Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of AUX/LAX, PIN, and ABCB Auxin Transporter Family Genes in Potato
2.2. Genome Distribution, Phylogenetic Tree Construction, and Promoter Analysis
2.3. Weighted Co-Expression Network Construction
2.4. Gene Structure and Tissue-Specific Expression Profiling Analysis
2.5. Three-Dimensional (3D) Structure Predictions and Subcellular Localization of Auxin Transporters in S. tuberosum
2.6. Plant Growth, Treatments, and Collection of Tissues
2.7. RNA Isolation and qRT-PCR Analysis
3. Results and Discussion
3.1. Genome-Wide Identification of Stlax, Stpin, and StABCB Auxin Transporter Genes in Potato
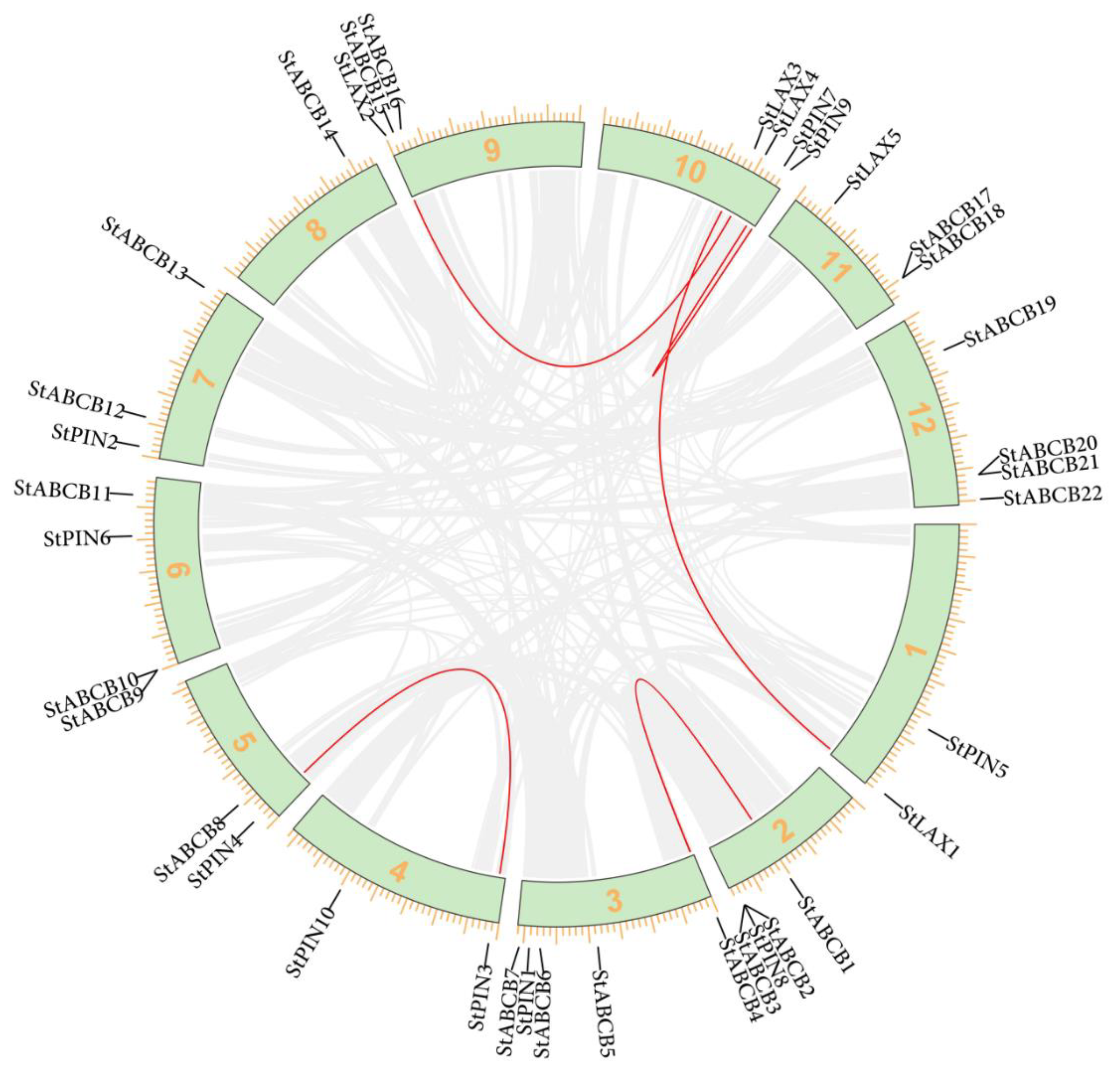
3.2. Chromosomal Distribution of S. tuberosum LAX, PIN, and ABCB Auxin Transporter Gene Families
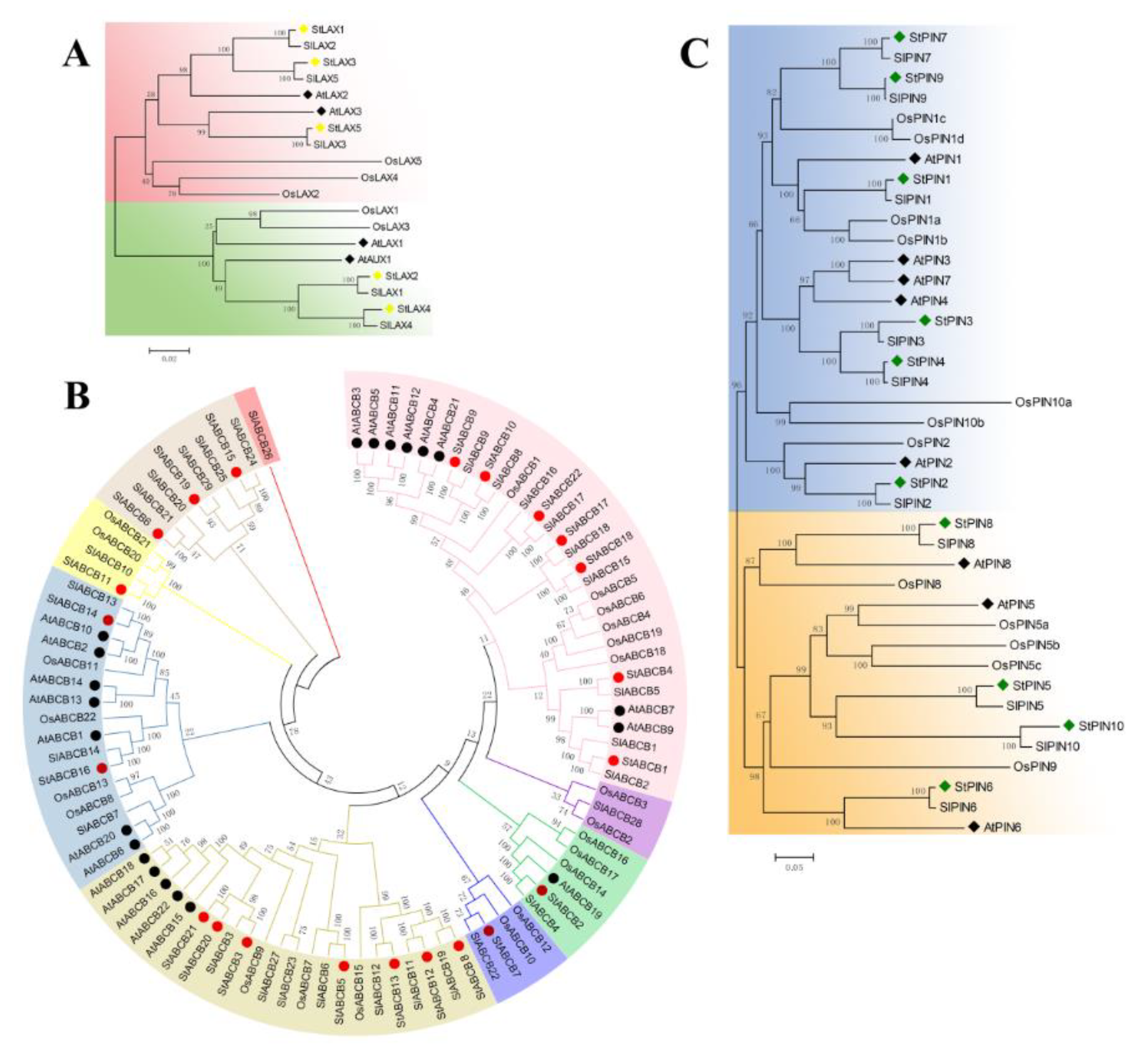
3.3. Phylogenetic Analysis of LAX, PIN, and ABCB Proteins in Arabidopsis, Rice, Tomato, and Potato
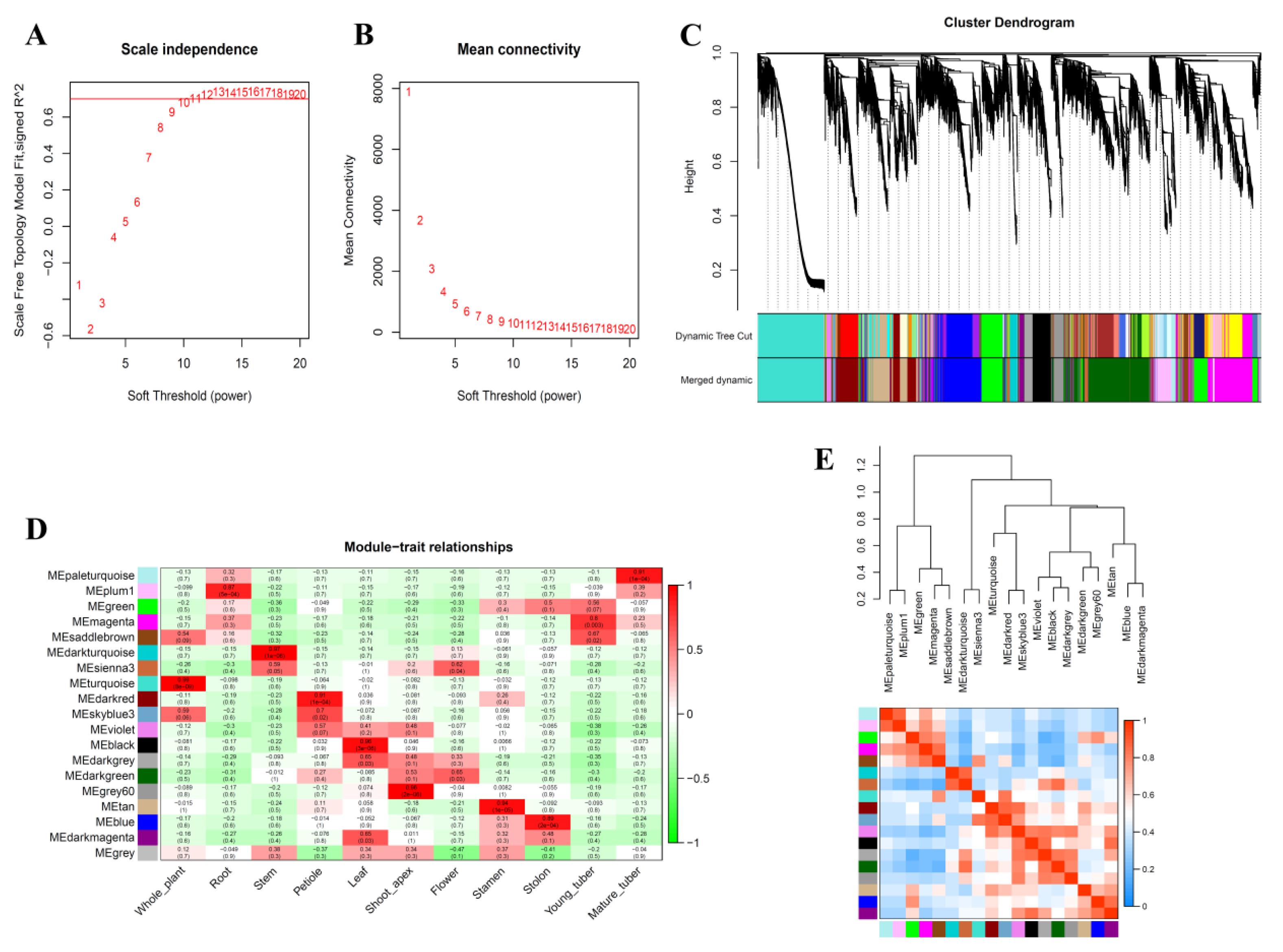
3.4. Weighted Co-Expression Network Analysis of S. tuberosum Genes
3.5. Gene Structure and Tissue-Specific Expression of StLAX, StPIN, and StABCB Family Genes
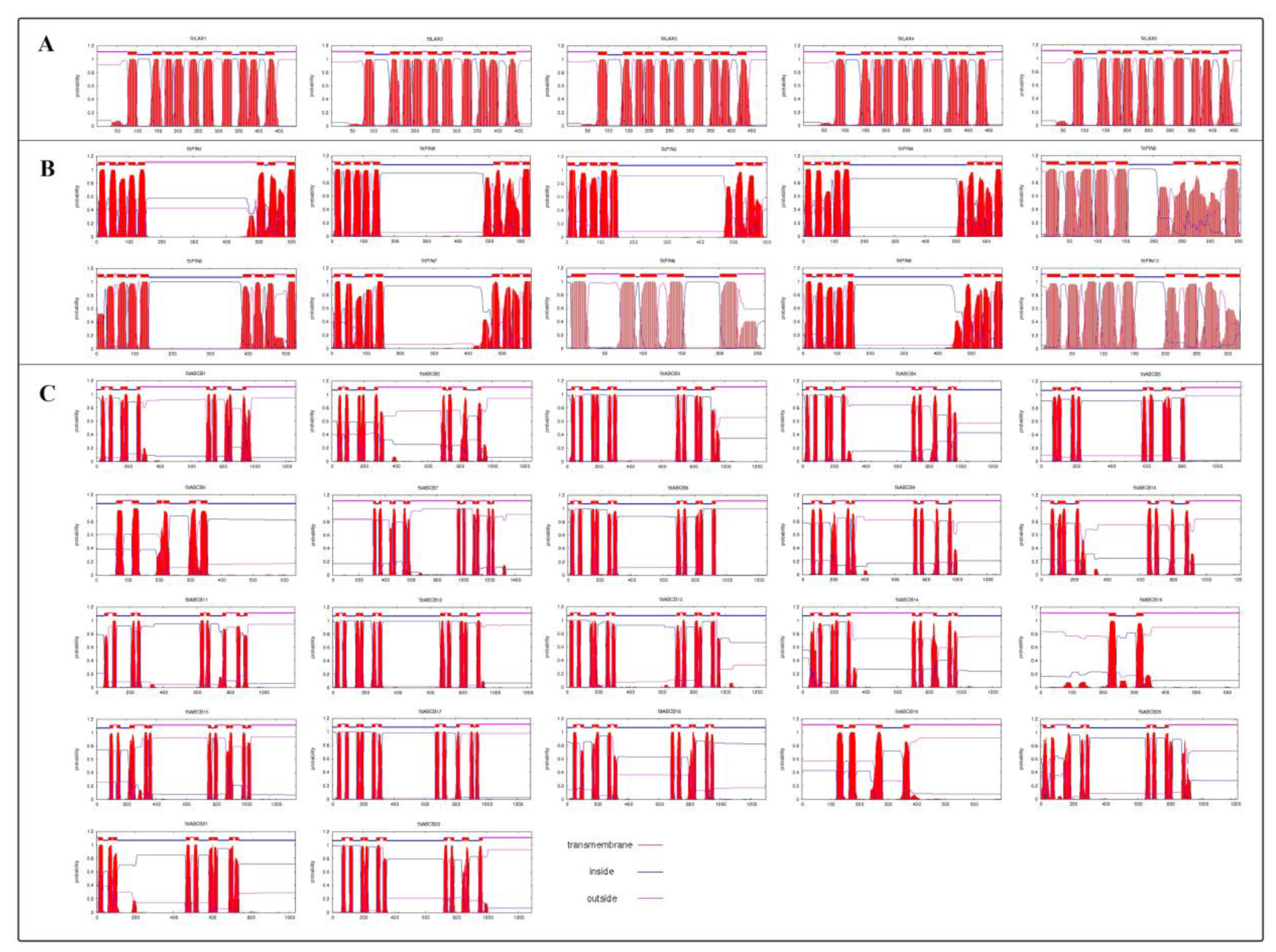
3.6. Analysis of StLAX, StPIN, and StABCB Protein Structures and Subcellular Location
3.7. Auxin Regulation of the StLAX, StPIN, and StABCB Genes
3.8. Expression Analysis of StLAX, StPIN, and StABCB Genes in Response to PATIs Treatments
3.9. Expression of StLAX, StPIN, and StABCB Genes in Response to ABA and Abiotic Stresses
3.10. Analysis of cis-Regulatory Elements in StLAX, StPIN, and StABCB Gene Promoters
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primer Name | Sequence (in 5′ → 3′ Order) | |
|---|---|---|
| Forward | Reverse | |
| StLAX1 | GGTTTGGCATTTAATTGTAC | CAGATTCGGTAGTTGTGA |
| StLAX2 | TGTTCTATGGTCTTATGGGTA | GGCAAGTCCTGCTGCTTT |
| StLAX3 | TTCACTGTTTACATTATCCCTG | TGTGCCTGCCCATCTTCC |
| StLAX4 | TATAGAACAACCTCTGCTCGAC | CAACAACTAAAACCCAGCCTAC |
| StLAX5 | CATTATTCCTGCCTTAGCACAC | AAAATTGACCATACTTGCCCAC |
| StPIN1 | TTTGCTATGGCTGTGAGAT | TTGTGGTAGAGCTGCCTGT |
| StPIN2 | AGGAGGTAGGAGTATGAGC | CACTGACACTGGCGTTGG |
| StPIN3 | TTGGTCCCTAATCTCGTAT | AAAGTAGCCACCGTGTTCC |
| StPIN4 | ATGATTTTCTCGAAACAGACGC | CGGATTCCTCGAAGAACTAAGA |
| StPIN5 | TGGCACATGTTTAAGCCAGA | GCCCATAAGACGAGGATCAG |
| StPIN6 | GAATCCGCATTTTCATCCTC | CCCGTTATGTAAAGGCGTGT |
| StPIN7 | CCTGATATACTTAGCACAGGGG | ACTCTCTTCAGTATCAAAGCCC |
| StPIN8 | ATCAGGAAGCAACCTCTACAAA | ACCAGACTAAACTCTGTAGTGC |
| StPIN9 | TCCTACCAATGCTGGAATCTTT | CATTACCATAGTCACCTCCTCC |
| StPIN10 | GTGGTGACCTTATTGCTAAAGC | CAGGAATTCCCATAACAAGTGC |
| StABCB1 | GGTGTGGTAACTTTGCTAGTTG | TGGAAGCATCAGTTGATAACCT |
| StABCB2 | AAACCGTCAGCCTAGCAC | CTCTTTCCCGACCCACTT |
| StABCB3 | CTTAAAAGTGCCAACGGGTAAA | GACTCACTAGACCCATTTGTGA |
| StABCB4 | GAATTGATTCAAGTAGCAGCGA | GCTCAAGCTCAAATCTCTGAAG |
| StABCB5 | TACCAAAATGACCGGTCGAATA | AGTGATCCTTTATCTGCATCGT |
| StABCB6 | GCAGGAAGATTACGAGAT | TTGGATAGTCAGAGTGGC |
| StABCB7 | GCTTTCTGTTGATTGTGTCAGT | GCTCCTAGAGTAAATGGTGTGA |
| StABCB8 | CTGTTTCTTGCAAGTCGTGTAA | TCATCAATTGCTATAACGTGCG |
| StABCB9 | GTTAAATGTCCCCATTAGTCGC | CCTAAGTTCTTCTGGTGGCTTA |
| StABCB10 | CAATGAAGGGCCATCTAG | GTATGAATGGTGGGAGCA |
| StABCB11 | GTATATGGCCTTCGCAACATTT | CAAACGTGCTACTTGTCTTTGA |
| StABCB12 | CAACAGATGCTAACATGGTACG | TTTTGGGACGTTTCTGACATTC |
| StABCB13 | GGTATGGCGGAAAACTATTGTC | CCTTATTTGTGTTCTCGTTGCA |
| StABCB14 | GCTCATCGCCTTTCTACC | GCACCAATACCGCTAACA |
| StABCB15 | AGGCAAACAATGCGGAGTC | TCAACAGAACCAGGGATG |
| StABCB16 | ATCAGAAATGCACATGTGATCG | TCTCCGTGTGTAAATCGTTGTA |
| StABCB17 | GAAAAGAATCCAGTTACTCGCC | GTACTGTTGCAAGAACCACAAT |
| StABCB18 | CACTCATGATTGTGGTTCTTGG | AGTTGCATCAGTAGAGAGCTTT |
| StABCB19 | TGATTTATTCTACATGGTGGGT | TTCAATGCGTCCTGCTAA |
| StABCB20 | GGGGAAATCGAATTCAAACACA | TCTCTGAAGCAGTGCTATTACC |
| StABCB21 | TTGAAGTCATTGAGGAACCAGA | CTTCTGATGCTAGCACTCCATA |
| StABCB22 | TGCTGCTGAAGTTCCATACTAT | CCAAACAAAACAGCCATCAAAG |
| ef1α | ATTGGAAACGGATATGCTCCA | TCCTTACCTGAACGCCTGTCA |
| StLAX | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | *** | 73.35 | 88.1 | 72.89 | 81.01 |
| 2 | 68.47 | *** | 72.21 | 89.12 | 73.39 |
| 3 | 82.16 | 68.3 | *** | 71.31 | 80.08 |
| 4 | 68.74 | 84.63 | 68.91 | *** | 73.2 |
| 5 | 72.24 | 70.4 | 72.99 | 69.11 | *** |
| StPIN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | *** | 58.81 | 42.54 | 62.95 | 27.6 | 48.14 | 66.13 | 14.63 | 69.82 | 21.01 |
| 2 | 63.77 | *** | 35.55 | 56.48 | 26.7 | 47.47 | 56.78 | 16.01 | 58.75 | 20.13 |
| 3 | 43.64 | 41.38 | *** | 59.94 | 23.13 | 30.62 | 41.57 | 15.67 | 42.74 | 18.32 |
| 4 | 62.76 | 59.06 | 58.52 | *** | 25.76 | 43.75 | 60.27 | 15.6 | 61.26 | 19.42 |
| 5 | 29.13 | 28.33 | 36.07 | 26.27 | *** | 28.62 | 28.35 | 31.83 | 28.36 | 45.63 |
| 6 | 54 | 51.92 | 43.38 | 50.38 | 32.58 | *** | 50.17 | 24.86 | 50.5 | 22.1 |
| 7 | 66.61 | 59.74 | 42.31 | 58.85 | 29.31 | 54.78 | *** | 16.16 | 84.14 | 21.64 |
| 8 | 21.51 | 20.92 | 26 | 19.62 | 36.65 | 23.63 | 23.27 | *** | 16.5 | 26.54 |
| 9 | 67.26 | 60.36 | 41.33 | 59.92 | 29.83 | 53.58 | 82.11 | 22.72 | *** | 21.72 |
| 10 | 25.89 | 23 | 33.52 | 24.19 | 57.77 | 29.69 | 26.06 | 38.12 | 26.04 | *** |
| StABCB | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | *** | 43.66 | 42.44 | 65.69 | 37.91 | 17.22 | 33.16 | 62.99 | 57.83 | 40.42 | 36.41 | 39.23 | 39.17 | 44.48 | 14.47 | 41.31 | 50.11 | 50.15 | 14.33 | 40.13 | 33.31 | 55.22 |
| 2 | 53.71 | *** | 45.35 | 42.31 | 40.50 | 17.21 | 35.40 | 44.45 | 40.93 | 43.17 | 39.56 | 40.71 | 42.86 | 50.87 | 14.46 | 50.26 | 38.19 | 38.88 | 14.68 | 43.68 | 36.07 | 41.97 |
| 3 | 51.19 | 53.92 | *** | 42.33 | 49.09 | 16.09 | 32.67 | 41.46 | 38.54 | 53.18 | 37.66 | 51.14 | 51.88 | 43.05 | 14.78 | 42.16 | 37.21 | 37.85 | 14.80 | 74.17 | 60.24 | 40.91 |
| 4 | 67.43 | 53.12 | 51.26 | *** | 39.67 | 16.67 | 32.22 | 59.78 | 54.33 | 41.33 | 35.27 | 39.34 | 40.88 | 43.92 | 13.08 | 40.30 | 47.13 | 47.33 | 15.78 | 41.40 | 33.97 | 51.77 |
