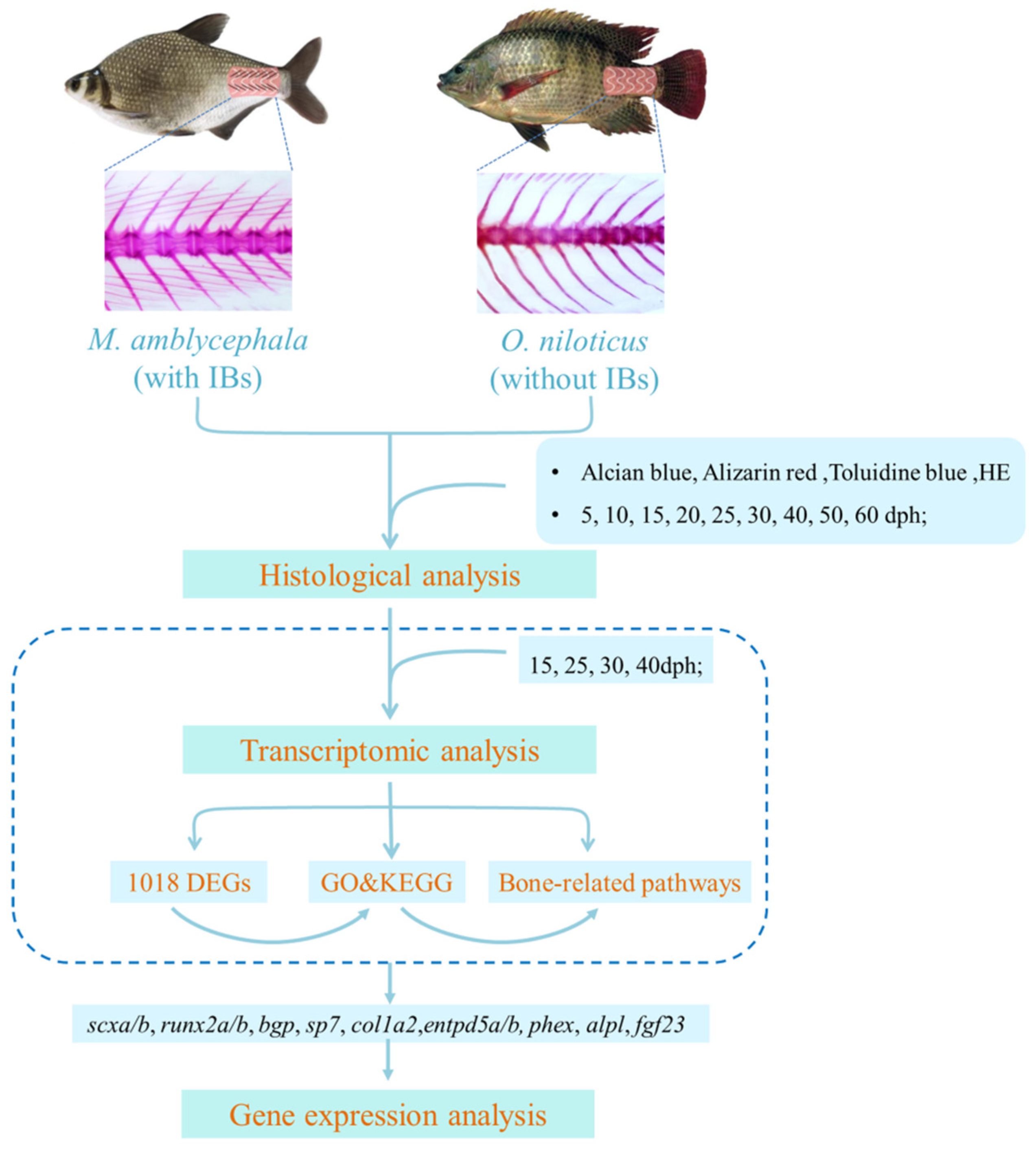
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. Histological Analysis
2.4. RNA Library Construction and Sequencing
2.5. Differential Expression and Functional Enrichment Analysis
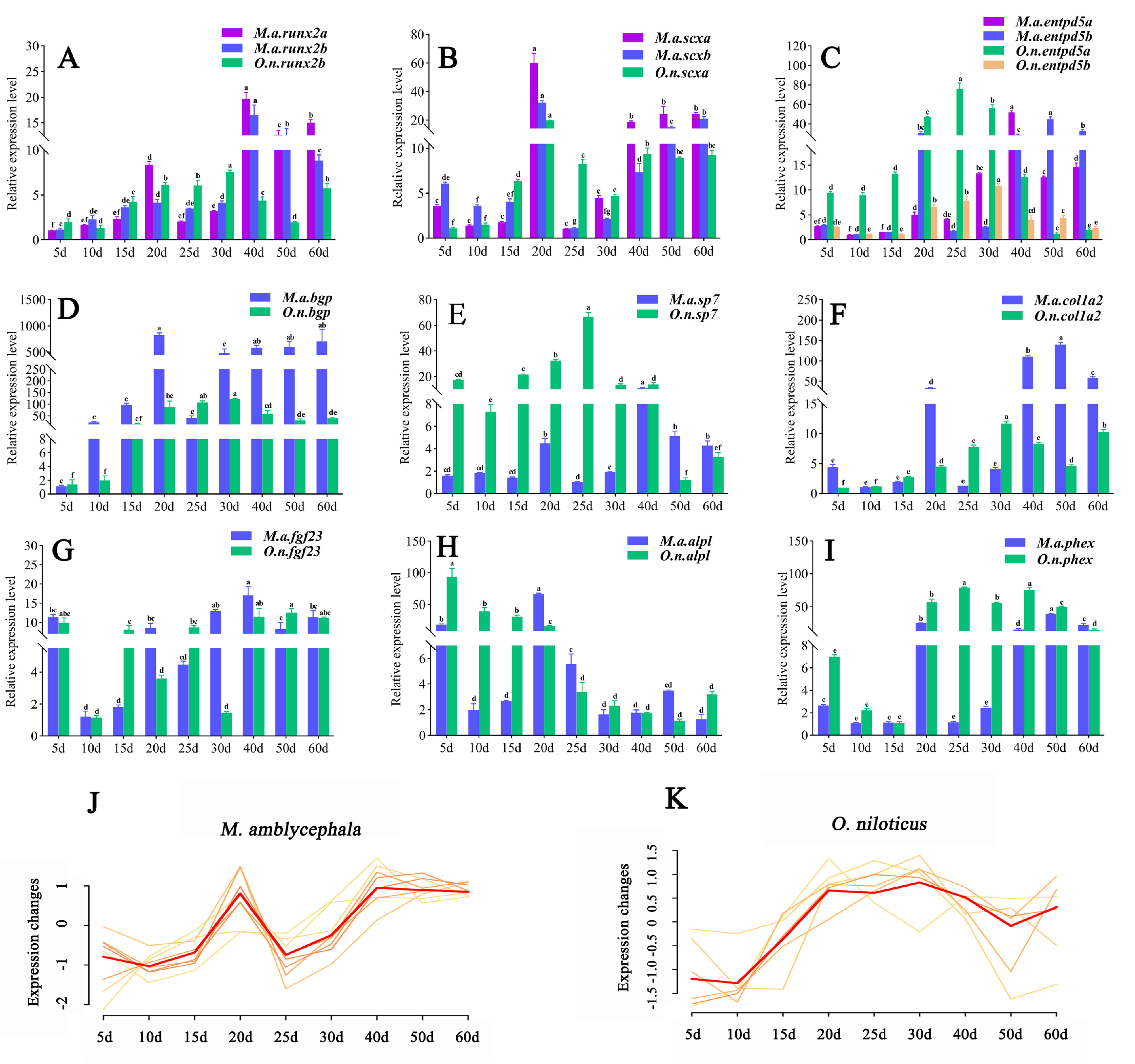
2.6. Gene Expression Analysis
3. Results
3.1. Histological Structure
3.2. Comparative Transcriptome Analysis
3.3. Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gemballa, S.; Britz, R. Homology of intermuscular bones in acanthomorph fishes. Am. Mus. Novit. 1988, 3241, 1–25. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2020. In Sustainability in Action; FAO: Rome, Italy, 2020; p. 206. [Google Scholar] [CrossRef]
- Nie, C.H.; Hilsdorf, A.W.S.; Wan, S.M.; Gao, Z.X. Understanding the development of intermuscular bones in teleost: Status and future directions for aquaculture. Rev. Aquac. 2020, 12, 759–772. [Google Scholar] [CrossRef]
- Li, L.; Zhong, Z.Z.; Zeng, M.; Liu, S.J.; Zhou, Y.; Xiao, J.; Wang, J.; Liu, Y. Comparative analysis of intermuscular bones in fish of different ploidies. Sci. China Life Sci. 2013, 56, 341–350. [Google Scholar] [CrossRef]
- Qian, Y.; Zheng, J.; Xu, X.; Luo, C. Normally grown and developed intermuscular bone-deficient mutant in grass carp, Ctenopharyngodon idellus. Chin. Sci. Bull. 2015, 60, 52–57. [Google Scholar] [CrossRef]
- Jiang, W.P.; Jia, Y.Y.; Liu, S.L.; Li, Q.; Li, T.; Gu, Z.M. Comparative analysis of intermuscular bone in hybrid F1, F2 of (C. alburnus) (♀) × (M. amblycephala) (♂) and its parents. Acta Hydrobiol. Sin. 2016, 40, 227–286. [Google Scholar]
- Xiong, X.-M.; Robinson, N.A.; Zhou, J.-J.; Chen, Y.-L.; Wang, W.M.; Wang, X.-B.; Gao, Z.-X. Genetic parameter estimates for intermuscular bone in blunt snout bream (Megalobrama amblycephala) based on a microsatellite-based pedigree. Aquaculture 2019, 502, 371–377. [Google Scholar] [CrossRef]
- Perazza, C.A.; de Menezes, J.T.B.; Ferraz, J.B.S.; Pinaffi, F.L.V.; Silva, L.A.; Hilsdorf, A. Lack of intermuscular bones in specimens of Colossoma macropomum: An unusual phenotype to be incorporated into genetic improvement programs. Aquaculture 2017, 472, 57–60. [Google Scholar] [CrossRef]
- Liu, H.; Chen, C.H.; Gao, Z.X.; Min, J.M.; Gu, Y.M.; Jian, J.B.; Jiang, X.W.; Cai, H.M.; Ebersberger, I.; Xu, M.; et al. The draft genome of blunt snout bream (Megalobrama amblycephala) reveals the development of intermuscular bone and adaptation to herbivorous diet. GigaScience 2017, 6, gix039. [Google Scholar] [CrossRef]
