IFP35 Is a Relevant Factor in Innate Immunity, Multiple Sclerosis, and Other Chronic Inflammatory Diseases: A Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. The bZIP Overview
3. IFP35
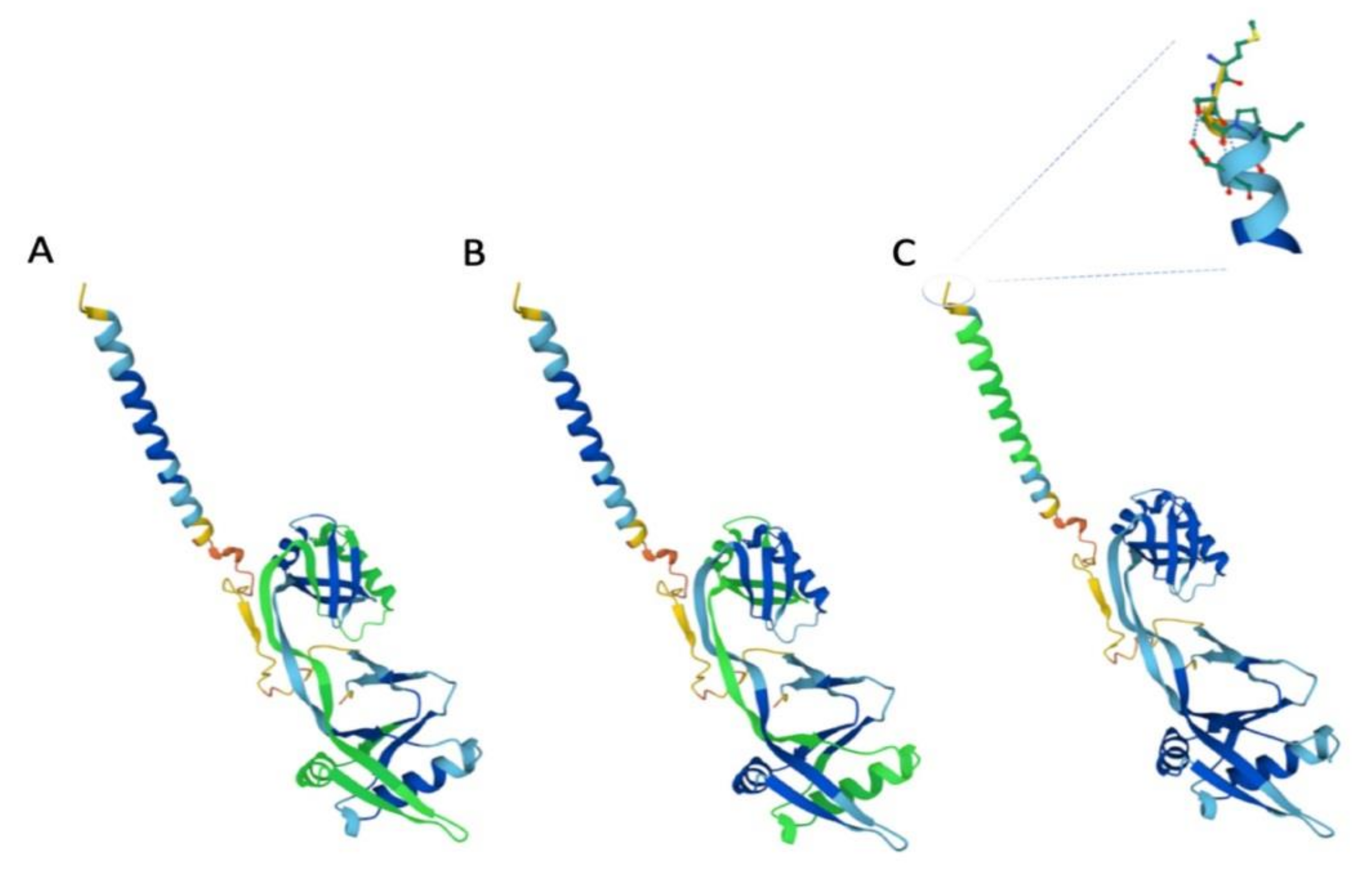
3.1. The Structure of IFP35
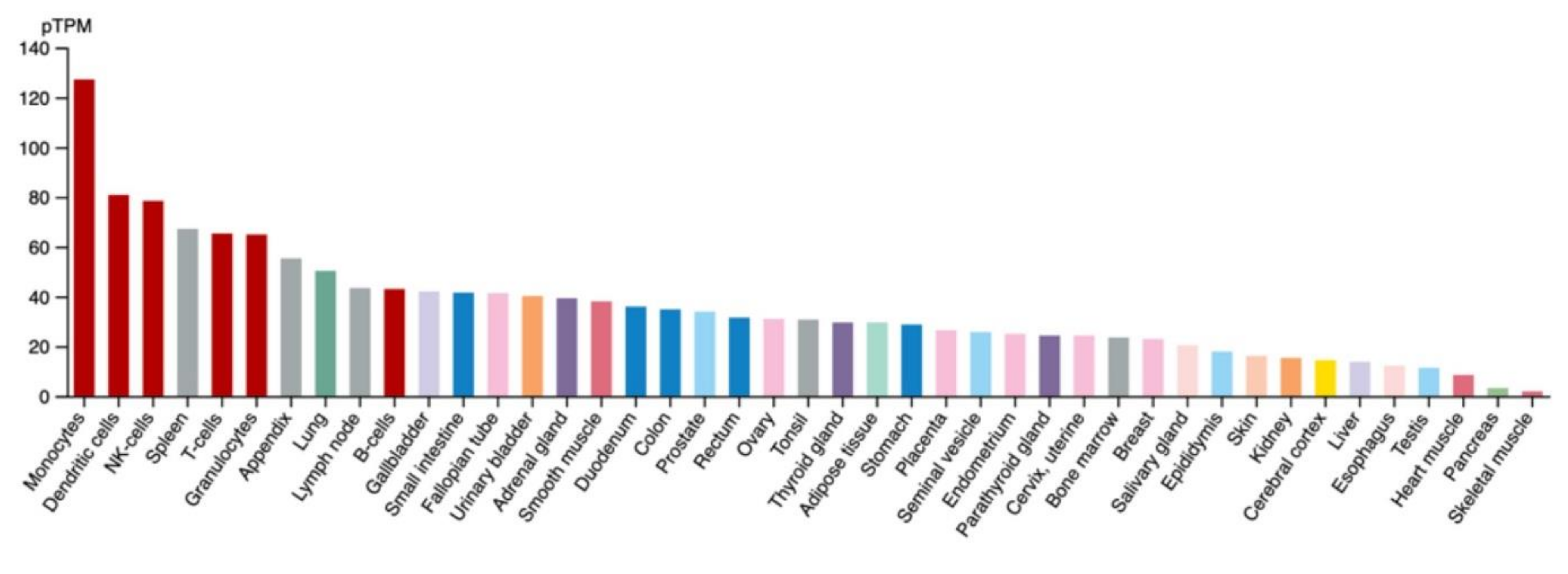
3.2. The Function of IFP35
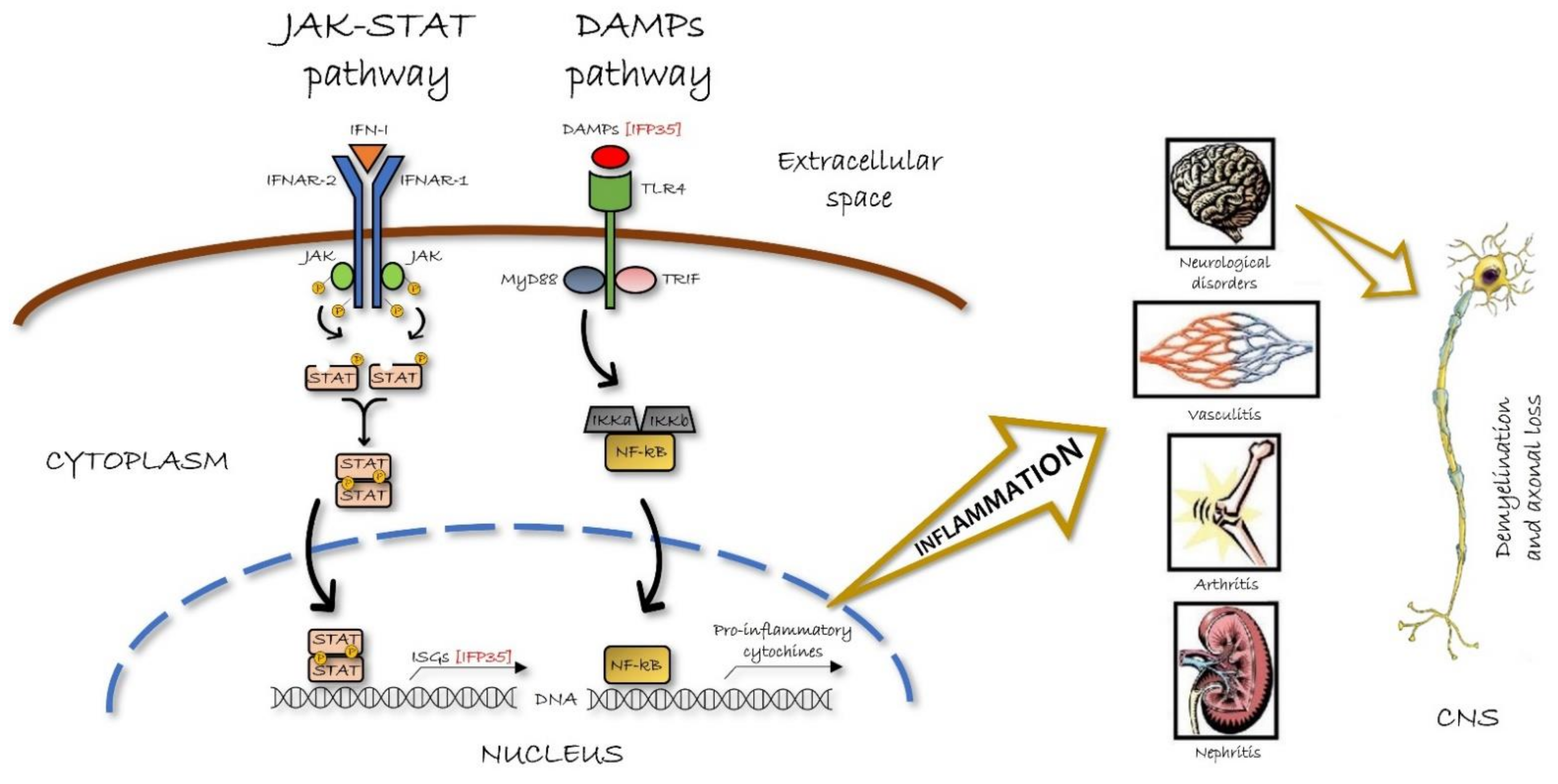
4. IFP35 in The Pathology
4.1. IFP35 in The Virus-Induced Pathologies
4.1.1. Bovines
4.1.2. Human Models of Influenza
4.1.3. Fish
4.1.4. Humans
4.2. IFP35 in The Organ-Specific Chronic Inflammatory Diseases
4.2.1. Rheumatoid Arthritis
4.2.2. Endothelial Inflammation
4.2.3. Drug-Associated Profiles of IFP35
4.2.4. Lupus Nephritis
4.2.5. Skin Diseases
4.2.6. Mice Models of Sepsis and Inflammation
4.2.7. Multiple Sclerosis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2DE | two-dimensional electrophoresis |
| AA | appendicitis and appendectomy |
| AD | atopic dermatitis |
| APC | antigen presenting cells |
| BAT | basic leucine zipper transcription factor |
| BFV | bovine foamy DNA virus |
| BR | basic region |
| BTas | transactivator bovine protein |
| bZIP | basic leucine zipper |
| CKIP-1 | casein kinase 2-interacting protein-1 |
| CNS | central nervous system |
| COPD | chronic obstructive pulmonary disease |
| CREB | cAMP response element-binding protein |
| DAMP | damage associate molecular pattern |
| DBA | Diamond Blackfan Anemia |
| DC | dendritic cells |
| DEP | differentially expressed protein |
| DMT | disease-modifying treatment |
| EAE | experimental autoimmune encephalomyelitis |
| EcLGP2 | laboratory of genetics and physiology 2 Epinephelus coioides |
| HLH | helix-loop-helix |
| IFN | interferon |
| IRF | interferon regulatory factor |
| ISG | interferon-stimulated gene |
| LTR | long terminal repeat |
| LZ | leucine zipper |
| Mafs | musculoaponeurotic fibrosarcoma protein |
| MDA5 | melanoma differentiation associated gene 5 |
| MHC | major histocompatibility complex |
| MS | Multiple Sclerosis |
| NIDs | NMI/IFP35 domains |
| NLS | nuclear localization signal |
| NMI | N-myc-interactor |
| PBMCs | peripheral blood mononuclear cells |
| Poly I:C | polyinosinic-polycytidylic acid |
| PRR | pattern recognition receptor |
| RA | Rheumatoid Arthritis |
| RGNNV | red spotted grouper nervous necrosis virus |
| RHC | Ractopamine hydrochloride |
| RIG-I | retinoic acid-inducible gene I |
| RLR | RIG-I-like receptors |
| RRMS | relapsing remitting MS patients |
| SGIV | Singapore grouper iridovirus |
| SHVV | snakehead vesiculovirus |
| SLE | Systemic Lupus Erythematous |
| SS | Sezary syndrome |
| STAT | signal transducer and activator of transcription |
| TFs | transcription factors |
| TLR | toll like receptors |
| TRIM21 | tripartite motif protein 21 |
| VSV | vesicular stomatitis virus |
References
- Bange, F.C.; Vogel, U.; Flohr, T.; Kiekenbeck, M.; Denecke, B.; Bottger, E.C. IFP35 is an interferon-induced leucine zipper protein that undergoes interferon-regulated cellular redistribution. J. Biol. Chem. 1994, 269, 1091–1098. [Google Scholar] [CrossRef]
- Brown, M.A.; Jones, K.A.; Nicolai, H.; Bonjardim, M.; Black, D.; McFarlane, R.; de Jong, P.; Quirk, J.P.; Lehrach, H.; Solomon, E. Physical mapping, cloning, and identification of genes within a 500-kb region containing BRCA1. Proc. Natl. Acad. Sci. USA 1995, 92, 4362–4366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiahou, Z.; Wang, X.; Shen, J.; Zhu, X.; Xu, F.; Hu, R.; Guo, D.; Li, H.; Tian, Y.; Liu, Y.; et al. NMI and IFP35 serve as proinflammatory DAMPs during cellular infection and injury. Nat. Commun. 2017, 8, 950. [Google Scholar] [CrossRef] [PubMed]