| 5 | 47.79 | 47.94 | 53.11 | 48.6 | *** | 16.17 | 31.05 | 38.50 | 39.67 | 45.74 | 34.03 | 44.47 | 45.04 | 38.72 | 15.53 | 37.90 | 36.58 | 35.92 | 15.05 | 48.77 | 43.41 | 36.56 |
| 6 | 24.17 | 23.53 | 23.16 | 24.19 | 25.07 | *** | 13.46 | 17.20 | 18.42 | 15.59 | 16.72 | 15.83 | 16.19 | 16.03 | 27.76 | 16.16 | 15.85 | 15.32 | 28.47 | 16.48 | 17.74 | 15.65 |
| 7 | 42.75 | 42.82 | 41.4 | 40.79 | 38.58 | 19.62 | *** | 34.28 | 31.44 | 31.53 | 28.93 | 30.62 | 31.64 | 32.55 | 10.58 | 34.53 | 31.01 | 30.97 | 11.42 | 31.07 | 24.56 | 33.35 |
| 8 | 64.04 | 52.14 | 50.59 | 61.58 | 46.88 | 23.50 | 43.21 | *** | 81.81 | 39.95 | 35.34 | 38.59 | 39.28 | 43.83 | 13.96 | 42.34 | 56.15 | 56.12 | 14.13 | 39.77 | 33.05 | 63.16 |
| 9 | 60.40 | 48.68 | 47.92 | 58.21 | 48.81 | 25.10 | 40.17 | 82.70 | *** | 37.65 | 35.66 | 34.88 | 37.71 | 40.36 | 14.50 | 39.63 | 51.60 | 52.80 | 15.81 | 37.59 | 34.70 | 57.82 |
| 10 | 50.74 | 52.31 | 56.92 | 52.33 | 51.70 | 23.91 | 41.64 | 49.40 | 48.58 | *** | 34.87 | 71.18 | 63.00 | 40.82 | 14.00 | 41.00 | 36.71 | 36.62 | 14.45 | 51.95 | 42.64 | 38.96 |
| 11 | 47.45 | 46.98 | 46.53 | 46.31 | 47.27 | 25.26 | 38.03 | 44.61 | 47.93 | 47.52 | *** | 34.41 | 33.92 | 37.84 | 13.79 | 36.07 | 33.03 | 32.95 | 14.97 | 37.53 | 33.84 | 34.67 |
| 12 | 51.43 | 50.69 | 56.28 | 51.62 | 50.72 | 24.19 | 40.34 | 49.28 | 48.25 | 74.49 | 47.37 | *** | 56.79 | 38.84 | 12.37 | 38.64 | 35.41 | 35.80 | 14.45 | 50.04 | 40.84 | 39.20 |
| 13 | 51 | 51.58 | 56.95 | 51.01 | 51.39 | 23.95 | 40.92 | 49.25 | 48.43 | 64.69 | 47.11 | 62.49 | *** | 40.40 | 13.55 | 42.12 | 36.85 | 36.49 | 14.24 | 49.41 | 40.10 | 37.96 |
| 14 | 53.55 | 55.60 | 51.29 | 52.18 | 47.02 | 23.97 | 41.27 | 52.07 | 48.45 | 51.24 | 47.38 | 50.80 | 50.35 | *** | 13.06 | 47.56 | 39.30 | 39.30 | 14.53 | 41.36 | 34.97 | 40.61 |
| 15 | 23.27 | 23.25 | 22.98 | 22.64 | 24.67 | 42.35 | 19.20 | 22.48 | 24.28 | 22.77 | 24.05 | 22.91 | 22.62 | 23.19 | *** | 12.80 | 12.69 | 13.11 | 23.50 | 15.20 | 17.44 | 14.03 |
| 16 | 50.53 | 54.56 | 50.57 | 49.48 | 44.69 | 22.82 | 43.30 | 50.02 | 45.58 | 49.98 | 45.50 | 48.69 | 48.57 | 54.25 | 22.05 | *** | 37.26 | 37.48 | 13.08 | 40.81 | 33.98 | 40.25 |
| 17 | 56.75 | 49.46 | 48.86 | 55.65 | 45.77 | 22.35 | 39.96 | 62.35 | 58.46 | 48.78 | 43.78 | 48.19 | 47.54 | 48.91 | 21.63 | 48.61 | *** | 72.55 | 13.50 | 38.19 | 30.67 | 49.22 |
| 18 | 56.37 | 49.85 | 48.50 | 55.35 | 45.62 | 23.53 | 40.10 | 61.55 | 57.82 | 49.24 | 45.30 | 47.89 | 47.10 | 49.64 | 22.36 | 48.76 | 82.66 | *** | 13.53 | 38.19 | 30.66 | 50.04 |
| 19 | 23.86 | 23.60 | 24.46 | 24.45 | 26.81 | 42.01 | 20.86 | 23.17 | 24.88 | 25.08 | 26.65 | 24.76 | 23.78 | 24.89 | 40.56 | 24.09 | 23.84 | 24.03 | *** | 14.80 | 17.42 | 16.02 |
| 20 | 50.59 | 52.28 | 78.32 | 50.34 | 53.90 | 24.49 | 40.70 | 48.88 | 48.42 | 56.78 | 46.81 | 56.34 | 55.46 | 50.45 | 23.35 | 49.10 | 48.43 | 47.58 | 24.79 | *** | 72.76 | 38.92 |
| 21 | 42.72 | 44.10 | 63.39 | 42.72 | 49.67 | 28.03 | 34.04 | 41.25 | 44.26 | 47.34 | 42.67 | 46.19 | 47.06 | 41.80 | 27.77 | 40.88 | 40.30 | 39.65 | 29.29 | 75.03 | *** | 31.76 |
| 22 | 60.64 | 50.90 | 50.78 | 57.77 | 46.27 | 23.52 | 42.91 | 66.67 | 60.38 | 48.63 | 44.53 | 49.09 | 49.37 | 51.62 | 21.89 | 50.68 | 56.48 | 56.77 | 24.06 | 48.82 | 40.55 | *** |
| Species | Gene Name | Locus ID |
|---|---|---|
| Arabidopsis | AtAUX1 | AT2G38120 |
| AtLAX1 | AT5G01240 | |
| AtLAX2 | AT2G21050 | |
| AtLAX3 | AT1G77690 | |
| AtPIN1 | AT1G73590 | |
| AtPIN2 | AT5G57090 | |
| AtPIN3 | AT1G70940 | |
| AtPIN4 | AT2G01420 | |
| AtPIN5 | AT5G16530 | |
| AtPIN6 | AT1G77110 | |
| AtPIN7 | AT1G23080 | |
| AtPIN8 | AT5G15100 | |
| AtABCB1 | AT2G36910 | |
| AtABCB2 | AT4G25960 | |
| AtABCB3 | AT4G01820 | |
| AtABCB4 | AT2G47000 | |
| AtABCB5 | AT4G01830 | |
| AtABCB6 | AT2G39480 | |
| AtABCB7 | AT5G46540 | |
| AtABCB9 | AT4G18050 | |
| AtABCB10 | AT1G10680 | |
| AtABCB11 | AT1G02520 | |
| AtABCB12 | AT1G02530 | |
| AtABCB13 | AT1G27940 | |
| AtABCB14 | AT1G28010 | |
| AtABCB15 | AT3G28345 | |
| AtABCB16 | AT3G28360 | |
| AtABCB17 | AT3G28380 | |
| AtABCB18 | AT3G28390 | |
| AtABCB19 | AT3G28860 | |
| AtABCB20 | AT3G55320 | |
| AtABCB21 | AT3G62150 | |
| AtABCB22 | AT3G28415 | |
| Solanum tuberosum | StLAX1 | PGSC0003DMT400004027 |
| StLAX2 | PGSC0003DMT400021923 | |
| StLAX3 | PGSC0003DMT400059693 | |
| StLAX4 | PGSC0003DMT400049377 | |
| StLAX5 | PGSC0003DMT400016760 | |
| StPIN1 | PGSC0003DMT400014752 | |
| StPIN2 | PGSC0003DMT400048251 | |
| StPIN3 | PGSC0003DMT400015267 | |
| StPIN4 | PGSC0003DMT400078330 | |
| StPIN5 | PGSC0003DMT400046253 | |
| StPIN6 | PGSC0003DMT400079013 | |
| StPIN7 | PGSC0003DMT400072459 | |
| StPIN8 | PGSC0003DMT400003569 | |
| StPIN9 | PGSC0003DMT400021600 | |
| StPIN10 | PGSC0003DMT400027309 | |
| StABCB1 | PGSC0003DMT400007960 | |
| StABCB2 | PGSC0003DMT400003590 | |
| StABCB3 | PGSC0003DMT400003546 | |
| StABCB4 | PGSC0003DMT400034908 | |
| StABCB5 | PGSC0003DMT400048379 | |
| StABCB6 | PGSC0003DMT400063067 | |
| StABCB7 | PGSC0003DMT400058977 | |
| StABCB8 | PGSC0003DMT400027962 | |
| StABCB9 | PGSC0003DMT400018820 | |
| StABCB10 | PGSC0003DMT400018812 | |
| StABCB11 | PGSC0003DMT400069516 | |
| StABCB12 | PGSC0003DMT400013988 | |
| StABCB13 | PGSC0003DMT400049576 | |
| StABCB14 | PGSC0003DMT400045176 | |
| StABCB15 | PGSC0003DMT400022893 | |
| StABCB16 | PGSC0003DMT400009924 | |
| StABCB17 | PGSC0003DMT400019156 | |
| StABCB18 | PGSC0003DMT400019085 | |
| StABCB19 | PGSC0003DMT400074962 | |
| StABCB20 | PGSC0003DMT400030345 | |
| StABCB21 | PGSC0003DMT400030342 | |
| StABCB22 | PGSC0003DMT400011930 | |
| Rice | OsLAX1 | LOC_Os01g63770 |
| OsLAX2 | LOC_Os03g14080 | |
| OsLAX3 | LOC_Os05g37470 | |
| OsLAX4 | LOC_Os10g05690 | |
| OsLAX5 | LOC_Os11g06820 | |
| OsPIN1a | LOC_Os06g12610 | |
| OsPIN1b | LOC_Os02g50960 | |
| OsPIN1c | LOC_Os11g04190 | |
| OsPIN1d | LOC_Os12g04000 | |
| OsPIN2 | LOC_Os06g44970 | |
| OsPIN5a | LOC_Os01g69070 | |
| OsPIN5b | LOC_Os08g41720 | |
| OsPIN5c | LOC_Os09g32770 | |