- Nie, C.-H.; Wan, S.-M.; Tomljanovic, T.; Treer, T.; Hsiao, C.-D.; Wang, W.-M.; Gao, Z.-X. Comparative proteomics analysis of teleost intermuscular bones and ribs provides insight into their development. BMC Genom. 2017, 18, 147. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.-H.; Wan, S.-M.; Liu, Y.; Liu, H.; Wang, W.-M.; Gao, Z.-X. Development of teleost intermuscular bones undergoing intramembranous ossification based on histological-transcriptomic-proteomic data. Int. J. Mol. Sci. 2019, 20, 4698. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, X.L.; Huang, X.; Huang, G.H.; Gao, Z.X.; Wang, W.M.; Liu, H. Genome-wide analysis of intermuscular bone development reveals changes of key genes expression and signaling pathways in blunt snout bream (Megalobrama amblycephala). Genomics 2021, 113, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, L.Y.; Chen, L.; Wang, L.; Ma, X.; Hu, C.C.; Kong, X.H.; Nie, G.X.; Li, X.J. The mRNA and protein expression of gene SOST during ossification process of intermuscular bone in crucian carp (Carassius auratus) in Qihe River. J. Fish. China 2016, 40, 673–680. [Google Scholar]
- Wang, X.D.; Nie, C.H.; Gao, Z.X. Research progress on molecular regulation mechanism and genetic selection of intermuscular bones in teleosts. Acta Hydrobiol. Sin. 2021, 45, 680–691. [Google Scholar]
- Lv, Y.P.; Yao, W.J.; Chen, J.; Bao, B.L. Newly identified gene muscle segment homeobox C may play a role in intermuscular bone development of Hemibarbus labeo. Genet. Mol. Res. 2015, 14, 11324–11334. [Google Scholar] [CrossRef]
- Nie, C.H.; Wan, S.M.; Chen, Y.L.; Zhu, D.J.; Wang, X.D.; Dong, X.R.; Gao, Z.-X. Loss of scleraxis leads to distinct reduction of mineralized intermuscular bone in zebrafish. Aquac. Fish. 2021, 6, 169–177. [Google Scholar] [CrossRef]
- Zhong, Z.; Niu, P.; Wang, M.; Huang, G.; Xu, S.; Sun, Y.; Xu, X.; Hou, Y.; Sun, X.; Yan, Y.; et al. Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci. Rep. 2016, 6, 22953. [Google Scholar] [CrossRef]
- Walia, B.; Huang, A.H. Tendon stem progenitor cells: Understanding the biology to inform therapeutic strategies for tendon repair. J. Orthop. Res. 2018, 37, 1270–1280. [Google Scholar] [CrossRef]
- Huang, Z.; Yin, Z.; Xu, J.; Fei, Y.; Heng, B.C.; Jiang, X.; Chen, W.; Shen, W. Tendon stem/progenitor cell subpopulations and their implications in tendon biology. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Salhotra, A.; Shah, H.N.; Levi, B.; Longaker, M.T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell Biol. 2020, 21, 696–711. [Google Scholar] [CrossRef]