- De Masi, R.; Orlando, S. IFI35 as a biomolecular marker of neuroinflammation and treatment response in multiple sclerosis. Life Sci. 2020, 259, 118233. [Google Scholar] [CrossRef]
- Katsuoka, F.; Yamamoto, M. Small Maf proteins (MafF, MafG, MafK): History, structure and function. Gene 2016, 586, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, M.A.; Goodman, R.H.; Brennan, R.G. The structure of a CREB bZIP middle dot Somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem. 2000, 275, 35242–35247. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.; Shuman, J.D.; Sebastian, T.; Dauter, Z.; Johnson, P.F. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. J. Biol. Chem. 2003, 278, 5178–15184. [Google Scholar] [CrossRef] [Green Version]
- Crick, F.H.C. The packaging of alpha-helices: Simple coiled-coils. Acta Cryst. 1953, 6, 689–697. [Google Scholar] [CrossRef] [Green Version]
- Receveur-Brechot, V.; Bourhis, J.M.; Uversky, V.N.; Canard, B.; Longhi, S. Assessing protein disorder and induced folding. Proteins 2006, 62, 24–45. [Google Scholar] [CrossRef] [PubMed]
- Seldeen, K.L.; Mcdonald, C.B.; Deegan, B.J.; Farooq, A. Coupling of folding and DNA-binding in the bZIP domains of Jun–Fos heterodimeric transcription factor. Arch. Biochem. Biophys. 2008, 473, 48–60. [Google Scholar] [CrossRef]
- Ryu, T.; Jung, J.; Lee, S.; Nam, H.J.; Hong, S.W.; Yoo, J.W.; Lee, D.K.; Lee, D. bZIPDB: A database of regulatory information for human bZIP transcription factors. BMC Genom. 2007, 8, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaikhali, J.; Noren, L.; de Dios Barajas-Lopez, J.; Srivastava, V.; Konig, J.; Sauer, U.H.; Wingsle, G.; Dietz, K.J.; Strand, A. Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in Arabidopsis. J. Biol. Chem. 2012, 287, 27510–27525. [Google Scholar] [CrossRef] [Green Version]
- Konig, P.; Richmond, T.J. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J. Mol. Biol. 1993, 233, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Miller, M. The importance of being flexible: The case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci. 2009, 10, 244–269. [Google Scholar] [CrossRef] [Green Version]
- Djamei, A.; Pitzschke, A.; Nakagami, H.; Rajh, I.; Hirt, H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 2007, 318, 453–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, S.; Yuasa, T.; Nakata, M.; Takahashi, Y. A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 2008, 20, 3273–3288. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.X.; Srivastava, R.; Howell, S.H. Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 2008, 31, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Srivastava, R.; Che, P.; Howell, S.H. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 2007, 19, 4111–4119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, Y.; Fedoroff, N.V.; Koizumi, N. Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 2008, 20, 3107–3121. [Google Scholar] [CrossRef] [Green Version]
- Alonso, R.; Onate-Sanchez, L.; Weltmeier, F.; Ehlert, A.; Diaz, I.; Dietrich, K.; Vicente-Carbajosa, J.; Droge-Laser, W. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 2009, 21, 1747–1761. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.G.; Price, J.; Lin, P.C.; Hong, J.C.; Jang, J.C. The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant 2010, 3, 361–373. [Google Scholar] [CrossRef]