| OsPIN8 | LOC_Os01g51780 | |
| OsPIN9 | LOC_Os01g58860 | |
| OsPIN10a | LOC_Os01g45550 | |
| OsPIN10b | LOC_Os05g50140 | |
| OsABCB1 | LOC_Os01g18670 | |
| OsABCB2 | LOC_Os01g34970 | |
| OsABCB3 | LOC_Os01g35030 | |
| OsABCB4 | LOC_Os01g50080 | |
| OsABCB5 | LOC_Os01g50100 | |
| OsABCB6 | LOC_Os01g50160 | |
| OsABCB7 | LOC_Os01g52550 | |
| OsABCB8 | LOC_Os01g74470 | |
| OsABCB9 | LOC_Os02g09720 | |
| OsABCB10 | LOC_Os02g21750 | |
| OsABCB11 | LOC_Os02g46680 | |
| OsABCB12 | LOC_Os03g08380 | |
| OsABCB13 | LOC_Os03g17180 | |
| OsABCB14 | LOC_Os04g38570 | |
| OsABCB15 | LOC_Os04g40570 | |
| OsABCB16 | LOC_Os04g54930 | |
| OsABCB18 | LOC_Os05g47490 | |
| OsABCB19 | LOC_Os05g47500 | |
| OsABCB20 | LOC_Os08g05690 | |
| OsABCB21 | LOC_Os08g05710 | |
| OsABCB22 | LOC_Os08g45030 | |
| Solanum lycopersicum | SlLAX1 | HQ671063 |
| SlLAX2 | HQ671064 | |
| SlLAX3 | HQ671065 | |
| SlLAX4 | HQ671066 | |
| SlLAX5 | HQ671067 | |
| SlPIN1 | HQ127074 | |
| SlPIN2 | HQ127077 | |
| SlPIN3 | HQ127079 | |
| SlPIN4 | HQ127078 | |
| SlPIN5 | HQ127080 | |
| SlPIN6 | HQ127082 | |
| SlPIN7 | HQ127076 | |
| SlPIN8 | HQ127083 | |
| SlPIN9 | HQ127075 | |
| SlPIN10 | HQ127081 | |
| SlABCB1 | Solyc02g071340.1 | |
| SlABCB2 | Solyc02g071350.2 | |
| SlABCB3 | Solyc02g087410.2 | |
| SlABCB4 | Solyc02g087870.2 | |
| SlABCB5 | Solyc03g005860.2 | |
| SlABCB6 | Solyc03g093650.2 | |
| SlABCB7 | Solyc04g010310.2 | |
| SlABCB8 | Solyc06g009280.1 | |
| SlABCB9 | Solyc06g009290.2 | |
| SlABCB10 | Solyc06g072960.1 | |
| SlABCB11 | Solyc07g018130.1 | |
| SlABCB12 | Solyc07g064120.1 | |
| SlABCB13 | Solyc08g076720.2 | |
| SlABCB14 | Solyc09g008240.2 | |
| SlABCB15 | Solyc11g067310.1 | |
| SlABCB16 | Solyc12g098840.1 | |
| SlABCB17 | Solyc12g098870.1 | |
| SlABCB18 | Solyc11g067300.1 | |
| SlABCB19 | Solyc05g013890.1 | |
| SlABCB20 | Solyc03g026310.2 | |
| SlABCB21 | Solyc03g114950.2 | |
| SlABCB22 | Solyc03g122050.1 | |
| SlABCB23 | Solyc03g122070.1 | |
| SlABCB24 | Solyc09g009910.2 | |
| SlABCB25 | Solyc09g055350.2 | |
| SlABCB26 | Solyc00g304030.1 | |
| SlABCB27 | Solyc12g049120.1 | |
| SlABCB28 | Solyc12g049130.1 | |
| SlABCB29 | Solyc12g070280.1 |
| Genes | Probes | Module Color |
|---|---|---|
| StABCB10 | PGSC0003DMT400018812 | black |
| StPIN5 | PGSC0003DMT400046253 | blue |
| StPIN6 | PGSC0003DMT400079013 | blue |
| StABCB1 | PGSC0003DMT400007960 | blue |
| StABCB5 | PGSC0003DMT400048379 | blue |
| StABCB6 | PGSC0003DMT400063067 | blue |
| StABCB9 | PGSC0003DMT400018820 | blue |
| StABCB15 | PGSC0003DMT400022893 | blue |
| StPIN1 | PGSC0003DMT400014752 | darkgreen |
| StABCB3 | PGSC0003DMT400003546 | darkgreen |
| StABCB16 | PGSC0003DMT400009924 | darkgrey |
| StLAX4 | PGSC0003DMT400049377 | darkmagenta |
| StLAX1 | PGSC0003DMT400004027 | darkred |
| StLAX3 | PGSC0003DMT400059693 | darkred |
| StPIN4 | PGSC0003DMT400078330 | darkred |
| StPIN7 | PGSC0003DMT400072459 | darkred |
| StPIN9 | PGSC0003DMT400021600 | darkred |
| StABCB19 | PGSC0003DMT400074962 | darkred |
| StPIN2 | PGSC0003DMT400048251 | grey60 |
| StPIN3 | PGSC0003DMT400015267 | grey60 |
| StABCB22 | PGSC0003DMT400011930 | grey60 |
| StABCB14 | PGSC0003DMT400045176 | magenta |
| StABCB17 | PGSC0003DMT400019156 | magenta |
| StLAX2 | PGSC0003DMT400021923 | turquoise |
| StLAX5 | PGSC0003DMT400016760 | violet |
| StABCB2 | PGSC0003DMT400003590 | violet |
| Gene | Flower | Leaf | Shoot Apex | Stem | Stolon | Root | Young Tuber | Mature Tuber |
|---|---|---|---|---|---|---|---|---|
| StLAX1 | 2.04 | 1.80 | 2.85 | 3.97 | 5.65 | 4.53 | 3.88 | 0.32 |
| StLAX2 | 6.48 | 5.78 | 4.12 | 6.42 | 5.12 | 5.48 | 6.86 | 2.96 |
| StLAX3 | −1.49 | −1.19 | 0.88 | 3.97 | 3.99 | −0.15 | 1.81 | −1.04 |
| StLAX4 | 2.20 | 1.68 | 0.36 | 4.62 | 2.32 | 4.39 | 3.56 | 0.62 |
| StLAX5 | 1.88 | 2.14 | −0.18 | 3.84 | 2.87 | 2.79 | 4.35 | −1.41 |
| StPIN1 | 2.42 | 1.26 | 3.92 | 3.78 | 3.96 | 2.77 | 4.05 | 2.34 |
| StPIN2 | −3.29 | −2.41 | −1.66 | −0.97 | −2.39 | −2.69 | 3.66 | −4.84 |
| StPIN3 | 3.17 | 1.51 | 1.74 | 3.16 | 1.98 | −0.28 | 5.37 | −0.41 |
| StPIN4 | 5.48 | 5.48 | 3.38 | 5.31 | 6.00 | 3.29 | 5.68 | 1.86 |
| StPIN5 | 2.69 | 2.61 | −0.10 | 2.70 | 1.12 | 4.88 | 0.50 | 0.56 |
| StPIN6 | −1.88 | −3.00 | −0.84 | 0.68 | 0.83 | 1.60 | 0.80 | −1.26 |
| StPIN7 | 1.24 | 1.28 | 1.49 | 0.69 | 3.53 | 0.84 | 2.39 | 0.72 |
| StPIN8 | 0.37 | −5.93 | 0.00 | 0.68 | −5.93 | −1.61 | 0.68 | −1.59 |
| StPIN9 | 0.80 | −1.68 | 0.70 | 3.18 | 4.36 | 3.62 | 3.62 | −0.43 |
| StPIN10 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 |
| StABCB1 | 1.95 | −1.13 | −2.80 | −0.07 | −2.14 | 2.70 | −1.70 | −2.75 |
| StABCB2 | 4.62 | 3.10 | 3.98 | 6.49 | 6.40 | 3.47 | 6.78 | 2.48 |
| StABCB3 | 0.29 | 0.23 | 2.73 | 2.50 | 2.46 | 2.61 | 2.52 | 0.08 |
| StABCB4 | −1.93 | −2.04 | −1.40 | −5.93 | 0.11 | −1.33 | −0.46 | −3.21 |
| StABCB5 | −2.51 | −4.21 | 0.86 | 1.82 | −2.60 | 5.35 | 0.53 | 1.53 |
| StABCB6 | 3.62 | 5.67 | 4.15 | 3.07 | 4.47 | 6.49 | 4.31 | 3.28 |
| StABCB7 | −4.54 | −3.07 | −5.93 | −1.10 | −4.64 | −2.53 | −5.93 | −5.93 |
| StABCB8 | 2.23 | 2.72 | 2.24 | 2.81 | 1.89 | 4.33 | 3.33 | 2.23 |
| StABCB9 | −1.18 | −0.50 | −1.55 | 2.26 | −0.29 | 1.25 | −0.63 | −1.56 |
| StABCB10 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 | −5.23 | −5.93 | −5.93 |
| StABCB11 | 0.14 | −1.48 | −1.11 | −0.69 | 0.20 | 0.97 | −0.45 | −1.45 |
| StABCB12 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 | −5.93 |
| StABCB13 | −3.25 | −5.93 | −5.93 | −4.27 | −3.35 | −4.23 | −5.93 | −5.93 |
| StABCB14 | 3.34 | 3.98 | −0.46 | 3.82 | 4.58 | 0.44 | 1.65 | −1.27 |
| StABCB15 | 2.66 | 2.75 | 2.23 | 2.10 | 2.72 | 3.28 | 2.07 | 1.81 |
| StABCB16 | 4.77 | 5.07 | 5.32 | 6.69 | 5.26 | 5.53 | 5.98 | 5.01 |
| StABCB17 | 0.42 | −0.15 | 0.81 | −0.05 | 1.24 | 0.96 | 0.34 | 0.34 |
| StABCB18 | −1.69 | −3.41 | −5.25 | −5.93 | −3.22 | −5.28 | −4.31 | −5.93 |
| StABCB19 | 3.63 | 4.94 | 2.90 | 4.03 | 4.44 | 1.41 | 2.86 | 1.82 |
| StABCB20 | −2.61 | −1.97 | −5.93 | −5.93 | −5.29 | −4.01 | −5.93 | −5.93 |