- Komori, T. Runx2, A multifunctional transcription factor in skeletal development. J. Cell. Biochem. 2002, 87, 1–8. [Google Scholar] [CrossRef]
- Su, S.; Dong, Z. Comparative expression analyses of bone morphogenetic protein 4 (BMP4) expressions in muscles of tilapia and common carp indicate that BMP4 plays a role in the intermuscular bone distribution in a dose-dependent manner. Gene Expr. Patterns 2018, 27, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.M.; Yi, S.K.; Zhong, J.; Nie, C.H.; Guan, N.N.; Zhang, W.Z.; Gao, Z.X. Dynamic mRNA and miRNA expression analysis in response to intermuscular bone development of blunt snout bream (Megalobrama amblycephala). Sci Rep. 2016, 6, 31050. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.B.; Kimmel, C.B. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef]
- Witten, P.E.; Huysseune, A. A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol. Rev. 2009, 84, 315–346. [Google Scholar] [CrossRef] [PubMed]
- Eames, B.F.; Yan, Y.-L.; Swartz, M.E.; Levic, D.S.; Knapik, E.W.; Postlethwait, J.H.; Kimmel, C.B. Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 2011, 7, e1002246. [Google Scholar] [CrossRef]
- Fisher, M.C.; Clinton, G.M.; Maihle, N.J.; Dealy, C.N. Requirement for ErbB2/ErbB signaling in developing cartilage and bone. Dev. Growth Differ. 2007, 49, 503–513. [Google Scholar] [CrossRef]
- Liu, Y.; Olsen, B.R. Distinct VEGF functions during bone development and homeostasis. Arch. Immunol. Ther. Exp. 2014, 62, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Sadeghpour, A.; Yousefi, B. The roles of signaling pathways in bone repair and regeneration. J. Cell. Physiol. 2018, 233, 2937–2948. [Google Scholar] [CrossRef]
- Ma, X.; Su, P.; Yin, C.; Lin, X.; Wang, X.; Gao, Y.; Patil, S.; War, A.R.; Qadir, A.; Tian, Y.; et al. The roles of Foxo transcription factors in regulation of bone cells function. Int. J. Mol. Sci. 2020, 21, 692. [Google Scholar] [CrossRef] [PubMed]
- Charvet, B.; Malbouyres, M.; Pagnon-Minot, A.; Ruggiero, F.; Le Guellec, D. Development of the zebrafish myoseptum with emphasis on the myotendinous junction. Cell Tissue Res. 2011, 346, 439–449. [Google Scholar] [CrossRef]