- Andronis, C.; Barak, S.; Knowles, S.M.; Sugano, S.; Tobin, E.M. The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant 2008, 1, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bensmihen, S.; Rippa, S.; Lambert, G.; Jublot, D.; Pautot, V.; Granier, F.; Giraudat, J.; Parcy, F. The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 2002, 14, 1391–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baena-Gonzalez, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Hummel, M.; Rahmani, F.; Smeekens, S.; Hanson, J. Sucrose-mediated translational control. Ann. Bot 2009, 104, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Dietrich, K.; Weltmeier, F.; Ehlert, A.; Weiste, C.; Stahl, M.; Harter, K.; Droge-Laser, W. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 2011, 23, 381–395. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.H.; Hong, J.W.; Kim, E.C.; Yoo, S.D. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2021, 158, 1955–1964. [Google Scholar] [CrossRef] [Green Version]
- Deppmann, C.D.; Thornton, T.M.; Utama, F.E.; Taparowsky, E.J. Phosphorylation of BATF regulates DNA binding: A novel mechanism for AP-1 (activator protein-1) regulation. Biochem. J. 2003, 374, 423–431. [Google Scholar] [CrossRef] [Green Version]
- Kirchler, T.; Briesemmeister, S.; Singer, M.; Schutze, K.; Keinath, M.; Kohlbacher, O.; Vicente-Carbajosa, J.; Teige, M.; Harter, K.; Chaban, C. The role of phosphorylatable serine residues in the DNA-binding domain of Arabidopsis bZIP transcription factors. Eur. J. Cell Biol. 2010, 89, 175–183. [Google Scholar] [CrossRef]
- Sirichandra, C.; Davanture, M.; Turk, B.E.; Zivy, M.; Valot, B.; Leung, J.; Merlot, S. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS ONE 2010, 5, e13935. [Google Scholar] [CrossRef] [Green Version]
- Hardtke, C.S.; Gohda, K.; Osterlund, M.T.; Oyama, T.; Okada, K.; Deng, X.W. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000, 19, 4997–5006. [Google Scholar] [CrossRef] [PubMed]
- Assuncao, A.G.L.; Herrero, E.; Lin, Y.F.; Huettel, B.; Talukdar, S.; Smaczniak, C.; Immink, R.G.; van Eldik, M.; Fiers, M.; Schat, H.; et al. Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 10296–10301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weltmeier, F.; Ehlert, A.; Mayer, C.S.; Dietrich, K.; Wang, X.; Schutze, K.; Alonso, R.; Harter, K.; Vicente-Carbajosa, J.; Droge-Laser, W. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 2006, 25, 3133–3143. [Google Scholar] [CrossRef] [Green Version]
- Wiese, A.; Elzinga, N.; Wobbes, B.; Smeekens, S. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 2004, 16, 1717–1729. [Google Scholar] [CrossRef] [Green Version]
- Weltmeier, F.; Rahmani, F.; Ehlert, A.; Dietrich, K.; Schutze, K.; Wang, X.; Chaban, C.; Hanson, J.; Teige, M.; Harter, K.; et al. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: Availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 2008, 69, 107–119. [Google Scholar] [CrossRef] [Green Version]
- Mallappa, C.; Singh, A.; Ram, H.; Chattopadhyay, S. GBF1, a transcription factor of blue light signaling in Arabidopsis, is degraded in the dark by a proteasome-mediated pathway independent of COP1 and SPA1. J. Biol. Chem. 2008, 283, 35772–35782. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.T.; Liu, H.; Stone, S.; Callis, J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013, 75, 965–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pontier, D.; Privat, I.; Trifa, Y.; Zhou, J.M.; Klessig, D.F.; Lam, E. Differential regulation of TGA transcription factors by post-transcriptional control. Plant J. 2002, 32, 641–653. [Google Scholar] [CrossRef]
