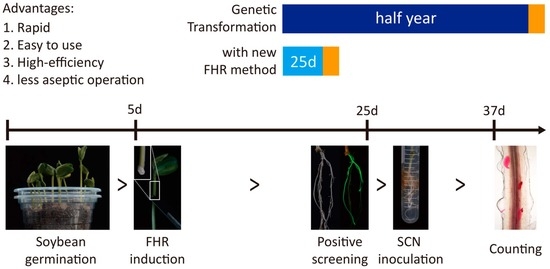
Fluorescent Soybean Hairy Root Construction and Its Application in the Soybean—Nematode Interaction: An Investigation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and Nematode Materials
2.2. A. Rhizogenes Strain K599 Materials
2.3. A. Rhizogenes-Mediated Soybean Hypocotyl Transformation
2.4. Incubation and Inoculation of SCNs
2.5. SCN Demographic Assays
2.6. Construction of the Plasmids
2.7. RNA Extraction and Real-Time qPCR
2.8. Detection of Phytohormones
3. Results
3.1. Construction of the Expression Vector for Transformation
3.2. Establishment of the FHR-SCN Pathosystem
3.3. Extension of the Application of the FHR-SCN Pathosystem in Multiple Cultivars
3.4. Overexpression of GmHS1pro-1 Enhances Resistance against H. Glycines Race 3
4. Discussion
4.1. The FHR-SCN Pathosystem Is a Rapid, Efficient and Visible Method for the Verification of Resistance Candidate Genes against SCNs
4.2. The Application of FHRs in Other Assays
4.3. Key Steps That Affect the Transformation Efficiency
4.4. GmHS1pro-1 Suppresses the Development of H. Glycines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hussey, R.S. Go where the science leads you. Annu. Rev. Phytopathol. 2010, 48, 1–19. [Google Scholar] [CrossRef]
- Mitchum, M.G. Soybean Resistance to the Soybean Cyst Nematode Heterodera glycines: An Update. Phytopathology 2016, 106, 1444–1450. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.E.; Lee, T.G.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.M.; Wang, J.; Hughes, T.J.; Willis, D.K.; Clemente, T.E.; et al. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Zhang, F.; Bao, A.; You, Q.; Li, Z.; Chen, J.; Cheng, Y.; Zhao, W.; Shen, X.; Zhou, X.; et al. The soybean Rhg1 amino acid transporter gene alters glutamate homeostasis and jasmonic acid-induced resistance to soybean cyst nematode. Mol. Plant Pathol. 2019, 20, 270–286. [Google Scholar] [CrossRef] [Green Version]
- Bayless, A.M.; Smith, J.M.; Song, J.; McMinn, P.H.; Teillet, A.; August, B.K.; Bent, A.F. Disease resistance through impairment of alpha-SNAP-NSF interaction and vesicular trafficking by soybean Rhg1. Proc. Natl. Acad. Sci. USA 2016, 113, E7375–E7382. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Hudson, M.E. WI12Rhg1 interacts with DELLAs and mediates soybean cyst nematode resistance through hormone pathways. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef]
- Liu, S.; Kandoth, P.K.; Warren, S.D.; Yeckel, G.; Heinz, R.; Alden, J.; Yang, C.; Jamai, A.; El-Mellouki, T.; Juvale, P.S.; et al. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 2012, 492, 256–260. [Google Scholar] [CrossRef]
- Concibido, V.C.; Diers, B.W.; Arelli, P.R. A Decade of QTL Mapping for Cyst Nematode Resistance in Soybean. Crop. Sci. 2004, 44, 1121–1131. [Google Scholar] [CrossRef] [Green Version]
- Meksem, K.; Pantazopoulos, P.; Njiti, V.N.; Hyten, L.D.; Arelli, P.R.; Lightfoot, D.A. ‘Forrest’ resistance to the soybean cyst nematode is bigenic: Saturation mapping of the Rhg1 and Rhg4 loci. Theor. Appl. Genet. 2001, 103, 710–717. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-H.; Kim, K.-S.; Riggs, R.D. Differential Subcellular Responses in Resistance Soybeans Infected with Soybean Cyst Nematode Races. Plant Pathol. J. 2010, 26, 154–158. [Google Scholar] [CrossRef] [Green Version]
- Bybd, D.W.; Kirkpatrick, T.; Barker, K.R. An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 1983, 15, 142–143. [Google Scholar] [PubMed]
