Bioactive Potential of Several Actinobacteria Isolated from Microbiologically Barely Explored Desert Habitat, Saudi Arabia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Actinomycetes
2.2. Extraction of DNA from the Most Potent Actinobacterial Isolates
2.3. Polymerase Chain Reaction (PCR) Amplifications of 16S rDNA
2.4. Sequencing and Phylogenetic Analysis
2.5. Biological Activity
2.6. Preparation of Actinomyces (ACT) Extract
2.7. Metabolites Determination
2.8. Biological Activity
2.9. In Vitro Cancer Cell Viability Assays
2.10. Determination of Lipoxygenase (LOX) and Cyclooxygenase (COX) Activities
2.11. Statistical Analyses
3. Results
3.1. Morphological and Chemical Characterization of the Isolated Actinomycetes
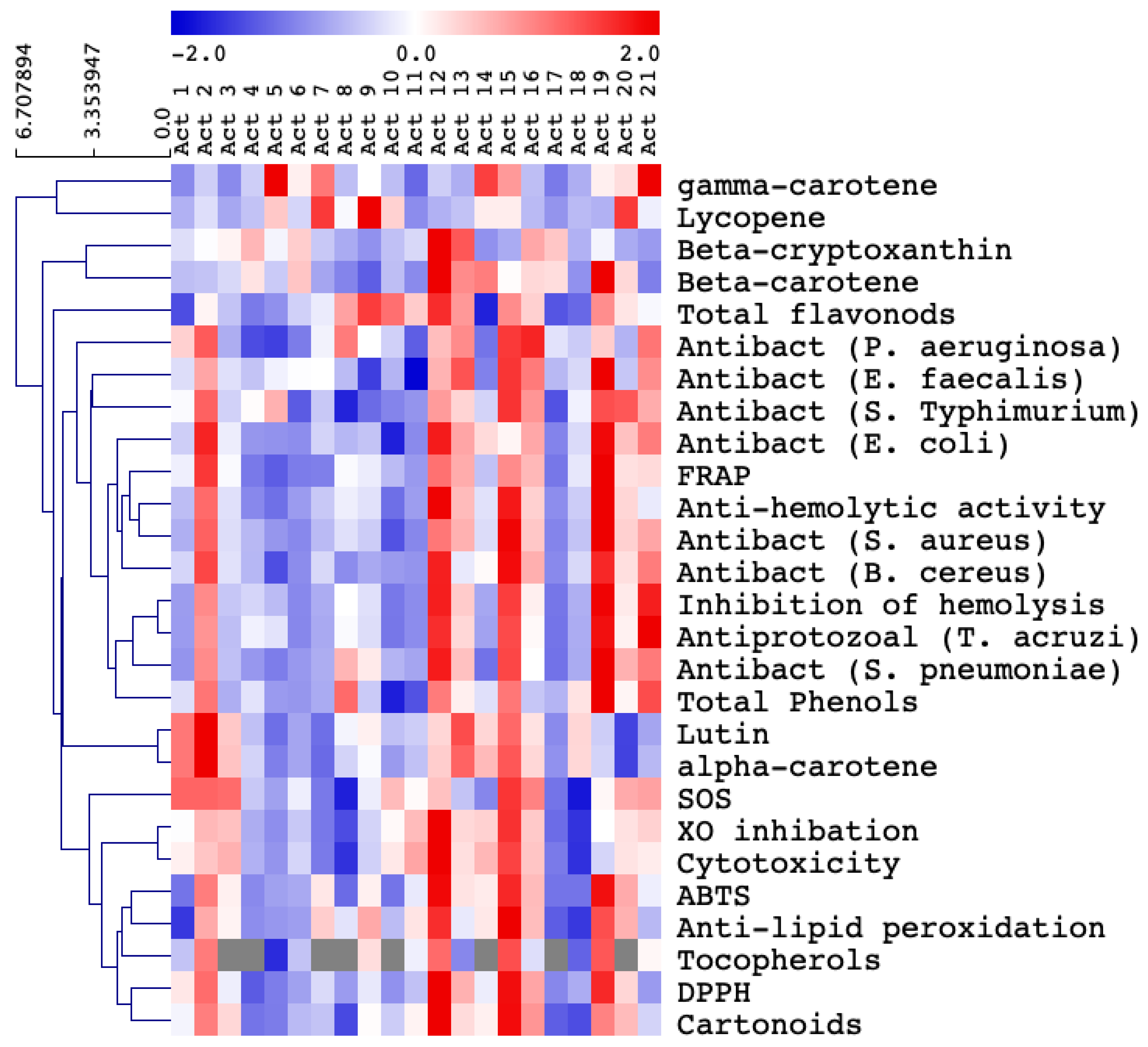
3.2. Selection of the Most Biologically Active Actinomycetes
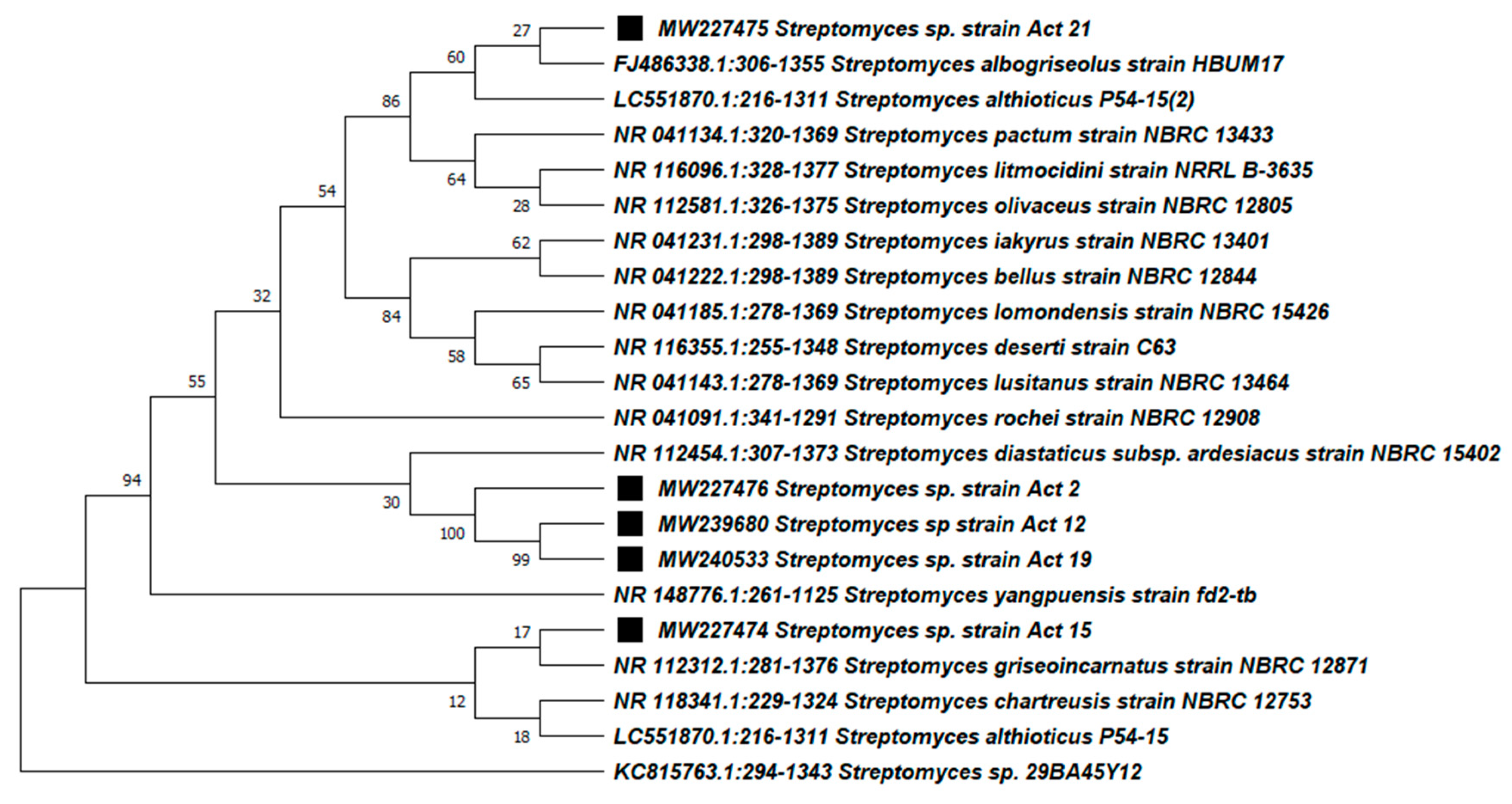
3.3. Molecular Identification and Phylogenetic Analysis of the Active Actinobacteria
3.4. Phenolic Profile of Selected Actinomycetes Isolates
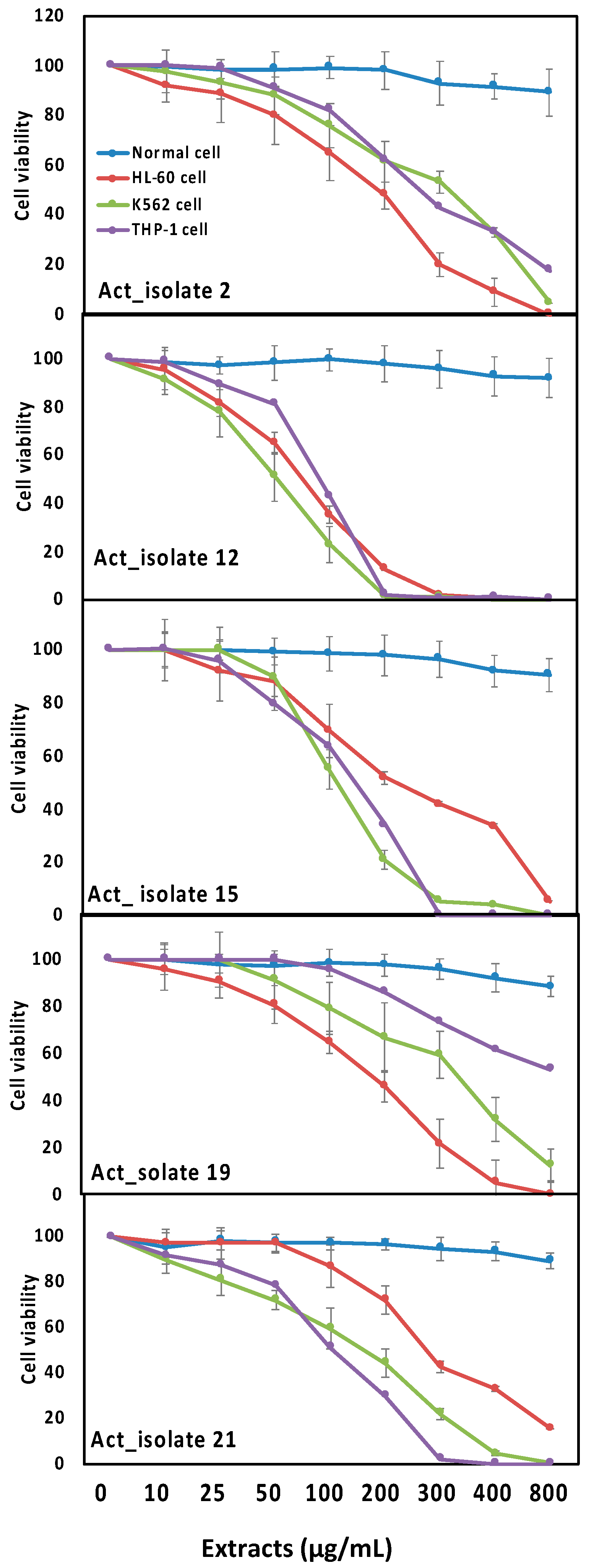
3.5. Biological Activities of Selected Actinomycetes Isolates towards Inflammation and Leukemia
4. Discussion
4.1. Characterization of the Isolates
4.2. Biological Activities and Secondary Metabolites of Actinomycetes Contribute to Determination of the Most Active Isolates
4.3. Production of Phenolic Compounds and Asparaginase by Actinomycetes Supports Their Anti-Leukemic Activities
4.4. Anti-Inflammatory Activities of Selected Actinomycetes Isolates
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Approval
References