- Meyerdierks, A.; Denecke, B.; Rohde, M.; Taparowsky, E.J.; Bottger, E.C. A cytoplasmic structure resembling large protein aggregates induced by interferons. J. Histochem. Cytochem. 1999, 47, 169–182. [Google Scholar] [CrossRef] [Green Version]
- De Masi, R.; Vergara, D.; Pasca, S.; Acierno, R.; Greco, M.; Spagnolo, L.; Blasi, E.; Sanapo, F.; Trianni, G.; Maffia, M. PBMCs protein expression profile in relapsing IFN-treated multiple sclerosis: A pilot study on relation to clinical findings and brain atrophy. J. Neuroimmunol. 2009, 210, 80–86. [Google Scholar] [CrossRef] [PubMed]
- De Masi, R.; Pasca, S.; Scarpello, R.; Idolo, A.; De Donno, A. The clinical potential of blood-proteomics in multiple sclerosis. BMC Neurol. 2013, 13, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreit, M.; Vertommen, D.; Gillet, L.; Michiels, T. The Interferon-Inducible Mouse Apolipoprotein L9 and Prohibitins Cooperate to Restrict Theiler’s Virus Replication. PLoS ONE 2015, 10, e0133190. [Google Scholar]
- Lee, N.D.; Chen, J.; Shpall, R.L.; Naumovski, L. Subcellular localization of interferon-inducible Myc/stat-interacting protein Nmi is regulated by a novel IFP 35 homologous domain. J. Interferon Cytokine Res. 1999, 19, 1245–1252. [Google Scholar] [CrossRef]
- Chen, J.; Shpall, R.L.; Meyerdierks, A.; Hagemeier, M.; Bottger, E.C.; Naumovski, L. Interferon-inducible Myc/STAT-interacting protein Nmi associates with IFP 35 into a high molecular mass complex and inhibits proteasome-mediated degradation of IFP 35. J. Biol. Chem. 2000, 275, 36278–36284. [Google Scholar] [CrossRef] [Green Version]
- Alber, T. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 1992, 2, 205–210. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, J.; Meyerdierks, A.; Feng, L.; Naumovski, L.; Bottger, E.C.; Omary, M.B. Interferon-alpha induces nmi-IFP35 heterodimeric complex formation that is affected by the phosphorylation of IFP35. J. Biol. Chem. 2000, 275, 21364–21371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fillmore, R.A.; Mitra, A.; Xi, Y.; Ju, J.; Scammell, J.; Shevde, L.A.; Samant, R.S. Nmi (N-Myc interactor) inhibits Wnt/beta-catenin signaling and retards tumor growth. Int. J. Cancer 2009, 125, 556–564. [Google Scholar] [CrossRef]
- Zhao, J.; Dong, Q.Z.; Zhong, F.; Cai, L.L.; Qin, Z.Y.; Liu, Y.; Lin, C.Z.; Qin, L.X.; He, F.C. NMI promotes hepatocellular carcinoma progression via BDKRB2 and MAPK/ERK pathway. Oncotarget 2017, 8, 12174–12185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Johansen, L.M.; Tae, H.J.; Taparowsky, E.J. IFP 35 forms complexes with B-ATF, a member of the AP1 family of transcription factors. Biochem. Biophys. Res. Commun. 1996, 229, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Schraml, B.U.; Hildner, K.; Ise, W.; Lee, W.L.; Smith, W.A.; Solomon, B.; Sahota, G.; Sim, J.; Mukasa, R.; Cemerski, S.; et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 2009, 460, 405–409. [Google Scholar] [CrossRef] [Green Version]
- Rhee, J.; Park, S.H.; Kim, S.K.; Kim, J.H.; Ha, C.W.; Chun, C.H.; Chun, J.S. Inhibition of BATF/JUN transcriptional activity protects against osteoarthritic cartilage destruction. Ann. Rheum. Dis. 2017, 76, 427–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Tang, Y.; Tie, Y.; Tian, C.; Wang, J.; Dong, Y.; Sun, Z.; He, F. The PH domain containing protein CKIP-1 binds to IFP35 and Nmi and is involved in cytokine signaling. Cell Signal. 2007, 19, 932–944. [Google Scholar] [CrossRef]
