3.1. The Rate of Loop Extrusion Is Controlled by the Friction Imposed by Cohesin
As we mentioned in the Introduction, there are several existing mechanisms currently considered to drive loop extrusion. It has been recently discovered that cohesin can exhibit a weak motor-like activity, actively extruding chromatin fibers. This activity has been shown in in vitro experiments on deproteinated DNA in the presence of Nipbl loader [
14]. This motor activity is relatively weak in terms of used energy but efficient in terms of extrusion rate of 2.1 kbps [
14]. Besides the proven ability of cohesin to act as a motor, there were other different mechanisms proposed to efficiently extrude the fiber and as such able to enhance the extrusion [
16,
17,
18]. These include purely diffusive extrusion of loops and transcriptionally driven extrusion. In our previous model for the transcriptionally driven extrusion, we showed that the accumulation of supercoiling generated during the transcription can be very efficient and powerful for extruding loops [
18]. Our previous model, however, inherently expected handcuff conformation of cohesin rings and mechanical pushing of the emerging supercoiling on the joint section of the handcuffs. Therefore, we wanted to test whether the model with transcriptionally induced supercoiling could still enhance loop extrusion in modified conditions. In these conditions, the cohesin is represented by a single ring embracing both fibers, but still inducing friction between the ring and the fibers. The levels of the friction between the fibers and cohesin are intended to be used in order to control the levels of accumulated supercoiling and possibly the rate of loop extrusion. Although the levels of supercoiling can be controlled by changing the speed of the motor, the speed of generation of supercoiling is a biologically fixed parameter of around 10 rotations per second generated by RNA polymerase [
39]. Therefore, we assume that varying the speed of the motor generating the supercoiling would not be correct. We consider varying the friction between cohesin and the fibers as the best option. The experimental work by Stigler et al. indicated that this friction can be very high, such that self-diffusion of the cohesin without the help of an active process would take too long for efficient loop extrusion in biologically reasonable times [
40]. On the other hand, the friction between cohesin and fibers not only would alter the release of the supercoiling by affecting axial rotations of the fiber but will also impose higher drag on the mutual translational movements of cohesin and fibers, i.e., diffusion of the rings. This makes the relation between the friction, accumulation of supercoiling and diffusion of cohesins rather non-trivial and needs to be explored by the simulations.
This determined the first step in the simulations, when after constructing the model, we performed series of simulations with different frictions between cohesin and fibers imposed. These simulations showed a clear effect and systematic dependence in terms of the extrusion rate as a function of the friction (
Figure 1a). First of all, we observe that the loops are extruded also by using a single ring embracing both fibers in so-called pseudo-topological binding [
14]. In the next step, we evaluated the sizes of the loops with the simulation time. The calculated loop size
ℓ with respect to the simulation time resulted in the loop extrusion rate that we show later in the discussion of the results (Figure 3). From the simulations, we observe that the loops are directionally extruded with a persistent motion of the ring in the direction from the motor towards the opposite side of the domain represented by the circular beaded chain to the point when the cohesin ring slips away from the beaded chain. This directional movement prevails even for very low friction imposed at the level of
γc = 2. In the case of very high friction between cohesin and the fiber, the movement is even more straightforward, with much less fluctuation of the instantaneous values of loop size (
Figure 1b). The extrusion also becomes very symmetric. The time of extrusion of the whole loop in the case of the highest
γc in the simulation was 15 times faster than the lowest
γc = 2. In the case of employing low gammas, the extrusion becomes more stochastic and asymmetric. The simulations also indicate that the dependence of the loop extrusion rate on the friction saturates when the increase of
γc from 2 to 20 speeds up the extrusion 4 times, but a further increase of
γc by a factor of 10 to
γc = 200 speeds extrusion only 2 times. This indicates that there exists an optimum value of friction mediating the loop extrusion, after which the loop extrusion rate reaches its maximum, and later, at much higher frictions, the extrusion would probably decrease or even stop. In our simulations, however, we do not explore frictions that high as this would require decreasing integration steps and would make the simulations unfeasible in terms of the computational time.