- Zhao, X.; Teng, W.; Li, Y.; Liu, D.; Cao, G.; Li, D.; Qiu, L.; Zheng, H.; Han, Y.; Li, W. Loci and candidate genes conferring resistance to soybean cyst nematode HG type 2.5.7. BMC Genom. 2017, 18, 462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, D.M.; Baltazar, B.M.; Rao-Arelli, A.P.; Schupp, J.; Clayton, K.; Keim, P.; Beavis, W.D. Genetic mapping of soybean cyst nematode race-3 resistance loci in the soybean PI 437.654. Theor. Appl. Genet. 1995, 91, 574–581. [Google Scholar] [CrossRef]
- Lakhssassi, N.; Patil, G.; Piya, S.; Zhou, Z.; Baharlouei, A.; Kassem, M.A.; Lightfoot, D.A.; Hewezi, T.; Barakat, A.; Nguyen, H.T.; et al. Genome reorganization of the GmSHMT gene family in soybean showed a lack of functional redundancy in resistance to soybean cyst nematode. Sci. Rep. 2019, 9, 1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, S.; Zhao, X.; Zhu, X.; Wang, Y.; Xuan, Y.; Liu, X.; Fan, H.; Chen, L.; Duan, Y. Modulation of (Homo)Glutathione Metabolism and H2O2 Accumulation during Soybean Cyst Nematode Infections in Susceptible and Resistant Soybean Cultivars. Int. J. Mol. Sci. 2020, 21, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, Q.; Fang, C.; Kong, L.; Yang, H.; Hou, Z.; Li, Y.; Nan, H.; Zhang, Y.; Chen, Q.; et al. Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol. Plant 2021. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Liu, X.; Liu, S.; Jamai, A.; Bendahmane, A.; Lightfoot, D.A.; Mitchum, M.G.; Meksem, K. Soybean cyst nematode resistance in soybean is independent of the Rhg4 locus LRR-RLK gene. Funct. Integr. Genom. 2011, 11, 539–549. [Google Scholar] [CrossRef]
- Wang, R.; Deng, M.; Yang, C.; Yu, Q.; Zhang, L.; Zhu, Q.; Guo, X. A Qa-SNARE complex contributes to soybean cyst nematode resistance via regulation of mitochondria-mediated cell death. J. Exp. Bot. 2021, 72, 7145–7162. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, X.; Li, H.; Liu, S.; Jin, L.; Lyu, Y.; Shi, M.; Liu, S.; Yang, X.; Lyu, S. Anthocyanin, a novel and user-friendly reporter for convenient, non-destructive, low cost, directly visual selection of transgenic hairy roots in the study of rhizobia-legume symbiosis. Plant Methods 2020, 16, 94. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, R.; Feng, Y.; Sikandar, A.; Zhu, X.; Fan, H.; Liu, X.; Chen, L.; Duan, Y. iTRAQ-Based Proteomic Analysis Reveals the Role of the Biological Control Agent, Sinorhizobium fredii Strain Sneb183, in Enhancing Soybean Resistance Against the Soybean Cyst Nematode. Front. Plant Sci. 2020, 11, 597819. [Google Scholar] [CrossRef]
- Carter, A.; Tenuta, A.; Rajcan, I.; Welacky, T.; Woodrow, L.; Eskandari, M.; Morris, B. Identification of quantitative trait loci for seed isoflavone concentration in soybean (Glycine max) against soybean cyst nematode stress. Plant Breed. 2018, 137, 721–729. [Google Scholar] [CrossRef]
- De La Torre, C.M.; Finer, J.J. The intron and 5’ distal region of the soybean Gmubi promoter contribute to very high levels of gene expression in transiently and stably transformed tissues. Plant Cell Rep. 2015, 34, 111–120. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Liu, J.; Geng, H.; Zhang, J.; Liu, Y.; Zhang, H.; Xing, S.; Du, J.; Ma, S.; Tian, Z. De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 2018, 61, 871–884. [Google Scholar] [CrossRef]
- Ste-Croix, D.T.; St-Marseille, A.G.; Lord, E.; Belanger, R.R.; Brodeur, J.; Mimee, B. Genomic Profiling of Virulence in the Soybean Cyst Nematode Using Single-Nematode Sequencing. Phytopathology 2021, 111, 137–148. [Google Scholar] [CrossRef]