- Fu, L.; Zhang, L. Physiological functions of CKIP-1: From molecular mechanisms to therapy implications. Ageing Res. Rev. 2019, 53, 100908. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, G.A.; Benke, S.; García-Sastre, A.; Rajsbaum, R. InTRIMsic immunity: Positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 2014, 25, 563–576. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Dinh, P.X.; Pattnaik, A.K. Trim21 regulates Nmi-IFI35 complex-mediated inhibition of innate antiviral response. Virology 2015, 485, 383–392. [Google Scholar] [CrossRef] [Green Version]
- James, L.C. Intracellular antibody immunity and the cytosolic Fc receptor TRIM21. Curr. Top. Microbiol. Immunol. 2014, 382, 51–66. [Google Scholar]
- McEwan, W.A.; James, L.C. TRIM21-dependent intracellular antibody neutralization of virus infection. Prog. Mol. Biol. Transl. Sci. 2015, 129, 167–187. [Google Scholar]
- Das, A.; Dinh, P.X.; Panda, D.; Pattnaik, A.K. Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. J. Virol. 2014, 88, 3103–3113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirai, K.; Shimada, T.; Yoshida, H.; Hayakari, R.; Matsumiya, T.; Tanji, K.; Murakami, M.; Tanaka, H.; Imaizumi, T. Interferon (IFN)-induced protein 35 (IFI35) negatively regulates IFN-β-phosphorylated STAT1-RIG-I-CXCL10/CCL5 axis in U373MG astrocytoma cells treated with polyinosinic-polycytidylic acid. Brain Res. 2017, 1658, 60–67. [Google Scholar] [CrossRef]
- Yu, Y.; Huang, Y.; Yang, Y.; Wang, S.; Yang, M.; Huang, X.; Qin, Q. Negative regulation of the antiviral response by grouper LGP2 against fish viruses. Fish Shellfish Immunol. 2016, 56, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Vaysburd, M.; Watkinson, R.E.; Cooper, H.; Reed, M.; O’Connell, K.; Smith, J.; Cruickshanks, J.; James, L.C. Intracellular antibody receptor TRIM21 prevents fatal viral infection. Proc. Natl. Acad. Sci. USA 2013, 110, 12397–12401. [Google Scholar] [CrossRef] [Green Version]
- Govindarajan, V.; de Rivero Vaccari, J.P.; Keane, R.W. Role of inflammasomes in multiple sclerosis and their potential as therapeutic targets. J. Neuroinflamm. 2020, 17, 260. [Google Scholar] [CrossRef]
- Sorci, G.; Bianchi, R.; Riuzzi, F.; Tubaro, C.; Arcuri, C.; Giambanco, I.; Donato, R. S100B Protein, A Damage-Associated Molecular Pattern Protein in the Brain and Heart, and Beyond. Cardiovasc. Psychiatry Neurol. 2010, 2010, 656481. [Google Scholar] [CrossRef]
- Kumar, V. Toll-like receptors in the pathogenesis of neuroinflammation. J. Neuroimmunol. 2019, 332, 16–30. [Google Scholar] [CrossRef]
- Krakauer, T. Inflammasomes, Autophagy, and Cell Death: The Trinity of Innate Host Defense against Intracellular Bacteria. Mediators Inflamm. 2019, 2019, 2471215. [Google Scholar] [CrossRef] [Green Version]
- Piccinini, A.M.; Midwood, K.S. DAMPening inflammation by modulating TLR signalling. Mediators Inflamm. 2010, 2010, 672395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef]
- Tan, J.; Qiao, W.; Wang, J.; Xu, F.; Li, Y.; Zhou, J.; Chen, Q.; Geng, Y. IFP35 is involved in the antiviral function of interferon by association with the viral tas transactivator of bovine foamy virus. J. Virol. 2008, 82, 4275–4283. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Wang, S.; Yu, Y.; Zhang, J.; Sun, Z.; Yan, Y.; Zhou, J. Subcellular proteomic analysis of human host cells infected with H3N2 swine influenza virus. Proteomics 2013, 13, 3309–3326. [Google Scholar] [CrossRef] [PubMed]