Differences are also found in the accumulation of supercoiling within the extruded loop as a function of imposed friction. We evaluated supercoiling within the extruded loop in terms of linking number and density of supercoiling (
Figure 2). For this, we computed values of the writhe and twist within the extruded loop bordered by the position
χc of the cohesin on the chromatin fiber for each frame of the simulation. The sum of the calculated writhe and twist gives the value of linking number ∆
Lk according to Fuller’s theorem [
41]. This can be used to calculate the density of supercoiling as
σ = (
Lk −
Lk0) /
Lk0 = ∆
Lk/
Lk0, where
Lk0 is the linking number of the relaxed state, and here, it will correspond to the number of turns of relaxed DNA
Lk0 = ~40 turns per bead [
42]. During extrusion of the loop, the supercoiling inside the loop is relaxed by the influx of relaxed portions of the fiber into the loop, but also by escaping of supercoiling through the ring by axial rotation of fibers. During the simulations, from several tens to hundreds of rotations of the motor are performed. This is consistent with the size of the loop, which is 60 kbp; hence, the extrusion of the whole fiber should take time in order of 10s of seconds. The total number of rotations of polymerase introducing supercoiling at a rate of 10 rotations per second should be also in the order of hundreds. The calculated linking numbers indicate that in the case of large gammas, about 87.5% of rotations are relaxed. In the case of simulations with low
γcs, the loop loses above 99% of rotations both due to axial rotations as well as by dissolving the supercoiling in the un-supercoiled portions of chromatin flowing through the cohesin ring into the loop. The fluctuations of the linking number in terms of its amplitude originate mostly from the fluctuation of the twist. The overall fluctuations are more intense in the case of the systems with lower settings of
γc. In the case of lower friction, the accumulation of supercoiling has a strong non-equilibrium character. On the other hand, the accumulation of supercoiling in the case of larger friction strongly limits the effusion of supercoiling by axial rotations, while the gradient of supercoiling between the motor and position of cohesin quickly re-establishes, yielding the linear dependence of ∆
Lk with time.
3.2. Mathematic Model of Transcriptionally Driven Loop Extrusion
In order to understand and explain the mechanism of the extrusion seen in the simulations, one may think of entropic competition of supercoiled and torsionally relaxed portions of the fiber. A similar problem has been studied by Orlandini et al. for two competing knots on a circular polymer chain separated by a sliplink [
43]. In their paper, the authors showed that increasing the topological complexity of the knotted part decreases the polymer’s conformational space. Consequently, the part of the molecule with a more complex knot tries to increase its entropy by pulling a larger portion of polymer through the border separated by the sliplink. Hence, in an analogical picture, one could think of the compacted supercoiled loop trying to restore its entropy by pulling the relaxed portion of the fiber through the cohesin ring. However, the model for the competition between the supercoiled and torsionally relaxed portions of fiber poses more difficulties for simulations. This is because, unlike the problem of Orlandini et al., our system is not topologically constrained. The supercoiling can escape by axial rotations. Even if we limit the axial rotations by friction, the diffusive motions of cohesin can still effectively erase the supercoiling from the extruded loop. Therefore, one needs to think of a more complex kinetic equilibrium. In the case of the kinetic equilibrium, the supercoiling is continuously introduced by the motor. In the model, the intrinsic minimum energy point would be when the cohesins are sitting just at the position of the polymerase. However, this is not possible due to the increase in excluded volume between the bead representing polymerase and the cohesin ring. Hence, the only way for the system to decrease its energy is by moving the cohesin ring ahead from the transcription site, which induces relaxed portions of the fiber into the loop. This causes a temporary drop in energy; however, the supercoiling is soon replenished; hence, the cohesin moves again to attain a new energy minimum state (
Video S1). In this way, the loop is extruded by cohesin movements, which follow the minimum energy path.