| StABCB21 | 0.64 | −4.06 | −5.93 | −3.96 | −5.93 | −5.93 | −5.93 | −5.93 |
| StABCB22 | 0.29 | −1.59 | −5.93 | 0.03 | −2.98 | −0.87 | 1.29 | −5.93 |
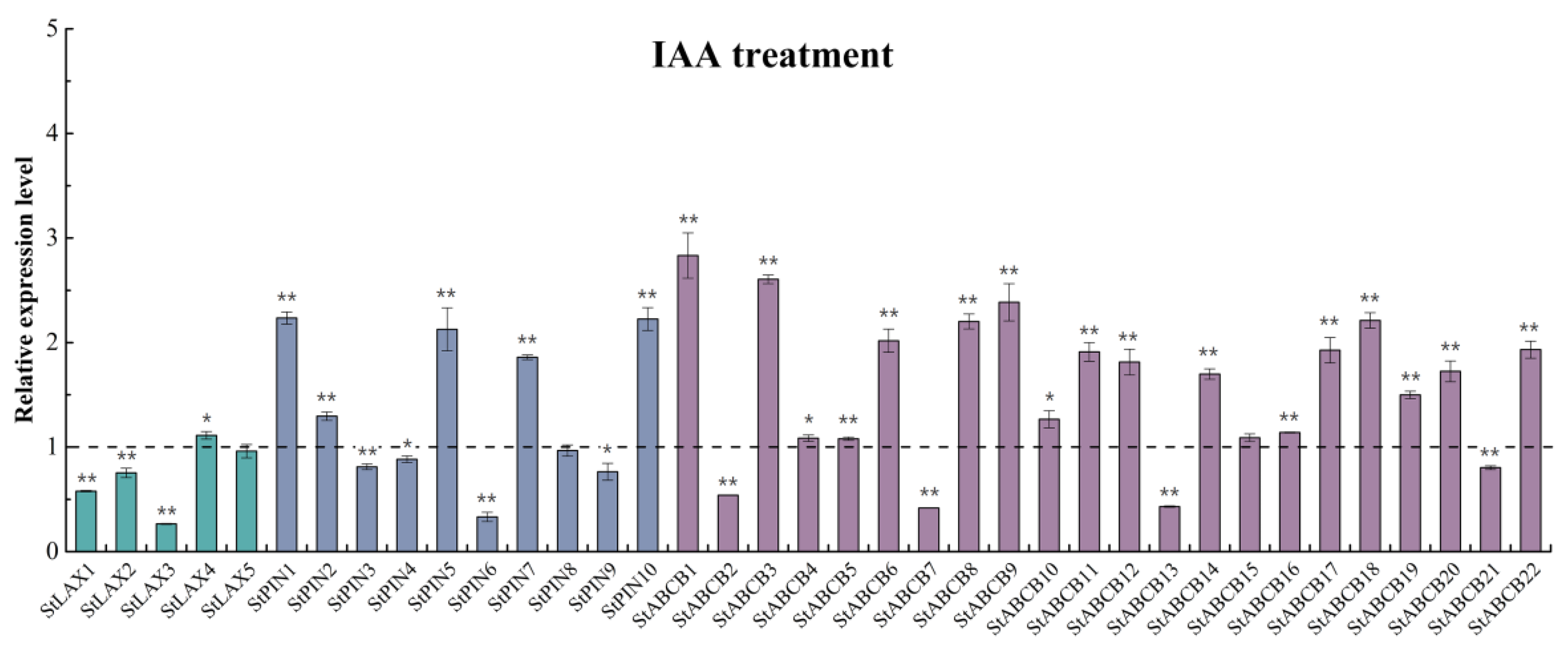
| Gene | Control | IAA | 1-NOA | TIBA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| StLAX1 | 1.00 | 0.59 | 0.57 | 0.58 | 0.58 ± 0.01 ** | 0.92 | 0.89 | 0.93 | 0.91 ± 0.01 ** | 1.09 | 1.12 | 1.14 | 1.12 ± 0.02 ** |
| StLAX2 | 1.00 | 0.69 | 0.73 | 0.84 | 0.75 ± 0.05 ** | 0.73 | 0.74 | 0.72 | 0.73 ± 0.01 ** | 0.78 | 0.83 | 0.80 | 0.80 ± 0.01 ** |
| StLAX3 | 1.00 | 0.27 | 0.25 | 0.27 | 0.26 ± 0.01 ** | 0.91 | 0.84 | 0.86 | 0.87 ± 0.01 ** | 1.43 | 1.37 | 1.41 | 1.40 ± 0.02 ** |
| StLAX4 | 1.00 | 1.05 | 1.11 | 1.17 | 1.11 ± 0.04 * | 0.82 | 0.86 | 0.88 | 0.85 ± 0.01 ** | 0.53 | 0.53 | 0.56 | 0.54 ± 0.01 ** |
| StLAX5 | 1.00 | 0.84 | 0.98 | 1.06 | 0.96 ± 0.06 | 0.65 | 0.66 | 0.71 | 0.67 ± 0.01 ** | 0.89 | 0.92 | 0.91 | 0.90 ± 0.01 ** |
| StPIN1 | 1.00 | 2.21 | 2.15 | 2.35 | 2.23 ± 0.06 ** | 1.36 | 1.23 | 1.32 | 1.30 ± 0.04 ** | 1.07 | 1.01 | 1.07 | 1.05 ± 0.02 |
| StPIN2 | 1.00 | 1.22 | 1.36 | 1.31 | 1.30 ± 0.04 ** | 1.40 | 1.48 | 1.35 | 1.41 ± 0.04 ** | 0.84 | 0.87 | 0.86 | 0.86 ± 0.01 ** |
| StPIN3 | 1.00 | 0.81 | 0.86 | 0.77 | 0.81 ± 0.03 ** | 0.64 | 0.64 | 0.58 | 0.62 ± 0.02 ** | 1.88 | 1.90 | 1.70 | 1.83 ± 0.06 ** |
| StPIN4 | 1.00 | 0.83 | 0.93 | 0.89 | 0.88 ± 0.03* | 0.28 | 0.28 | 0.31 | 0.29 ± 0.01 ** | 0.46 | 0.50 | 0.52 | 0.49 ± 0.02 ** |
| StPIN5 | 1.00 | 1.71 | 2.32 | 2.35 | 2.13 ± 0.21 ** | 1.13 | 1.28 | 1.29 | 1.24 ± 0.05 * | 0.55 | 0.55 | 0.58 | 0.56 ± 0.01 ** |
| StPIN6 | 1.00 | 0.42 | 0.26 | 0.32 | 0.33 ± 0.04 ** | 0.64 | 0.42 | 0.45 | 0.50 ± 0.07 ** | 0.83 | 0.56 | 0.57 | 0.65 ± 0.09 * |
| StPIN7 | 1.00 | 1.88 | 1.88 | 1.81 | 1.86 ± 0.02 ** | 1.03 | 1.13 | 1.15 | 1.11 ± 0.04 | 0.84 | 0.91 | 0.92 | 0.89 ± 0.02 * |
| StPIN8 | 1.00 | 0.87 | 0.97 | 1.06 | 0.97 ± 0.05 | 0.90 | 0.90 | 0.81 | 0.87 ± 0.03 * | 0.77 | 0.81 | 0.66 | 0.75 ± 0.04 ** |
| StPIN9 | 1.00 | 0.70 | 0.92 | 0.67 | 0.76 ± 0.08 * | 1.64 | 1.57 | 1.19 | 1.47 ± 0.14 * | 1.86 | 1.86 | 1.35 | 1.69 ± 0.17 * |
| StPIN10 | 1.00 | 2.20 | 2.43 | 2.05 | 2.23 ± 0.11 ** | 1.35 | 1.31 | 1.24 | 1.30 ± 0.03 ** | 0.66 | 0.70 | 0.64 | 0.67 ± 0.02 ** |
| StABCB1 | 1.00 | 2.58 | 2.65 | 3.26 | 2.83 ± 0.22 ** | 5.03 | 5.18 | 5.10 | 5.11 ± 0.04 ** | 1.30 | 1.30 | 1.32 | 1.31 ± 0.01 ** |
| StABCB2 | 1.00 | 0.54 | 0.54 | 0.54 | 0.54 ± 0.00 ** | 0.61 | 0.63 | 0.65 | 0.63 ± 0.01 ** | 0.98 | 0.99 | 0.99 | 0.98 ± 0.00 |
| StABCB3 | 1.00 | 2.63 | 2.66 | 2.53 | 2.60 ± 0.04 ** | 1.16 | 1.27 | 1.20 | 1.21 ± 0.03 ** | 0.58 | 0.59 | 0.55 | 0.57 ± 0.01 ** |
| StABCB4 | 1.00 | 1.06 | 1.05 | 1.14 | 1.08 ± 0.03 * | 1.31 | 1.30 | 1.31 | 1.31 ± 0.00 ** | 1.44 | 1.40 | 1.34 | 1.39 ± 0.03 ** |
| StABCB5 | 1.00 | 1.06 | 1.07 | 1.11 | 1.08 ± 0.02 ** | 1.24 | 1.40 | 1.40 | 1.35 ± 0.05 ** | 0.98 | 0.99 | 1.02 | 1.00 ± 0.01 |
| StABCB6 | 1.00 | 2.23 | 1.97 | 1.85 | 2.02 ± 0.11 ** | 1.38 | 1.24 | 1.26 | 1.29 ± 0.04 ** | 0.65 | 0.61 | 0.56 | 0.60 ± 0.03 ** |
| StABCB7 | 1.00 | 0.42 | 0.41 | 0.42 | 0.42 ± 0.00 ** | 1.88 | 2.07 | 2.07 | 2.00 ± 0.06 ** | 0.54 | 0.55 | 0.60 | 0.56 ± 0.02 ** |
| StABCB8 | 1.00 | 2.34 | 2.11 | 2.15 | 2.20 ± 0.07 ** | 2.20 | 1.91 | 2.02 | 2.04 ± 0.08 ** | 2.46 | 2.19 | 2.19 | 2.28 ± 0.09 ** |
| StABCB9 | 1.00 | 2.03 | 2.58 | 2.55 | 2.39 ± 0.18 ** | 1.37 | 1.37 | 1.29 | 1.34 ± 0.03 ** | 0.96 | 0.96 | 0.91 | 0.95 ± 0.02* |
| StABCB10 | 1.00 | 1.11 | 1.29 | 1.40 | 1.27 ± 0.08* | 0.92 | 0.94 | 0.97 | 0.94 ± 0.02 * | 0.52 | 0.49 | 0.51 | 0.51 ± 0.01 ** |
| StABCB11 | 1.00 | 1.90 | 2.07 | 1.76 | 1.91 ± 0.09 ** | 0.51 | 0.50 | 0.43 | 0.48 ± 0.03 ** | 0.53 | 0.55 | 0.50 | 0.53 ± 0.02 ** |
| StABCB12 | 1.00 | 2.06 | 1.68 | 1.69 | 1.81 ± 0.12 ** | 1.49 | 1.35 | 1.27 | 1.37 ± 0.06 ** | 0.12 | 0.10 | 0.10 | 0.11 ± 0.01 ** |
| StABCB13 | 1.00 | 0.43 | 0.42 | 0.44 | 0.43 ± 0.01 ** | 0.74 | 0.78 | 0.78 | 0.76 ± 0.01 ** | 0.18 | 0.19 | 0.22 | 0.20 ± 0.01 ** |
| StABCB14 | 1.00 | 1.65 | 1.80 | 1.65 | 1.70 ± 0.05 ** | 0.95 | 0.92 | 1.05 | 0.97 ± 0.04 | 0.85 | 0.82 | 0.80 | 0.82 ± 0.02 ** |
| StABCB15 | 1.00 | 1.14 | 1.11 | 1.02 | 1.09 ± 0.04 | 0.51 | 0.50 | 0.55 | 0.52 ± 0.02 ** | 0.75 | 0.76 | 0.70 | 0.74 ± 0.02 ** |
| StABCB16 | 1.00 | 1.14 | 1.15 | 1.13 | 1.14 ± 0.00 ** | 0.88 | 0.89 | 0.97 | 0.92 ± 0.03 * | 1.52 | 1.52 | 1.50 | 1.51 ± 0.01 ** |
| StABCB17 | 1.00 | 2.16 | 1.86 | 1.76 | 1.93 ± 0.12 ** | 1.31 | 1.24 | 1.11 | 1.22 ± 0.06 * | 0.98 | 0.83 | 0.77 | 0.86 ± 0.06 |
| StABCB18 | 1.00 | 2.08 | 2.22 | 2.34 | 2.21 ± 0.07 ** | 1.01 | 1.14 | 1.07 | 1.07 ± 0.04 | 0.63 | 0.70 | 0.70 | 0.68 ± 0.02 ** |
| StABCB19 | 1.00 | 1.43 | 1.54 | 1.53 | 1.50 ± 0.04 ** | 0.96 | 0.98 | 1.04 | 0.99 ± 0.03 | 1.17 | 1.19 | 1.18 | 1.18 ± 0.01 ** |
| StABCB20 | 1.00 | 1.71 | 1.90 | 1.56 | 1.72 ± 0.10 ** | 1.03 | 1.06 | 0.98 | 1.03 ± 0.02 | 0.75 | 0.76 | 0.65 | 0.72 ± 0.03 ** |
| StABCB21 | 1.00 | 0.84 | 0.79 | 0.78 | 0.80 ± 0.02 ** | 1.87 | 1.71 | 1.60 | 1.72 ± 0.08 ** | 0.33 | 0.31 | 0.29 | 0.31 ± 0.01 ** |
| StABCB22 | 1.00 | 1.88 | 1.82 | 2.09 | 1.93 ± 0.08 ** | 1.86 | 1.94 | 2.06 | 1.95 ± 0.06 ** | 1.85 | 1.79 | 1.96 | 1.86 ± 0.05 ** |
| Gene | Control | ABA | Salt | Drought | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| StLAX1 | 1.00 | 0.33 | 0.35 | 0.36 | 0.35 ± 0.01 ** | 0.45 | 0.45 | 0.44 | 0.45 ± 0.00 ** | 0.29 | 0.31 | 0.32 | 0.31 ± 0.01 ** |
| StLAX2 | 1.00 | 0.64 | 0.67 | 0.68 | 0.66 ± 0.01 ** | 1.13 | 1.05 | 1.05 | 1.07 ± 0.03 * | 1.14 | 1.19 | 1.17 | 1.17 ± 0.01 ** |
| StLAX3 | 1.00 | 0.25 | 0.38 | 0.38 | 0.34 ± 0.04 ** | 0.24 | 0.27 | 0.30 | 0.27 ± 0.02 ** | 0.08 | 0.12 | 0.14 | 0.11 ± 0.02 ** |
| StLAX4 | 1.00 | 0.69 | 0.75 | 0.79 | 0.74 ± 0.03 ** | 1.79 | 2.14 | 1.98 | 1.97 ± 0.10 ** | 1.24 | 1.48 | 1.34 | 1.35 ± 0.07 ** |
| StLAX5 | 1.00 | 0.54 | 0.69 | 0.69 | 0.64 ± 0.05 ** | 0.56 | 0.55 | 0.48 | 0.53 ± 0.03 ** | 0.84 | 0.91 | 0.77 | 0.84 ± 0.04* |