- Gounder, A.P.; Yokoyama, C.C.; Jarjour, N.N.; Bricker, T.L.; Edelson, B.T.; Boon, A.C.M. Interferon induced protein 35 exacerbates H5N1 influenza disease through the expression of IL-12p40 homodimer. PLoS Pathog. 2018, 14, e1007001. [Google Scholar] [CrossRef]
- Li, L.; Chen, S.N.; Li, N.; Nie, P. Transcriptional and subcellular characterization of interferon induced protein-35 (IFP35) in mandarin fish, Siniperca chuatsi. Dev. Comp. Immunol. 2021, 115, 103877. [Google Scholar] [CrossRef]
- Perera, N.C.N.; Godahewa, G.I.; Nam, B.H.; Lee, J. Molecular structure and immune-stimulated transcriptional modulation of the first teleostean IFP35 counterpart from rockfish (Sebastes schlegelii). Fish Shellfish Immunol. 2016, 56, 496–505. [Google Scholar] [CrossRef]
- Liu, X.; Qin, Z.; Babu, V.S.; Zhao, L.; Li, J.; Zhang, X.; Lin, L. Transcriptomic profiles of striped snakehead cells (SSN-1) infected with snakehead vesiculovirus (SHVV) identifying IFI35 as a positive factor for SHVV replication. Fish Shellfish Immunol. 2018, 86, 46–52. [Google Scholar] [CrossRef]
- Liu, J.; Huang, Y.; Huang, X.; Li, C.; Ni, S.W.; Yu, Y.; Qin, Q. Grouper DDX41 exerts antiviral activity against fish iridovirus and nodavirus infection. Fish Shellfish Immunol. 2019, 91, 40–49. [Google Scholar] [CrossRef]
- Cai, J.; Zou, Z.; Wei, S.; Zheng, Q.; Xu, Y.; Lu, Y.; Wu, Z.; Qin, Q.; Jian, J. Identification of Beclin-1 from orange-spotted grouper (Epinephelus coioides) involved in viral infection. Fish Shellfish Immunol. 2019, 94, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Estrabaud, E.; Appourchaux, K.; Bièche, I.; Carrat, F.; Lapalus, M.; Lada, O.; Martinot-Peignoux, M.; Boyer, N.; Marcellin, P.; Vidaud, M.; et al. IFI35, mir-99a and HCV genotype to predict sustained virological response to pegylated-interferon plus ribavirin in chronic hepatitis C. PLoS ONE 2015, 10, e0121395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanin, N.; Viaris de Lesegno, C.; Lamaze, C.; Blouin, C.M. Interferon Receptor Trafficking and Signaling: Journey to the Cross Roads. Front. Immunol. 2021, 11, 615603. [Google Scholar] [CrossRef] [PubMed]
- Raison, C.L.; Miller, A.H. Pathogen-Host Defense in the Evolution of Depression: Insights into Epidemiology, Genetics, Bioregional Differences and Female Preponderance. Neuropsychopharmacology 2017, 42, 5–27. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Li, X.; Wang, J.; Li, X.; Cao, H.; Wang, Y.; Zeng, Q.; Zheng, S.J. A critical role of interferon-induced protein IFP35 in the type I interferon response in cells induced by foot-and-mouth disease virus (FMDV) protein 2C. Arch. Virol. 2014, 159, 2925–2935. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Gack, M.U. Ubiquitination in the antiviral immune response. Virology 2015, 479, 52–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumiya, T.; Stafforini, D.M. Function and regulation of retinoic acid-inducible gene-I. Crit. Rev. Immunol. 2010, 30, 489–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaya-Amaya, J.; Rojas-Villarraga, A.; Mantilla, R.D.; Anaya, J.M. Rheumatoid arthritis. In Autoimmunity: From Bench to Bedside [Internet]; Anaya, J.M., Shoenfeld, Y., Rojas-Villarraga, A., Levy, R.A., Cervera, R., Eds.; El Rosario University Press: Bogota, Colombia, 2013; Chapter 24. [Google Scholar]