For a mathematical description of this picture one would need to describe the non-equilibrium evolution of supercoiling along the fiber. For this purpose, we use Fick’s second law of non-equilibrium diffusion and balancing the supercoiling along the fiber as suggested by Brackley et al. in their stochastic model for supercoiling [
44]:
where
σ denotes the density of supercoiling along the fiber at the distance
x, represented by the position of the bead on the simulated chain and at the simulated time
τ. Although the diffusivity of supercoiling is denoted here as a variable of the position
, its value in terms of
γR imposed on the periaxial beads in the simulations along the fiber is the same, and set to
γR = 1 for all
x except the position of cohesin
xc. The value is increased at the position of cohesin
xc, such that
Dc =
=
kBT/
γc =
γR/
γc with
ε =
kBT = 1. Initially, at the time
τ = 0, the fiber is torsionally relaxed
. For times
τ > 0, the supercoiling is continuously introduced at the position of
x = 0, which is mathematically treated as a setting of the boundary condition
, where the speed of production is given as
10 rotations per second. The equation is solved within the region
x ϵ <0,
xc >, which represents the size of the loop. Because the movement of cohesin is one of the most prominent features of our proposed model, this needs to be included in the solution. Hence, we need to describe the movement of cohesin and solve the diffusion of supercoiling as a problem with moving boundary set as
[
45]. The movement of cohesin can be described by the equation for displacement from the overdamped Langevin equation, where we neglect the noisy part of the equation.
The term on the right side of the equation originally describes the change of energy potential for a particle moving in a field. The
γc reflects the dissipation of the energy during the movement. It should be noted that the value of friction between the fiber and the rings is not the result of simulations but is introduced as a parameter setting into the model. Simulating the friction starting from first principles would require the employment of a fully atomistic model, and it would encounter several issues starting from the computational complexity of the problem [
46]. The atomistic computer simulations of chromatin fiber with a comparable size to ours were undertaken recently, while the system of 83 kbp consisted of one billion atoms [
47]. However, such simulations were only possible thanks to the employment of 100 thousand CPUs, while the time scales of seconds or minutes of real time were far beyond the reach of the atomistic simulations, as we could not take advantage of coarse-grained time units. If one was able to undertake the simulations of this kind of magnitude though, the friction coefficient could be explored as a measured property dependent also on cohesin ring mass, its hydrodynamic drag, etc., which would be then a part of Equation (2). Instead, in our simulations, we explore the behavior of supercoiling and loop extrusion rate for a wide range of frictions, which is a parameter setting. Hence, the rings themselves could be even fixed, and we would still obtain the same simulation for a given setting of friction. The wide range of frictions employed is plausible based on the experimental work of Stigler et al. [
40], which suggested that friction can be very high. Physically, the friction arises from a combination of inter-surface adhesion, surface roughness and deformation. Based on the results reported by Stigler et al. [
40], the molecular surface of chromatin appears to be very rugged for a diffusing cohesin due to the presence of individual nucleosomes, nucleosome arrays and other protein obstacles and machinery. In our model, we suppose a uniform distribution of the obstacles that are represented by a given averaged setting of the friction parameter
γc. In principle, however, the beaded model also provides the possibility for implementing a site-specific friction within the resolution given by the particular level of coarse-graining employed in the beaded model. This would be interesting, e.g., to investigate the effects of local modulation of friction due to chromatin folding and the presence of protein/DNA regulatory hubs such as those identified at specific sites on micro-C contact maps [
48]. At the same time, we consider the right term of Equation (2) to be the change of internal energy associated with the drop in energy of supercoiling as new relaxed portions of fiber infuse into the loop once the cohesin advances in movement. In our mathematical model, we assume that the temporary drop of energy of supercoiling induced by cohesin movement propels the cohesin movement and loop extrusion. The energy of supercoiling can be expressed as
u =
K.∆
Lk2, where the constant
K represents energy of fiber’s force interactions such as bond stretching, angle bending and torsional flexibility [
42].
The second-order differential equation describing non-equilibrium diffusion is notorious for having analytical solutions only for a limited group of problems [
45]. Hence, the system of differential equations with the moving boundary proposed above needs to be solved numerically. The equations are solved and fitted over loop sizes and linking numbers obtained along the simulated trajectories. The parameters
and
K were obtained by a concatenated fit over all trajectories with a particular value of
γc. The fitted dependencies are shown in
Figure 1 and
Figure 2, together with the values of loop size and linking number obtained by coarse-grained simulations. The values of model parameters are obtained per two fibers going through one ring. As our set of equations treats the problem in 1 dimension with one motor on one side and cohesin boundary on the other side, the coefficients obtained should be therefore halved in order to properly grasp the fact that effusion occurs through two fibers embraced by a single ring. As underlined in the paper by Bonato et al., using a similar approach to mathematical modeling in order to describe the movement of cohesin ring along the fiber, this aspect of the mathematical treatment does not qualitatively affect the results [
33].