- Al Kashgry, N.A.T.; Abulreesh, H.H.; El-Sheikh, I.A.; Almaroai, Y.A.; Salem, R.; Mohamed, I.; Waly, F.R.; Osman, G.; Mohamed, M.S.M. Utilization of a recombinant defensin from Maize (Zea mays L.) as a potential antimicrobial peptide. AMB Express 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Siegel, S.E.; Stock, W.; Johnson, R.H.; Advani, A.; Muffly, L.; Douer, D.; Reed, D.; Lewis, M.; Freyer, D.R.; Shah, B. Pediatric-inspired treatment regimens for adolescents and young adults with philadelphia chromosome–negative acute lymphoblastic leukemia: A review. JAMA Oncol. 2018, 4, 725–734. [Google Scholar] [CrossRef]
- Jeha, S.; Kantarjian, H.; O’Brien, S.; Huh, Y.; Pisa, P.; Ordonez, N.; Beran, M. Growth and biologic properties of karyotypically defined subcategories of adult acute lymphocytic leukemia in mice with severe combined immunodeficiency. Blood 1995, 86, 4278–4285. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlier, C.; Michaux, C. Dual inhibition of cyclooxygenase-2 (COX-2) and 5-lipoxygenase (5-LOX) as a new strategy to provide safer non-steroidal anti-inflammatory drugs. Eur. J. Med. Chem. 2003, 38, 645–659. [Google Scholar] [CrossRef]
- Demain, A.L.; Vaishnav, P. Natural products for cancer chemotherapy. Microb. Biotechnol. 2011, 4, 687–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AbdElgawad, H.; Abuelsoud, W.; Madany, M.M.Y.; Selim, S.; Zinta, G.; Mousa, A.S.M.; Hozzein, W.N. Actinomycetes Enrich Soil Rhizosphere and Improve Seed Quality as well as Productivity of Legumes by Boosting Nitrogen Availability and Metabolism. Biomolecules 2020, 10, 1675. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.-K. Marine actinobacterial metabolites: Current status and future perspectives (Retraction Article, vol 168, pg 311, 2013) (Retraction of Vol 6, Pg 311, 2013). Microbiol. Res. 2018, 211, 69. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olano, C.; Méndez, C.; Salas, J.A. Antitumor compounds from actinomycetes: From gene clusters to new derivatives by combinatorial biosynthesis. Nat. Prod. Rep. 2009, 26, 628–660. [Google Scholar] [CrossRef]
- Lin, S.; Chang, C.; Hsu, C.; Tsai, M.; Cheng, H.; Leong, M.K.; Sung, P.; Chen, J.; Weng, C. Natural compounds as potential adjuvants to cancer therapy: Preclinical evidence. Br. J. Pharmacol. 2020, 177, 1409–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janardhan, A.; Kumar, A.P.; Viswanath, B.; Saigopal, D.V.R.; Narasimha, G. Production of bioactive compounds by actinomycetes and their antioxidant properties. Biotechnol. Res. Int. 2014, 2014, 217030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Wen, Y.; Li, M.; Chen, Z.; Guo, J.; Song, Y.; Li, J. A new strain of Streptomyces avermitilis produces high yield of oligomycin A with potent anti-tumor activity on human cancer cell lines in vitro. Appl. Microbiol. Biotechnol. 2009, 81, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, C.; Sangeetha, B.; Duraipandiyan, V.; Raj, M.K.; Ignacimuthu, S.; Al-Dhabi, N.A.; Balakrishna, K.; Parthasarathy, K.; Arulmozhi, N.M.; Arasu, M.V. A flavonoid isolated from Streptomyces sp.(ERINLG-4) induces apoptosis in human lung cancer A549 cells through p53 and cytochrome c release caspase dependant pathway. Chem. Biol. Interact. 2014, 224, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Waksman, S.A. The Actinomycetes. Vol. II. Classification, identification and descriptions of genera and species. In The Actinomycetes. Vol. II. Classification, Identification and Descriptions of Genera and Species; Baillière, Tindall & Cox, Ltd.: London, UK, 1961; p. ix+363. [Google Scholar]