- Fiedler, I.A.K.; Zeveleva, S.; Duarte, A.; Zhao, X.; Depalle, B.; Cardoso, L.; Jin, S.; Berteau, J.P. Microstructure, mineral and mechanical properties of teleost intermuscular bones. J. Biomech. 2019, 94, 59–66. [Google Scholar] [CrossRef]
- Schlesinger, P.H.; Blair, H.C.; Stolz, D.B.; Riazanski, V.; Ray, E.C.; Tourkova, I.L.; Nelson, D.J. Cellular and extracellular matrix of bone, with principles of synthesis and dependency of mineral deposition on cell membrane transport. Am. J. Physiol. Cell Physiol. 2020, 318, C111–C124. [Google Scholar] [CrossRef]
- Sharrow, A.C.; Li, Y.; Micsenyi, A.; Griswold, R.D.; Wells, A.; Monga, S.S.P.; Blair, H.C. Modulation of osteoblast gap junction connectivity by serum, TNFα, and TRAIL. Exp. Cell Res. 2008, 314, 297–308. [Google Scholar] [CrossRef]
- Wongdee, K.; Pandaranandaka, J.; Teerapornpuntakit, J.; Tudpor, K.; Thongbunchoo, J.; Thongon, N.; Jantarajit, W.; Krishnamra, N.; Charoenphandhu, N. Osteoblasts express claudins and tight junction-associated proteins. Histochem. Cell Biol. 2008, 130, 79–90. [Google Scholar] [CrossRef]
- Dbouk, H.A.; Mroue, R.M.; El-Sabban, M.E.; Talhouk, R.S. Connexins: A myriad of functions extending beyond assembly of gap junction channels. Cell Commun. Signal. 2009, 7, 4. [Google Scholar] [CrossRef]
- Bhat, A.A.; Uppada, S.; Achkar, I.W.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight junction proteins and signaling pathways in cancer and inflammation: A functional crosstalk. Front. Physiol. 2019, 9, 1942. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.S.; Baumann, F.; Daday, C.; Redondo, P.; Durner, E.; Jobst, M.A.; Milles, L.F.; Mercadante, D.; Pippig, D.A.; Gaub, H.E.; et al. Structural and mechanistic insights into mechanoactivation of focal adhesion kinase. Proc. Natl. Acad. Sci. USA 2019, 116, 6766–6774. [Google Scholar] [CrossRef] [PubMed]
- Tonnesen, M.G.; Feng, X.; Clark, R.A. Angiogenesis in wound healing. J. Investig. Dermatol. Symp. Proc. 2000, 5, 40–46. [Google Scholar] [CrossRef]
- Campbell, H.K.; Maiers, J.L.; DeMali, K.A. Interplay between tight junctions & adherens junctions. Exp. Cell Res. 2017, 358, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.J.; Nelson, W.J. Secret handshakes: Cell–cell interactions and cellular mimics. Curr. Opin. Cell Biol. 2018, 50, 14–19. [Google Scholar] [CrossRef]
- Lui, P.P.Y.; Chan, K.M. Tendon-Derived Stem Cells (TDSCs): From basic science to potential roles in tendon pathology and tissue engineering applications. Stem Cell Rev. Rep. 2011, 7, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cheng, B. Tendon-derived stem cells as a new cell source for tendon tissue engineering. Front. Biosci. 2013, 18, 756–764. [Google Scholar]