| StPIN1 | 1.00 | 0.88 | 0.80 | 0.94 | 0.87 ± 0.04 * | 0.40 | 0.36 | 0.31 | 0.35 ± 0.03 ** | 0.35 | 0.33 | 0.28 | 0.32 ± 0.02 ** |
| StPIN2 | 1.00 | 1.26 | 1.28 | 1.35 | 1.30 ± 0.03 ** | 0.62 | 0.59 | 0.57 | 0.60 ± 0.02 ** | 0.53 | 0.49 | 0.47 | 0.50 ± 0.02 ** |
| StPIN3 | 1.00 | 0.69 | 0.74 | 0.62 | 0.68 ± 0.03 ** | 1.65 | 1.89 | 1.79 | 1.78 ± 0.07 ** | 1.19 | 1.35 | 1.15 | 1.23 ± 0.06 * |
| StPIN4 | 1.00 | 0.47 | 0.49 | 0.47 | 0.47 ± 0.01 ** | 1.29 | 1.29 | 1.08 | 1.22 ± 0.07 * | 1.26 | 1.47 | 1.29 | 1.34 ± 0.07 ** |
| StPIN5 | 1.00 | 0.95 | 1.24 | 1.34 | 1.18 ± 0.12 | 1.45 | 1.26 | 1.20 | 1.30 ± 0.08 * | 0.77 | 0.69 | 0.66 | 0.71 ± 0.03 ** |
| StPIN6 | 1.00 | 0.42 | 0.27 | 0.27 | 0.32 ± 0.05 ** | 0.11 | 0.15 | 0.17 | 0.14 ± 0.02 ** | 0.13 | 0.19 | 0.21 | 0.18 ± 0.02 ** |
| StPIN7 | 1.00 | 0.87 | 0.96 | 0.93 | 0.92 ± 0.03 | 1.30 | 1.28 | 1.20 | 1.26 ± 0.03 ** | 1.09 | 1.32 | 1.06 | 1.16 ± 0.08 |
| StPIN8 | 1.00 | 1.41 | 1.50 | 1.21 | 1.37 ± 0.09 * | 1.72 | 1.76 | 1.71 | 1.73 ± 0.02 ** | 0.80 | 0.87 | 0.94 | 0.87 ± 0.04 * |
| StPIN9 | 1.00 | 0.56 | 0.69 | 0.57 | 0.61 ± 0.04 ** | 0.44 | 0.53 | 0.50 | 0.49 ± 0.03 ** | 0.12 | 0.18 | 0.18 | 0.16 ± 0.02 ** |
| StPIN10 | 1.00 | 0.51 | 0.58 | 0.51 | 0.53 ± 0.02 ** | 1.88 | 1.97 | 2.25 | 2.04 ± 0.11 ** | 1.32 | 1.44 | 1.84 | 1.54 ± 0.16 * |
| StABCB1 | 1.00 | 1.40 | 1.69 | 1.76 | 1.62 ± 0.11 ** | 3.82 | 3.45 | 2.83 | 3.36 ± 0.29 ** | 5.11 | 3.97 | 3.43 | 4.17 ± 0.49 ** |
| StABCB2 | 1.00 | 0.77 | 0.77 | 0.79 | 0.78 ± 0.01 ** | 1.23 | 1.15 | 1.14 | 1.17 ± 0.03 ** | 0.99 | 0.90 | 0.78 | 0.89 ± 0.06 |
| StABCB3 | 1.00 | 0.64 | 0.70 | 0.72 | 0.68 ± 0.03 ** | 0.84 | 0.84 | 0.85 | 0.84 ± 0.00 ** | 1.81 | 2.01 | 2.03 | 1.95 ± 0.07 ** |
| StABCB4 | 1.00 | 0.46 | 0.51 | 0.50 | 0.49 ± 0.02 ** | 0.35 | 0.28 | 0.28 | 0.31 ± 0.02 ** | 1.08 | 0.87 | 0.90 | 0.95 ± 0.07 |
| StABCB5 | 1.00 | 0.33 | 0.41 | 0.44 | 0.39 ± 0.03 ** | 0.97 | 1.03 | 0.91 | 0.97 ± 0.03 | 0.24 | 0.24 | 0.21 | 0.23 ± 0.01 ** |
| StABCB6 | 1.00 | 1.25 | 1.15 | 1.09 | 1.16 ± 0.05 * | 1.71 | 1.33 | 1.39 | 1.48 ± 0.12* | 1.08 | 1.03 | 1.09 | 1.07 ± 0.02 * |
| StABCB7 | 1.00 | 0.40 | 0.42 | 0.45 | 0.42 ± 0.02 ** | 1.32 | 1.33 | 1.33 | 1.33 ± 0.00 ** | 1.57 | 1.72 | 1.76 | 1.69 ± 0.06 ** |
| StABCB8 | 1.00 | 1.16 | 1.05 | 1.29 | 1.16 ± 0.07 | 0.36 | 0.41 | 0.36 | 0.37 ± 0.02 ** | 0.26 | 0.26 | 0.27 | 0.26 ± 0.00 ** |
| StABCB9 | 1.00 | 0.61 | 0.71 | 0.74 | 0.68 ± 0.04 ** | 0.59 | 0.60 | 0.62 | 0.61 ± 0.01 ** | 0.45 | 0.54 | 0.55 | 0.52 ± 0.03 ** |
| StABCB10 | 1.00 | 0.72 | 0.75 | 0.95 | 0.81 ± 0.07 | 2.74 | 2.53 | 2.19 | 2.49 ± 0.16 ** | 1.95 | 1.85 | 1.63 | 1.81 ± 0.09 ** |
| StABCB11 | 1.00 | 0.40 | 0.49 | 0.42 | 0.44 ± 0.03 ** | 1.87 | 1.91 | 1.93 | 1.90 ± 0.02 ** | 3.00 | 3.29 | 3.22 | 3.17 ± 0.09 ** |
| StABCB12 | 1.00 | 1.31 | 1.29 | 1.19 | 1.26 ± 0.04 ** | 1.53 | 1.56 | 1.60 | 1.57 ± 0.02 ** | 1.10 | 1.16 | 1.15 | 1.14 ± 0.02 ** |
| StABCB13 | 1.00 | 0.32 | 0.31 | 0.37 | 0.34 ± 0.02 ** | 1.75 | 1.64 | 1.55 | 1.65 ± 0.06 ** | 0.58 | 0.62 | 0.57 | 0.59 ± 0.02 ** |
| StABCB14 | 1.00 | 0.95 | 0.91 | 0.94 | 0.93 ± 0.01 ** | 2.59 | 2.81 | 2.61 | 2.67 ± 0.07 ** | 4.13 | 4.53 | 3.97 | 4.21 ± 0.17 ** |
| StABCB15 | 1.00 | 0.67 | 0.77 | 0.72 | 0.72 ± 0.03 ** | 1.77 | 2.23 | 1.77 | 1.92 ± 0.15 ** | 3.49 | 3.52 | 2.80 | 3.27 ± 0.24 ** |
| StABCB16 | 1.00 | 0.89 | 1.06 | 1.02 | 0.99 ± 0.05 | 0.45 | 0.41 | 0.42 | 0.43 ± 0.01 ** | 0.84 | 0.76 | 0.77 | 0.79 ± 0.02 ** |
| StABCB17 | 1.00 | 1.26 | 1.14 | 1.06 | 1.16 ± 0.06 | 1.31 | 1.31 | 1.32 | 1.31 ± 0.00 ** | 1.09 | 1.05 | 1.05 | 1.06 ± 0.01 * |
| StABCB18 | 1.00 | 1.04 | 1.24 | 1.18 | 1.15 ± 0.06 | 3.22 | 3.15 | 3.29 | 3.22 ± 0.04 ** | 3.09 | 3.08 | 3.31 | 3.16 ± 0.07 ** |
| StABCB19 | 1.00 | 1.03 | 1.04 | 1.06 | 1.05 ± 0.01* | 1.09 | 1.19 | 1.18 | 1.15 ± 0.03 ** | 1.32 | 1.31 | 1.34 | 1.32 ± 0.01 ** |
| StABCB20 | 1.00 | 0.92 | 0.97 | 0.80 | 0.90 ± 0.05 | 3.27 | 2.55 | 2.43 | 2.75 ± 0.26 ** | 2.70 | 2.21 | 2.12 | 2.34 ± 0.18 ** |
| StABCB21 | 1.00 | 0.99 | 0.97 | 0.91 | 0.96 ± 0.03 | 0.53 | 0.52 | 0.49 | 0.51 ± 0.01 ** | 0.25 | 0.27 | 0.25 | 0.26 ± 0.01 ** |
| StABCB22 | 1.00 | 0.74 | 0.76 | 0.76 | 0.75 ± 0.01 ** | 2.70 | 2.77 | 2.70 | 2.72 ± 0.02 ** | 1.46 | 1.46 | 1.41 | 1.44 ± 0.02 ** |
| cis-Element | Core Sequences | PLACE Entries | Function |
|---|---|---|---|
| AuxRE | TGTCTC | S000270 | auxin−responsive element |
| TGACG | S000024 | bZIP response elements | |
| ACTCAT | S000450 | ||
| MACCWAMC | S000167 | Myb response element | |
| ABRE | ACGTG | S000414 | ABA−responsive element |
| MYBR | CTAACCA | S000175 | MYB recognition site |
| MYCR | CACATG | S000174 | MYC recognition site |
| DRE | TACCGACAT | S000152 | dehydration responsive element |
| CE1 | TGCCACCGG | S000014 | ABA responsive complex |
| W-box | TTGAC | S000390 | |
| TGACT | S000442 | ||
| GCC box | GCCGCC | S000430 | |
| HB | CAATNATTG | S000371 | Homeodomein−leucine zipper |
| EE | AAAATATCT | S000385 | Evening element |
References
- Dubrovsky, J.G.; Sauer, M.; Napsucialy-Mendivil, S.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Celenza, J.; Benkova, E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 2008, 105, 8790–8794. [Google Scholar] [CrossRef] [Green Version]
- Gallavotti, A. The role of auxin in shaping shoot architecture. J. Exp. Bot. 2013, 64, 2593–2608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novak, S.D.; Luna, L.J.; Gamage, R.N. Role of auxin in orchid development. Plant Signal. Behav. 2014, 9, e972277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, A. Auxin: A regulator of cold stress response. Physiol. Plant. 2013, 147, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Ghanashyam, C.; Jain, M. Role of auxin-responsive genes in biotic stress responses. Plant Signal. Behav. 2009, 4, 846–848. [Google Scholar] [CrossRef]