- Haefner, B. Drugs from the deep: Marine natural products as drug candidates. Drug Discov. Today 2003, 8, 536–544. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Warrad, M.; Hassan, Y.M.; Mohamed, M.S.M.; Hagagy, N.; Al-Maghrabi, O.A.; Selim, S.; Saleh, A.M.; AbdElgawad, H. A bioactive fraction from Streptomyces sp. enhances maize tolerance against drought stress. J. Microbiol. Biotechnol. 2020, 30, 1156–1168. [Google Scholar] [CrossRef]
- Shaaban, M.T.; El-Sabbagh, S.M.M.; Alam, A. Studies on an actinomycete producing a melanin pigment inhibiting aflatoxin B1 production by Aspergillus flavus. Life Sci. J. 2013, 10, 1437–1448. [Google Scholar]
- Mohamed, M.S.M.; Saleh, A.M.; Abdel-Farid, I.B.; El-Naggar, S.A. Growth, hydrolases and ultrastructure of Fusarium oxysporum as affected by phenolic rich extracts from several xerophytic plants. Pestic. Biochem. Physiol. 2017, 141, 57–64. [Google Scholar] [CrossRef]
- Saleh, A.M.; Madany, M.M.Y.; González, L. The effect of coumarin application on early growth and some physiological parameters in Faba Bean (Vicia faba L.). J. Plant Growth Regul. 2015, 34, 233–241. [Google Scholar] [CrossRef]
- Hamad, I.; Abdelgawad, H.; Al Jaouni, S.; Zinta, G.; Asard, H.; Hassan, S.; Hegab, M.; Hagagy, N.; Selim, S. Metabolic analysis of various date palm fruit (Phoenix dactylifera L.) cultivars from Saudi Arabia to assess their nutritional quality. Molecules 2015, 20, 13620–13641. [Google Scholar] [CrossRef] [PubMed]