As for the obtained value of diffusivity of supercoiling,
= 1.6, agrees well with the value obtained by the study of plectoneme dynamics of braided polymers like DNA by Forte et al. [
49]. In their study, the authors observed that the dynamics of twist is faster than that of writhe, while the obtained decoupled diffusivities were
DTw = 2.0 and
DWr = 0.1, giving the value of diffusivity of supercoiling as a weighted average with contributions of each based on whether the system accommodates a stable plectonemic or straight conformation. In our simulations, we also see the fluctuations of ∆
Lk affected in greater part by the fluctuations of twist, implying that the dynamics of twist is much faster. Additionally, we note that the diffusivity of supercoiling in real units obtained from transforming
σ2/
τ will be much larger than the diffusivity of whole plectonemes observed by Dekker et al. to be
D = 0.1 kbp2/s [
50]. However, the diffusivity of supercoiling along the fiber agrees well with expected rotational mobility of DNA that was experimentally observed to be in order of thousands of rotations per second [
51]. As observed from the simulations and supported also by mathematical modeling, in the case of large gammas, the equilibrium state of the gradient of supercoiling along the fiber is relatively quickly re-established after each movement of cohesin, as the high friction with cohesin limits losses of supercoiling (
Video S2). As result, the value of the average supercoiling of the loop increases rather linearly over time. On the other hand, in the case of low gammas, the gradient of supercoiling along the fiber is more time-dependent as the semipermeable boundary formed by cohesin allows escaping of supercoiling by axial rotations to a much greater extent (
Video S3). As result, the average value of ∆
Lk starts to increase more intensely in later stages of the loop extrusion. This is consistent with the expectation that in an infinitely long loop, the average density of supercoiling would approach a ratio of turns introduced by polymerase versus the average loop extrusion rate. Additionally, we would like to stress, the purpose of the mathematical model presented in this section is not to provide a full quantitative treatment for describing precisely the loop extrusion in vivo by two parameters; rather, we aimed to provide an additional support in terms of qualitative agreement between the simulated data and fits from the mathematical model that would support the proposed mechanism of the transcriptionally driven loop extrusion mediated by friction between cohesin rings and chromatin fibers.
3.3. Towards Entropically Driven Loop Extrusion, Osmotic Pressure and Other Models
As for the value
K, the energy constant of supercoiling, we obtained a value from the fits of
Ksim = 3.4 × 10
3. The halved value per arm is 1.7 × 10
3, which is similar to the value of the energy constant given by Vologodskii as
K = 1100, and it is referred to as a dimensionless constant [
52]. On the other hand, it was pointed out that the dimension of the constant is probably per bp [
42]. In our case, we use arbitrary units in terms of unitless beads on both sides of the equation; hence, a change of the units would not change the constant obtained from the fit. The difference of the fitted value of the energy constant is most likely given due to the internal settings of the force field in our model and can be improved in future works. In order to decouple contributions of other forces acting on the extrusion, one would need to go into a more detailed description of the energy term. For example, Bonato et al. have shown that chromatin stiffness and compaction play a crucial role in enhancing diffusive loop extrusion [
33]. In their model and subsequent mathematical description, they proposed that the change of the loop’s internal energy follows the equation
, where the first term on the right side of the equation represents energy penalty due to the chromatin stiffness that would drop as the loop size grows, AND the second term represents the entropic cost of looping. When these terms are added to our energy term given by the energy of supercoiling
u =
K.∆
Lk2 in (Equation (2)), the fitted value of
K drops to 1550 (
c = 0.03); however, the quality of the fit remains the same (