- Ljung, K.; Hull, A.K.; Kowalczyk, M.; Marchant, A.; Celenza, J.; Cohen, J.D.; Sandberg, G. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 2002, 50, 309–332. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, S.; Benjamin, P. AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, M.; Murphy, A.S. The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett. 2006, 580, 1094–1102. [Google Scholar] [CrossRef] [Green Version]
- Péret, B.; Swarup, K.; Ferguson, A.; Seth, M.; Yang, Y.; Dhondt, S.; James, N.; Casimiro, I.; Paula, P.; Syed, A. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 2012, 24, 2874–2885. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Hammes, U.Z.; Taylor, C.G.; Schachtman, D.P.; Nielsen, E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 2006, 16, 1123–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarup, R. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001, 15, 2648–2653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarup, R. Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 2004, 16, 3069–3083. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Swarup, R.; Bennett, M.; Schaller, G.E.; Kieberet, J.J. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 2013, 23, 1979–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarup, K.; Benková, E.; Swarup, R.; Casimiro, I.; Péret, B.; Yang, Y.; Parry, G.; Nielsen, E.; Smet, I.; Vanneste, S. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Yue, R.; Tie, S.; Sun, T.; Zhang, L.; Yang, Y.; Qi, J.; Yan, S.; Han, X.; Wang, H.; Shem, C. Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE 2015, 10, e0118751. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Bai, Y.; Wang, S.; Zhang, S.; Wu, Y.; Chen, M.; Jiang, D.; Qi, Y. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 2010, 277, 2954–2969. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Tisdaleorr, T.E.; Clouse, R.M.; Anne, S.K.; Rachel, S. Diversification and expression of the PIN, AUX/LAX, and ABCB families of putative auxin transporters in Populus. Front. Plant Sci. 2012, 3, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.; Dong, W.; Zhan, Y.; Huang, Z.A.; Li, Z.; Kim, I.S.; Zhang, C. Genome-wide identification and expression analysis of ClLAX, ClPIN and ClABCB genes families in Citrullus lanatus under various abiotic stresses and grafting. BMC Genet. 2017, 18, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.W.; Lyu, S.W.; Tang, J.; Zhou, D.Y.; Bonnema, G.; Xiao, D.; Hou, X.L.; Zhang, C.W. Genome-wide analysis of auxin transport genes identifies the hormone responsive patterns associated with leafy head formation in Chinese cabbage. Sci. Rep. 2017, 7, 42229. [Google Scholar] [CrossRef]
- Sawchuk, M.G.; Scarpella, E. Control of vein patterning by intracellular auxin transport. Plant Signal. Behav. 2013, 8, e27205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paponov, I.A.; Teale, W.D.; Trebar, M.; Blilou, I.; Palme, K. The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 2005, 10, 170–177. [Google Scholar] [CrossRef]
- GäLweiler, L.; Guan, C.; Müller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef] [Green Version]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.; Guan, C.; Gälweiler, L.; Tänzler, P.; Huijser, P.; Marchant, A.; Parry, G.; Bennett, M.; Wisman, E.; Palme, K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 2014, 17, 6903–6911. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, C.; Gaxiola, R.A.; Grisafi, P.; Fink, G.R. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998, 12, 2175–2187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Hilson, P.; Sedbrook, J.; Rosen, E.; Caspar, T.; Masson, P.H. The Arabidopsis thaliana AGRAVITROPIC1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 1998, 95, 15112–15117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [Green Version]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Palme, K. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Krecek, P.; Bielach, A.; Petrásek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.D. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Skůpa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klíma, P.; Čarná, M.; Rolčík, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.L.; Fekete, M.L.; Klinkenberg, P.M.; Hampton, M.; Bauer, B.; Malecha, M.; Lindgren, K.; Maki, J.A.; Perera, M.A.D.N.; Nikolau, B.J. PIN6 is required for nectary auxin response and short stamen development. Plant J. 2013, 74, 893–904. [Google Scholar] [CrossRef]
- Nisar, N.; Cuttriss, A.J.; Pogson, B.J.; Cazzonelli, C.I. The promoter of the Arabidopsis PIN6 auxin transporter enabled strong expression in the vasculature of roots, leaves, floral stems and reproductive organs. Plant Signal. Behav. 2014, 9, e27898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pěník, A.; Xu, C.; Tejos, R. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawchuk, M.G.; Alexander, E.; Enrico, S. Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet. 2013, 9, e1003294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodoulou, F.L. Plant ABC transporters. Biochim. Biophys. Acta 2000, 1465, 79–103. [Google Scholar] [CrossRef] [Green Version]
- Cho, M.; Cho, H.T. The function of ABCB transporters in auxin transport. Plant Signal. Behav. 2013, 8, e22990. [Google Scholar] [CrossRef] [Green Version]
- Titapiwatanakun, B.; Murphy, A.S. Post-transcriptional regulation of auxin transport proteins: Cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J. Exp. Bot. 2009, 60, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Dudler, R.; Hertig, C. Structure of an mdr-like gene from Arabidopsis thaliana. Evolutionary implications. J. Biol. Chem. 1992, 267, 5882–5888. [Google Scholar] [CrossRef]
- Sidler, M.; Hassa, P.; Hasan, S.; Ringli, C.; Dudleret, R. Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell 1998, 10, 1623–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noh, B.; Murphy, A.S.; Spalding, E.P. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 2001, 13, 2441–2454. [Google Scholar] [PubMed] [Green Version]