- van der Pouw Kraan, T.C.; Wijbrandts, C.A.; van Baarsen, L.G.; Voskuyl, A.E.; Rustenburg, F.; Baggen, J.M.; Ibrahim, S.M.; Fero, M.; Dijkmans, B.A.; Tak, P.P.; et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: Assignment of a type I interferon signature in a subpopulation of patients. Ann. Rheum. Dis. 2007, 66, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- Castaneda-Delgado, J.E.; Bastián-Hernandez, Y.; Macias-Segura, N.; Santiago-Algarra, D.; Castillo-Ortiz, J.D.; Alemán-Navarro, A.L.; Martínez-Tejada, P.; Enciso-Moreno, L.; Garcia-De Lira, Y.; Olguín-Calderón, D.; et al. Type I Interferon Gene Response Is Increased in Early and Established Rheumatoid Arthritis and Correlates with Autoantibody Production. Front. Immunol. 2017, 8, 285. [Google Scholar] [CrossRef] [Green Version]
- Weix, J.; Haupl, T.; Raio, L.; Villiger, P.M.; Forger, F. The physiologic increase in expression of some type I IFN-inducible genes during pregnancy is not associated with improved disease activity in pregnant patients with rheumatoid arthritis. Transl. Res. 2013, 161, 505–512. [Google Scholar] [CrossRef]
- Bandman, O.; Coleman, R.T.; Loring, J.F.; Seilhamer, J.J.; Cocks, B.G. Complexity of inflammatory responses in endothelial cells and vascular smooth muscle cells determined by microarray analysis. Ann. N. Y. Acad. Sci. 2002, 975, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Jian, D.; Wang, W.; Zhou, X.; Jia, Z.; Wang, J.; Yang, M.; Zhao, W.; Jiang, Z.; Hu, X.; Zhu, J. Interferon-induced protein 35 inhibits endothelial cell proliferation, migration and re-endothelialization of injured arteries by inhibiting the nuclear factor-kappa B pathway. Acta Physiol. 2018, 223, e13037. [Google Scholar] [CrossRef]
- Sui, X. Inhibition of the NF-kB signaling pathway on endothelial cell function and angiogenesis in mice with acute cerebral infarction. J. Biol. Regul. Homeost. Agents 2019, 33, 375–384. [Google Scholar]
- Burrack, R.M.; Duffy, E.M.; Yates, D.T.; Schmidt, T.B.; Petersen, J.L. Whole blood transcriptome analysis in feedlot cattle after 35 days of supplementation with a β1-adrenergic agonist. J. Appl. Genet. 2020, 61, 117–121. [Google Scholar] [CrossRef]
- Cheng, K.; Ma, C.; Guo, X.; Huang, Y.; Tang, R.; Karrow, N.A.; Wang, C. Vitamin D3 modulates yellow catfish (Pelteobagrus fulvidraco) immune function in vivo and in vitro and this involves the vitamin D3/VDR-type I interferon axis. Dev. Comp. Immunol. 2020, 107, 103644. [Google Scholar] [CrossRef]
- Obeidat, M.; Fishbane, N.; Nie, Y.; Chen, V.; Hollander, Z.; Tebbutt, S.J.; Bossé, Y.; Ng, R.T.; Miller, B.E.; McManus, B.; et al. The Effect of Statins on Blood Gene Expression in COPD. PLoS ONE 2015, 10, e0140022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urosevic, M.; Dummer, R.; Conrad, C.; Beyeler, M.; Laine, E.; Burg, G.; Gilliet, M. Disease-independent skin recruitment and activation of plasmacytoid predendritic cells following imiquimod treatment. J. Natl. Cancer Inst. 2005, 97, 1143–1153. [Google Scholar] [CrossRef] [Green Version]
- Mohty, M.; Vialle-Castellano, A.; Nunes, J.A.; Isnardon, D.; Olive, D.; Gaugler, B. IFN-alpha skews monocyte differentiation into Toll-like receptor 7-expressing dendritic cells with potent functional activities. J. Immunol. 2003, 171, 3385–3393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Mi, W.; Luo, H.; Chen, T.; Liu, S.; Raman, I.; Zuo, X.; Li, Q.Z. Whole-genome transcription and DNA methylation analysis of peripheral blood mononuclear cells identified aberrant gene regulation pathways in systemic lupus erythematosus. Arthritis Res. Ther. 2016, 18, 162. [Google Scholar] [CrossRef] [Green Version]