Figure S1). In general, adding more terms into the functional decreases degrees of freedom and keeps the quality of fits the same if not improved. Additionally, one may think of other energy contributions acting in favor of the loop extrusion, such as opening the folded conformation of the cohesin rings in the early stages of the simulations. On one hand, incorporating the energy functional used by the diffusive model into our model provides a perspective for merging both models that both exhibit enhancement of the loop extrusion by entropic mechanism. On the other hand, providing a rigorous description of the energy term and decoupling energy contributions arising from bending, stretching, unfolding the cohesin, motor activity that acts as a constant speed, i.e., infinite energy motor, etc. is beyond the scope of the present paper. Moreover, the levels of coarse-graining in our work and in Bonato et al.’s work are currently different, which makes a direct comparison of the results complicated. Thus, we instead intend to show that the proposed mechanism is still able to drive loop extrusion at varying levels of friction, accumulated supercoiling and non-topologically bound cohesins, with proof-of-concept simulations supported by a mathematical description. As the energy term ∂
u/∂
x is a definition of chemical potential
μ, the loop extrusion driven by the change of the chemical potential can be considered as an entropic process. The difference of the chemical potentials on the interface determined by the moving cohesin,
μ(
σ,
p + Π) on supercoiled side and
μ(
σ = 0) on relaxed side, corresponds to the definition of osmotic pressure. Hence, one may think of an analogy of the osmotic process when the difference of concentrations on the interface drives the solvent into the concentrated phase. In our case, the solvent would be represented by relaxed portions of the fiber that “dissolve” the accumulated supercoiling within the extruded loop. The similar influx of chromatin fiber works in the mechanism of loop extrusion by an osmotic ratchet [
53], where the fiber “dissolves” the concentration of cohesin rings loaded on the fiber.
3.4. Biological Contexts and Implications
As we mentioned earlier, the existence of loops and their formation by loop extrusion mechanism involving the presence of specialized proteins is nowadays generally accepted. Less consensus has been attained about the role of SMC proteins and mechanisms by which they can perform the extrusion. We know now that cohesin is capable of self-motional motor activity. This was shown to be relatively weak in terms of consumed ATP energy, ca. 1.7 ATP molecules per cohesin per second, but very effective in terms of extruding rate, 2 kbp [
14]. On the other hand, before the motor activity of cohesin was experimentally shown, other biological and biophysical processes that can enhance loop extrusion were discovered by computer simulations [
16,
17,
18]. In the case of transcriptionally driven loop extrusion, RNA polymerase is known to be one of the strongest biomolecular motors [
54]. As we have shown earlier, negative supercoiling generated by RNAP+TOP1 complex can be a very effective driving force for the loop extrusion at high levels of supercoiling conserved in the loop [
16]. In
Section 3.1, we showed that even if much of the supercoiling is continuously relaxed to low levels by axial rotations, the energy of generated supercoiling still represents an effective force driving the loop extrusion.
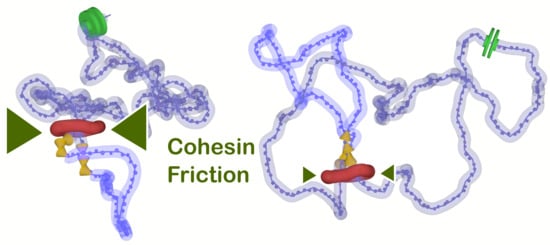
Figure 3a shows loop extrusion rates obtained as averages over five independent runs for each setting of friction between chromatin fiber and cohesin ring for the values of
γc = 2, 5, 20 and 200 that were the main scenarios discussed in this paper. The loop extrusion rates are shown as a function of a product of the imposed friction. As the figure shows, the loop extrusion rate still increases with increasing friction as the energy of accumulated supercoiling increases, but it follows a logarithmic relation, showing the saturating effect on the loop extrusion rate. Biologically, the speeds should be such that the entire human genome could be extruded within minutes [
14]. We may see that biologically relevant rates consistent with experimental measurements between 0.5–2 kbps [
14] are achieved mainly for lower settings of
γc ≤ 20.