- Nagashima, A.; Suzuki, G.; Uehara, Y.; Saji, K.; Sakai, T. Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. Plant J. 2008, 53, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Santelia, D.; Vincenzetti, V.; Azzarello, E.; Bovet, L.; Fukao, Y.; Düchtig, P.; Mancuso, S.; Martinoia, E.; Geisler, M. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 2005, 579, 5399–5406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balzan, S.; Johal, G.S.; Carraro, N. The role of auxin transporters in monocots development. Front. Plant Sci. 2014, 5, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, C.; Yue, R.; Bai, Y.; Feng, R.; Sun, T.; Wang, X.; Yang, Y.; Shuanggui, T.; Wang, H. Identification and analysis of Medicago truncatula auxin transporter gene families uncover their roles in responses to Sinorhizobium meliloti infection. Plant Cell Physiol. 2015, 56, 1930. [Google Scholar] [CrossRef] [Green Version]
- Chai, C.; Subudhi, P. Comprehensive analysis and expression profiling of the OsLAX and OsABCB auxin transporter gene families in rice (Oryza sativa) under phytohormone stimuli and abiotic stresses. Front. Plant Sci. 2016, 7, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; Wang, J. The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [PubMed] [Green Version]
- Roumeliotis, E.; Kloosterman, B.; Oortwijn, M.; Visser, R.G.F.; Bachem, C.W.B. The PIN family of proteins in potato and their putative role in tuberization. Front. Plant Sci. 2013, 4, 524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Kelley, L.; Mezulis, S.; Yates, C.; Wass, M.; Sternberg, M. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Nie, T.; Wang, D.; Ma, X.; Chen, Y.; Chen, Q. Analysis of key genes involved in potato anthocyanin biosynthesis based on genomics and transcriptomics data. Front. Plant Sci. 2019, 10, 603. [Google Scholar]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. Biotechniques 1998, 24, 318–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Zhou, Y.; Zhao, Y.; Liu, Y.; Ke, Y.; Jin, X.; Ma, H. Internal reference gene selection for quantitative real-time RT-PCR normalization in potato tissues. Phyton Int. J. Exp. Bot. 2020, 89, 329–344. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Petrasek, J.; Zadnikova, P.; Hoyerová, K.; Pesek, B.; Raz, V.; Swarup, R.; Bennett, M.; Zazimalova, E.; Benkova, E. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 2010, 137, 597–606. [Google Scholar] [CrossRef] [Green Version]
- Kube, M.; Yang, H.; Richter, G.L.; Cheng, Y.; Młodzińska, E.; Wang, X.; Blakeslee, J.J.; Carraro, N.; Petrášek, J.; Zažímalová, E. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 2012, 69, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.R.; Wu, G.; Ljung, K.; Spalding, E.P. Auxin transport into cotyledons and cotyledon growth depend similarly on the ABCB19 multidrug resistance-like transporter. Plant J. 2009, 60, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Hoyerova, K.; Perry, L.; Hand, P.; Lanková, M.; Kocábek, T.; May, S.; Kottova, J.; Paces, J.; Napier, R.; Zazimalova, E. Functional characterization of PaLAX1, a putative auxin permease, in heterologous plant systems. Plant Physiol. 2008, 146, 1128–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tom, B.; Brockington, S.F.; Carl, R.; Graham, S.W.; Dennis, S.; Toni, K.; Megan, R.; Philip, T.; Ka-Shu, W.G.; Ottoline, L. Paralogous radiations of PIN proteins with multiple origins of noncanonical PIN structure. Mol. Biol. Evol. 2014, 31, 2042–2060. [Google Scholar]
- Kamimoto, Y.; Terasaka, K.; Hamamoto, M.; Takanashi, K.; Fukuda, S.; Shitan, N.; Akifumi, S.; Hideyuki, S.; Daisuke, S.; Wang, B. Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol. 2012, 53, 2090–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Choi, Y.; Burla, B.; Kim, Y.Y.; Jeon, B.; Maeshima, M.; Yoo, J.Y.; Martinoia, E.; Lee, Y. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 2008, 10, 1217–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, R.; Wang, H. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 2005, 138, 949–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneda, M.; Schuetz, M.; Lin, B.S.P.; Chanis, C.; Hamberger, B.; Western, T.L.; Ehlting, J.; Samuels, A.L. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot. 2011, 62, 2063–2077. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Dhonukshe, P.; Brewer, P.B.; Friml, J. Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 2006, 63, 2738–2754. [Google Scholar] [CrossRef]
- Hawkins, C.; Liu, Z. A model for an early role of auxin in Arabidopsis gynoecium morphogenesis. Front. Plant Sci. 2014, 5, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Wang, D.; Zhang, C.; Kong, N.; Ma, H.; Chen, Q. Comparative analysis of the PIN auxin transporter gene family in different plant species: A focus on structural and expression profiling of PINs in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 3270. [Google Scholar] [CrossRef] [Green Version]
- Lomax, T.L. Auxin transport. Plant Hormones 1995, 51, 494–500. [Google Scholar]
- Zhang, K.X.; Xu, H.H.; Yuan, T.T.; Zhang, L.; Lu, Y.T. Blue-light-induced PIN3 polarization for root negative phototropic response in Arabidopsis. Plant J. 2013, 76, 308–321. [Google Scholar]
- Mehrotra, R.; Bhalothia, P.; Bansal, P.; Basantani, M.K.; Bharti, V.; Mehrotra, S. Abscisic acid and abiotic stress tolerance-different tiers of regulation. J. Plant Physiol. 2014, 171, 486–496. [Google Scholar] [CrossRef]
- Kemal, K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 2013, 112, 1655–1665. [Google Scholar]
- Wang, Y.; Li, K.; Li, X. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. J. Plant Physiol. 2009, 166, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhang, W.; Hu, H.; Li, B.; Wang, Y.; Zhao, Y.; Li, K.; Liu, M.; Li, X. Salt modulates gravity signaling pathway to regulate growth direction of primary roots in Arabidopsis. Plant Physiol. 2008, 146, 178–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terasaka, K.; Blakeslee, J.J.; Titapiwatanakun, B.; Peer, W.A.; Bandyopadhyay, A.; Makam, S.N.; Lee, B.O.R.; Richards, B.E.L.; Murphy, A.S.; Sato, F. PGP4, an ATP binding cassette p-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 2005, 17, 2922–2939. [Google Scholar] [CrossRef] [PubMed]