- Cserjesi, P.; Brown, D.; Ligon, K.L.; Lyons, G.E.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Olson, E.N. Scleraxis: A basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 1995, 121, 1099–1110. [Google Scholar] [CrossRef]
- Schweitzer, R.; Chyung, J.H.; Murtaugh, L.C.; Brent, A.E.; Rosen, V.; Olson, E.N.; Lassar, A.; Tabin, C.J. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 2001, 128, 3855–3866. [Google Scholar] [CrossRef]
- Maruyama, Z.; Yoshida, C.A.; Furuichi, T.; Amizuka, N.; Ito, M.; Fukuyama, R.; Miyazaki, T.; Kitaura, H.; Nakamura, K.; Fujita, T.; et al. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev. Dyn. 2007, 236, 1876–1890. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.; Jagadeeswaran, P.; Halpern, M.E. Radiographic analysis of zebrafish skeletal defects. Dev. Biol. 2003, 264, 64–76. [Google Scholar] [CrossRef]
- Vimalraj, S. Alkaline phosphatase: Structure, expression and its function in bone mineralization. Gene 2020, 754, 144855. [Google Scholar] [CrossRef]
- Xu, B.; Ju, Y.; Cui, Y.; Song, G. Carbon nanotube array inducing osteogenic differentiation of human mesenchymal stem cells. Mater. Sci. Eng. C 2015, 51, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B. Normal Bone Anatomy and Physiology. Clin. J. Am. Soc. Nephrol. 2008, 3, S131–S139. [Google Scholar] [CrossRef]
- Liang, W.; Li, X.; Gao, B.; Gan, H.; Lin, X.; Liao, L.; Li, C. Observing the development of the temporomandibular joint in embryonic and post-natal mice using various staining methods. Exp. Ther. Med. 2015, 11, 481–489. [Google Scholar] [CrossRef]
- Pico, M.J.; Hashemi, S.; Xu, F.; Nguyen, K.H.; Donnelly, R.; Moran, E.; Flowers, S. Glucocorticoid receptor-mediated cis-repression of osteogenic genes requires BRM-SWI/SNF. Bone Rep. 2016, 5, 222–227. [Google Scholar] [CrossRef]
- Komori, T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem. Cell Biol. 2018, 149, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Huitema, L.F.A.; Apschner, A.; Logister, I.; Spoorendonk, K.M.; Bussmann, J.; Hammond, C.; Schulte-Merker, S. Entpd5 is essential for skeletal mineralization and regulates phosphate homeostasis in zebrafish. Proc. Natl. Acad. Sci. USA 2012, 109, 21372–21377. [Google Scholar] [CrossRef] [PubMed]