The corresponding average levels of accumulated supercoiling observed in the simulations in terms of linking number ∆
Lk over the distance of 60 kbp range from −2 for the lowest friction
γc = 2 to −10 for the largest friction
γc = 200 employed. These values, however, represent the total value along the chain, while in reality, the supercoiling creates a gradient from the position from transcription site (TSS)
x = 0 towards the site of release of supercoiling at the position of cohesin,
x =
xc. The site-specific linking number ∆
Lk calculated at a given bead can be considered as the bead’s local density of the supercoiling
σ. In this way, for
γc = 200, the values measured and modeled at the position of the bead representing transcription site in our simulations reach
σ(
x = 0) = −12%. At the same time, the supercoiling drops towards the position of cohesin, where the value is maintained by the semipermeable boundary at
σ(
xc) = −5%. The evolution of ∆
Lk along the chain and simulation time obtained from the mathematical model is given in
Videos S2 and S3. The profile of ∆
Lk obtained as average over the simulation run is given in
Figure S2. The total value of
Lk is the integral under the curve. In the case of low friction, the values range from
σ = −7% at the TSS to
σ(
xc) = −0.2%, where the supercoiling drops more dramatically along the fiber, at the position of cohesin. The values of local supercoiling at the transcription site are a bit lower than predicted by the stochastic model of supercoiling-dependent transcription by Brackley et al. [
44]. However, the values are in good agreement with the local DNA supercoiling determined for genes with low, medium and high expression around the position of TSS in terms of crosslinks (CLs) by psoralen photobinding in vivo by Kouzine et al. [
55]. The faster decay of supercoiling in the simulations with lower
γcs is consistent with the profiles measured by Kouzine et al.
Figure 3.
The higher friction between cohesin ring and fiber induces higher levels of supercoiling and extrudes loops at higher rates. (
a) Loop extrusion rate for different settings of
γc imposing friction between cohesin ring and chromatin fiber, resulting in different levels of accumulated supercoiling. The loop extrusion is shown as a function of imposed friction between cohesin and fibers. The insets show snapshots from the end of the simulations at the lowest and highest settings of the friction. (
b) The contact maps are calculated during the loop extrusion. The contact maps were calculated along the trajectories as the average over 10 simulation runs. The maps show the loss of ant-diagonal when the loop extrusion is more stochastic, i.e., random, when lower settings of friction between cohesin ring and chromatin fiber allow the fiber to sample larger conformational space. The data acquisition for the contact maps calculation stopped once the ring slipped away from the chromatin fiber. Biologically, the rings should stay stuck to CTCF’s proteins at borders of the domain [
56], which would emphasize the loss of the anti-diagonal in simulations with lower friction.
Figure 3.
The higher friction between cohesin ring and fiber induces higher levels of supercoiling and extrudes loops at higher rates. (
a) Loop extrusion rate for different settings of
γc imposing friction between cohesin ring and chromatin fiber, resulting in different levels of accumulated supercoiling. The loop extrusion is shown as a function of imposed friction between cohesin and fibers. The insets show snapshots from the end of the simulations at the lowest and highest settings of the friction. (
b) The contact maps are calculated during the loop extrusion. The contact maps were calculated along the trajectories as the average over 10 simulation runs. The maps show the loss of ant-diagonal when the loop extrusion is more stochastic, i.e., random, when lower settings of friction between cohesin ring and chromatin fiber allow the fiber to sample larger conformational space. The data acquisition for the contact maps calculation stopped once the ring slipped away from the chromatin fiber. Biologically, the rings should stay stuck to CTCF’s proteins at borders of the domain [
56], which would emphasize the loss of the anti-diagonal in simulations with lower friction.
![Biology 10 00130 g003]()
The lower levels of supercoiling also allow for higher conformational statistics and fluctuations of chromatin fiber than in the systems with strong supercoiling. The insets of
Figure 3a show two corresponding snapshots from molecular simulations comparing the resulting structures with lower and higher levels of supercoiling. The higher conformational statistics within the extrusion are also reflected in the contact maps by the disappearance of the anti-diagonal feature that is prominent in the case of extrusion with strong supercoiling and which creates plectonemic conformation of the loop (
Figure 3b). The contact maps on
Figure 3b were obtained as averages from 10 trajectories. One has to note, that in our simulations the fiber does not contain CTCF proteins at the end of the domain, and the simulation stops after the cohesin reaches the end of the loop and unloads. In a more biological setting, the cohesin would stick to CTCFs and stay there while the simulation would continue for a period equivalent to 20 min of biological time [
56]. This would enhance the appearance of the anti-diagonal feature on the simulated contact maps and emphasize the difference between the contact maps obtained for systems with low and high levels of supercoiling.