- Siepel, A.; Arbiza, L. Cis-regulatory elements and human evolution. Curr. Opin. Genet. Dev. 2014, 29, 81–89. [Google Scholar] [CrossRef]
- Weirauch, M.T.; Hughes, T.R. Conserved expression without conserved regulatory sequence: The more things change, the more they stay the same. Trends Genet. 2010, 26, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]





| Gene a | Locus ID a | ORF Length (bp) a | No.of Extrons | Chromosome Location (bp) a | Deducted Polypeptid b | No. of Transmembrane c | Subcellular Localization d | ||
|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | MI wt (Da) | pI | |||||||
| StLAX1 | PGSC0003DMT400004027 | 1485 | 8 | ch01:87287332..87290998 (−) | 494 | 55,706.95 | 8.5797 | 10 | cyto |
| StLAX2 | PGSC0003DMT400021923 | 1446 | 7 | ch09:210800..214038 (−) | 481 | 54,328.39 | 8.1707 | 10 | PM |
| StLAX3 | PGSC0003DMT400059693 | 1467 | 8 | ch10:46747590..46751792 (−) | 488 | 54,971.25 | 8.7078 | 10 | PM |
| StLAX4 | PGSC0003DMT400049377 | 1458 | 7 | ch10:50480526..50485783 (−) | 485 | 54,606.6 | 7.8706 | 10 | PM |
| StLAX5 | PGSC0003DMT400016760 | 1407 | 8 | ch11:10042232..10046787 (−) | 468 | 53,199.12 | 8.7862 | 10 | PM |
| StPIN1 | PGSC0003DMT400014752 | 1845 | 6 | ch03:58350702..58354249 (+) | 614 | 67,134.08 | 9.3381 | 8 | chlo |
| StPIN2 | PGSC0003DMT400048251 | 1896 | 7 | ch07:2647114..2649884 (+) | 631 | 68,666.48 | 9.4981 | 9 | PM |
| StPIN3 | PGSC0003DMT400015267 | 1395 | 6 | ch04:2170473..2172957 (−) | 601 | 65,908.59 | 7.0298 | 8 | PM |
| StPIN4 | PGSC0003DMT400078330 | 1965 | 6 | ch05:4250058..4253070 (−) | 654 | 71,290.07 | 7.3826 | 9 | chlo/PM |
| StPIN5 | PGSC0003DMT400046253 | 1068 | 5 | ch01:64013966..64017139 (−) | 355 | 39,264.82 | 9.2151 | 9 | PM |
| StPIN6 | PGSC0003DMT400079013 | 1587 | 7 | ch06:41187368..41193747 (−) | 528 | 57,652.42 | 8.8456 | 9 | PM |
| StPIN7 | PGSC0003DMT400072459 | 1764 | 6 | ch10:57054506..57057784 (+) | 587 | 63,906.07 | 8.8949 | 8 | chlo |
| StPIN8 | PGSC0003DMT400003569 | 783 | 4 | ch02:46450539..46452728 (+) | 260 | 28,407.25 | 9.6913 | 5 | PM |
| StPIN9 | PGSC0003DMT400021600 | 1785 | 6 | ch10:59284672..59287550 (−) | 594 | 64,304.33 | 9.4377 | 9 | chlo |
| StPIN10 | PGSC0003DMT400027309 | 966 | 3 | ch04:49481160..49482767 (+) | 321 | 35,909.45 | 7.2746 | 9 | vacu |
| StABCB1 | PGSC0003DMT400007960 | 3789 | 12 | ch02:30568338..30574681 (+) | 1262 | 136,961.15 | 7.3521 | 9 | PM |
| StABCB2 | PGSC0003DMT400003590 | 3750 | 10 | ch02:46284463..46291628 (+) | 1249 | 136,236.51 | 8.0288 | 9 | PM |
| StABCB3 | PGSC0003DMT400003546 | 3792 | 7 | ch02:46615100..46621182 (+) | 1263 | 137,488.42 | 8.0619 | 11 | PM |
| StABCB4 | PGSC0003DMT400034908 | 3780 | 12 | ch03:822878..830598 (+) | 1259 | 136,332.61 | 8.7596 | 10 | PM |
| StABCB5 | PGSC0003DMT400048379 | 3414 | 8 | ch03:37579500..37585781 (+) | 1137 | 124,433.16 | 8.2246 | 9 | PM |
| StABCB6 | PGSC0003DMT400063067 | 1917 | 17 | ch03:55025184..55031533 (+) | 638 | 68,411.61 | 8.8478 | 6 | PM |
| StABCB7 | PGSC0003DMT400058977 | 4584 | 11 | ch03:61365905..61372730 (+) | 1527 | 167,888.19 | 9.0608 | 12 | PM |
| StABCB8 | PGSC0003DMT400027962 | 3765 | 7 | ch05:11042954..11047763 (−) | 1254 | 137,231.31 | 9.1918 | 11 | PM |
| StABCB9 | PGSC0003DMT400018820 | 3864 | 12 | ch06:336919..342710 (+) | 1287 | 138,575.71 | 7.9275 | 9 | PM |
| StABCB10 | PGSC0003DMT400018812 | 3639 | 10 | ch06:344414..349662 (+) | 1212 | 130,723.67 | 7.799 | 8 | PM |
| StABCB11 | PGSC0003DMT400069516 | 3561 | 9 | ch06:53569963..53576584 (+) | 1186 | 130,633.57 | 7.4101 | 9 | PM |
| StABCB12 | PGSC0003DMT400013988 | 3681 | 8 | ch07:11605714..11610849 (−) | 1226 | 134,428.48 | 8.6851 | 11 | PM |
| StABCB13 | PGSC0003DMT400049576 | 3780 | 7 | ch07:54289854..54295427 (+) | 1259 | 137,937.76 | 9.2144 | 12 | PM |
| StABCB14 | PGSC0003DMT400045176 | 3774 | 12 | ch08:49324870..49334350 (−) | 1257 | 137,797.09 | 8.6611 | 10 | PM |
| StABCB15 | PGSC0003DMT400022893 | 1920 | 10 | ch09:2609568..2618204 (+) | 639 | 69,814.41 | 8.7886 | 2 | chlo |
| StABCB16 | PGSC0003DMT400009924 | 4002 | 10 | ch09:5129170..5136543 (+) | 1333 | 145,939.27 | 7.8141 | 11 | PM |
| StABCB17 | PGSC0003DMT400019156 | 3864 | 14 | ch11:40225813..40235522 (+) | 1287 | 141,384.58 | 9.5827 | 11 | PM |
| StABCB18 | PGSC0003DMT400019085 | 3885 | 13 | ch11:40240421..40248719 (−) | 1294 | 142,465.24 | 8.3389 | 12 | PM |
| StABCB19 | PGSC0003DMT400074962 | 2091 | 17 | ch12:13618482..13637049 (−) | 696 | 78,066.69 | 8.8706 | 4 | PM |
| StABCB20 | PGSC0003DMT400030345 | 3651 | 7 | ch12:52311070..52319362 (+) | 1216 | 133,255.89 | 8.8183 | 8 | PM |
| StABCB21 | PGSC0003DMT400030342 | 3096 | 6 | ch12:52333684..52340071 (+) | 1031 | 112,725.71 | 8.785 | 9 | PM |
| StABCB22 | PGSC0003DMT400011930 | 3864 | 12 | ch12:59696990..59702667 (−) | 1287 | 139,652.46 | 7.1317 | 11 | PM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Wang, D.; Zhang, C.; Ye, M.; Kong, N.; Ma, H.; Chen, Q. Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology 2021, 10, 127. https://doi.org/10.3390/biology10020127
Yang C, Wang D, Zhang C, Ye M, Kong N, Ma H, Chen Q. Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology. 2021; 10(2):127. https://doi.org/10.3390/biology10020127
Chicago/Turabian StyleYang, Chenghui, Dongdong Wang, Chao Zhang, Minghui Ye, Nana Kong, Haoli Ma, and Qin Chen. 2021. "Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses" Biology 10, no. 2: 127. https://doi.org/10.3390/biology10020127
APA StyleYang, C., Wang, D., Zhang, C., Ye, M., Kong, N., Ma, H., & Chen, Q. (2021). Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology, 10(2), 127. https://doi.org/10.3390/biology10020127