- Imaizumi, T.; Yano, C.; Numata, A.; Tsugawa, K.; Hayakari, R.; Matsumiya, T.; Yoshida, H.; Watanabe, S.; Tsuruga, K.; Kawaguchi, S.; et al. Interferon (IFN)-Induced Protein 35 (IFI35), a Type I Interferon-Dependent Transcript, Upregulates Inflammatory Signaling Pathways by Activating Toll-Like Receptor 3 in Human Mesangial Cells. Kidney Blood Press. Res. 2016, 41, 635–642. [Google Scholar] [CrossRef]
- Rebane, A.; Zimmermann, M.; Aab, A.; Baurecht, H.; Koreck, A.; Karelson, M.; Abram, K.; Metsalu, T.; Pihlap, M.; Meyer, N.; et al. Mechanisms of IFN-γ-induced apoptosis of human skin keratinocytes in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2012, 129, 1297–12306. [Google Scholar] [CrossRef]
- Pomerantz, R.G.; Mirvish, E.D.; Erdos, G.; Falo, L.D.; Geskin, L.J. Novel approach to gene expression profiling in Sézary syndrome. Br. J. Dermatol. 2010, 163, 1090–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trautmann, A.; Akdis, M.; Kleemann, D.; Altznauer, F.; Simon, H.U.; Graeve, T.; Noll, M.; Brocker, E.B.; Blaser, K.; Akdis, C.A. T cell-mediated Fas-induced keratinocyte apoptosis plays a key pathogenetic role in eczematous dermatitis. J. Clin. Investig. 2000, 106, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yang, B.; Hu, Y.; Zheng, Y.; Zhou, H.; Wang, Y.; Ma, Y.; Mao, K.; Yang, L.; Lin, G.; et al. Negative regulation of Nmi on virus-triggered type I IFN production by targeting IRF7. J. Immunol. 2013, 191, 3393–3399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheluvappa, R.; Eri, R.; Luo, A.S.; Grimm, M.C. Modulation of interferon activity-associated soluble molecules by appendicitis and appendectomy limits colitis-identification of novel anti-colitic targets. J. Interferon Cytokine Res. 2015, 35, 108–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajaj, S.; Alam, S.I.; Ahmad, B.; Farooqi, H.; Gupta, M.L. Combination of podophyllotoxin and rutin modulate radiation-induced alterations of jejunal proteome in mice. Int. J. Radiat. Biol. 2020, 96, 879–893. [Google Scholar] [CrossRef]
- Pesciotta, E.N.; Lam, H.S.; Kossenkov, A.; Ge, J.; Showe, L.C.; Mason, P.J.; Bessler, M.; Speicher, D.W. In-Depth, Label-Free Analysis of the Erythrocyte Cytoplasmic Proteome in Diamond Blackfan Anemia Identifies a Unique Inflammatory Signature. PLoS ONE 2015, 10, e0140036. [Google Scholar] [CrossRef]
- Jing, X.; Yao, Y.; Wu, D.; Hong, H.; Feng, X.; Xu, N.; Liu, Y.; Liang, H. IFP35 family proteins promote neuroinflammation and multiple sclerosis. Proc. Natl. Acad. Sci. USA 2021, 118, e2102642118. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Masi, R.; Orlando, S.; Bagordo, F.; Grassi, T. IFP35 Is a Relevant Factor in Innate Immunity, Multiple Sclerosis, and Other Chronic Inflammatory Diseases: A Review. Biology 2021, 10, 1325. https://doi.org/10.3390/biology10121325
De Masi R, Orlando S, Bagordo F, Grassi T. IFP35 Is a Relevant Factor in Innate Immunity, Multiple Sclerosis, and Other Chronic Inflammatory Diseases: A Review. Biology. 2021; 10(12):1325. https://doi.org/10.3390/biology10121325
Chicago/Turabian StyleDe Masi, Roberto, Stefania Orlando, Francesco Bagordo, and Tiziana Grassi. 2021. "IFP35 Is a Relevant Factor in Innate Immunity, Multiple Sclerosis, and Other Chronic Inflammatory Diseases: A Review" Biology 10, no. 12: 1325. https://doi.org/10.3390/biology10121325
APA StyleDe Masi, R., Orlando, S., Bagordo, F., & Grassi, T. (2021). IFP35 Is a Relevant Factor in Innate Immunity, Multiple Sclerosis, and Other Chronic Inflammatory Diseases: A Review. Biology, 10(12), 1325. https://doi.org/10.3390/biology10121325