- Apschner, A.; Huitema, L.F.; Ponsioen, B.; Peterson-Maduro, J.; Schulte-Merker, S. Zebrafish enpp1 mutants exhibit pathological mineralization, mimicking features of generalized arterial calcification of infancy (GACI) and pseudoxanthoma elasticum (PXE). Dis. Model. Mech. 2014, 7, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Song, Z.Y.; Yang, J.J.; Huang, J.; Jiang, H.B. Sp7/osterix positively regulates dlx2b and bglap to affect tooth development and bone mineralization in zebrafish larvae. J. Biosci. 2019, 44, 127. [Google Scholar] [CrossRef]
- Zhang, C.; Tang, W.; Li, Y.; Yang, F.; Dowd, D.R.; Macdonald, P.N. Osteoblast-specific transcription factor osterix increases vitamin d receptor gene expression in osteoblasts. PLoS ONE 2011, 6, e26504. [Google Scholar] [CrossRef]
- Simon, S.; Resch, H.; Klaushofer, K.; Roschger, P.; Zwerina, J.; Kocijan, R. Hypophosphatasia: From diagnosis to treatment. Curr. Rheumatol. Rep. 2018, 20, 69. [Google Scholar] [CrossRef]
- Marsell, R.; Jonsson, K.B. The phosphate regulating hormone fibroblast growth factor-23. Acta Physiol. 2010, 200, 97–106. [Google Scholar] [CrossRef]
- Takeda, E.; Taketani, Y.; Sawada, N.; Sato, T.; Yamamoto, H. The regulation and function of phosphate in the human body. BioFactors 2004, 21, 345–355. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.-J.; Chang, Y.-J.; Chen, Y.-L.; Wang, X.-D.; Liao, Q.; Shi, R.-H.; Gao, Z.-X. Comparison of Myosepta Development and Transcriptome Profiling between Blunt Snout Bream with and Tilapia without Intermuscular Bones. Biology 2021, 10, 1311. https://doi.org/10.3390/biology10121311
Zhou J-J, Chang Y-J, Chen Y-L, Wang X-D, Liao Q, Shi R-H, Gao Z-X. Comparison of Myosepta Development and Transcriptome Profiling between Blunt Snout Bream with and Tilapia without Intermuscular Bones. Biology. 2021; 10(12):1311. https://doi.org/10.3390/biology10121311
Chicago/Turabian StyleZhou, Jia-Jia, Yong-Jie Chang, Yu-Long Chen, Xu-Dong Wang, Qing Liao, Rui-Hui Shi, and Ze-Xia Gao. 2021. "Comparison of Myosepta Development and Transcriptome Profiling between Blunt Snout Bream with and Tilapia without Intermuscular Bones" Biology 10, no. 12: 1311. https://doi.org/10.3390/biology10121311
APA StyleZhou, J.-J., Chang, Y.-J., Chen, Y.-L., Wang, X.-D., Liao, Q., Shi, R.-H., & Gao, Z.-X. (2021). Comparison of Myosepta Development and Transcriptome Profiling between Blunt Snout Bream with and Tilapia without Intermuscular Bones. Biology, 10(12), 1311. https://doi.org/10.3390/biology10121311