- Lian, Y.; Guo, J.; Li, H.; Wu, Y.; Wei, H.; Wang, J.; Li, J.; Lu, W. A New Race (X12) of Soybean Cyst Nematode in China. J. Nematol. 2017, 49, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium Using the Freeze-Thaw Method. Cold Spring Harbor. Protoc. 2006, 2006. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Govindarajulu, M.; Elmore, J.M.; Fester, T.; Taylor, C.G. Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol. Plant Microbe. Interact. 2008, 21, 1027–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Chen, Y.; Zhu, X.; Wang, Y.; Jung, K.H.; Chen, L.; Xuan, Y.; Duan, Y. The transcriptomic changes of Huipizhi Heidou (Glycine max), a nematode-resistant black soybean during Heterodera glycines race 3 infection. J. Plant Physiol. 2018, 220, 96–104. [Google Scholar] [CrossRef]
- Cai, D.; Kleine, M.; Kifle, S.; Harloff, H.J.; Sandal, N.N.; Marcker, K.A.; Klein-Lankhorst, R.M.; Salentijn, E.M.; Lange, W.; Stiekema, W.J.; et al. Positional cloning of a gene for nematode resistance in sugar beet. Science 1997, 275, 832–834. [Google Scholar] [CrossRef]
- Zhong, X.; Zhou, Q.; Cui, N.; Cai, D.; Tang, G. BvcZR3 and BvHs1 (pro-1) Genes Pyramiding Enhanced Beet Cyst Nematode (Heterodera schachtii Schm.) Resistance in Oilseed Rape (Brassica napus L.). Int. J. Mol. Sci. 2019, 20, 1740. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Zhou, G.; Li, Y.; Wang, K.; Wang, Z.; Li, X.; Chang, R.; Qiu, L. Cloning and sequence diversity analysis of GmHs1 pro-1 in Chinese domesticated and wild soybeans. Mol. Breed. 2008, 22, 593–602. [Google Scholar] [CrossRef]
- Jiang, H.; Li, K.; Gai, J. Agrobacterium rhizogenes-induced soybean hairy roots versus Soybean mosaic virus (ARISHR-SMV) is an efficient pathosystem for studying soybean-virus interactions. Plant Methods 2019, 15, 56. [Google Scholar] [CrossRef]
- Fan, Y.L.; Zhang, X.H.; Zhong, L.J.; Wang, X.Y.; Jin, L.S.; Lyu, S.H. One-step generation of composite soybean plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. BMC Plant Biol. 2020, 20, 208. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.F.; Beard, H.; Brewer, E.; Kabir, S.; MacDonald, M.H.; Youssef, R.M. Arabidopsis genes, AtNPR1, AtTGA2 and AtPR-5, confer partial resistance to soybean cyst nematode (Heterodera glycines) when overexpressed in transgenic soybean roots. BMC Plant Biol. 2014, 14, 96. [Google Scholar] [CrossRef] [Green Version]
- Ithal, N.; Recknor, J.; Nettleton, D.; Hearne, L.; Maier, T.; Baum, T.J.; Mitchum, M.G. Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol. Plant Microbe Interact. 2007, 20, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Gleason, C.; Leelarasamee, N.; Meldau, D.; Feussner, I. OPDA Has Key Role in Regulating Plant Susceptibility to the Root-Knot Nematode Meloidogyne hapla in Arabidopsis. Front. Plant Sci. 2016, 7, 1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Li, S.; Yang, X.; Zhu, X.; Fan, H.; Xuan, Y.; Chen, L.; Liu, X.; Wang, Y.; Duan, Y. Fluorescent Soybean Hairy Root Construction and Its Application in the Soybean—Nematode Interaction: An Investigation. Biology 2021, 10, 1353. https://doi.org/10.3390/biology10121353
Yang R, Li S, Yang X, Zhu X, Fan H, Xuan Y, Chen L, Liu X, Wang Y, Duan Y. Fluorescent Soybean Hairy Root Construction and Its Application in the Soybean—Nematode Interaction: An Investigation. Biology. 2021; 10(12):1353. https://doi.org/10.3390/biology10121353
Chicago/Turabian StyleYang, Ruowei, Shuang Li, Xiaowen Yang, Xiaofeng Zhu, Haiyan Fan, Yuanhu Xuan, Lijie Chen, Xiaoyu Liu, Yuanyuan Wang, and Yuxi Duan. 2021. "Fluorescent Soybean Hairy Root Construction and Its Application in the Soybean—Nematode Interaction: An Investigation" Biology 10, no. 12: 1353. https://doi.org/10.3390/biology10121353
APA StyleYang, R., Li, S., Yang, X., Zhu, X., Fan, H., Xuan, Y., Chen, L., Liu, X., Wang, Y., & Duan, Y. (2021). Fluorescent Soybean Hairy Root Construction and Its Application in the Soybean—Nematode Interaction: An Investigation. Biology, 10(12), 1353. https://doi.org/10.3390/biology10121353