- AbdElgawad, H.; Farfan-Vignolo, E.R.; de Vos, D.; Asard, H. Elevated CO2 mitigates drought and temperature-induced oxidative stress differently in grasses and legumes. Plant Sci. 2015, 231, 1–10. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Aruoma, O.I.; Laughton, M.J.; Halliwell, B. Carnosine, homocarnosine and anserine: Could they act as antioxidants in vivo? Biochem. J. 1989, 264, 863–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AbdElgawad, H.; De Vos, D.; Zinta, G.; Domagalska, M.A.; Beemster, G.T.S.; Asard, H. Grassland species differentially regulate proline concentrations under future climate conditions: An integrated biochemical and modelling approach. New Phytol. 2015, 208, 354–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, A.M.; Abdel-Mawgoud, M.; Hassan, A.R.; Habeeb, T.H.; Yehia, R.S.; AbdElgawad, H. Global metabolic changes induced by arbuscular mycorrhizal fungi in oregano plants grown under ambient and elevated levels of atmospheric CO2. Plant Physiol. Biochem. 2020, 151, 255–263. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C.; Aruoma, O.I. The deoxyribose method: A simple “test-tube” assay for determination of rate constants for reactions of hydroxyl radicals. Anal. Biochem. 1987, 165, 215–219. [Google Scholar] [CrossRef]
- Selim, S.A.; Aziz, M.H.A.; Mashait, M.S.; Warrad, M.F. Antibacterial activities, chemical constitutes and acute toxicity of Egyptian Origanum majorana L., Peganum harmala L. and Salvia officinalis L. essential oils. Afr. J. Pharm. Pharmacol. 2013, 7, 725–735. [Google Scholar]
- Mohammadipanah, F.; Momenilandi, M. Potential of rare actinomycetes in the production of metabolites against multiple oxidant agents. Pharm. Biol. 2018, 56, 51–59. [Google Scholar] [CrossRef]
- Almuhayawi, M.; AbdElgawad, H.; Al Jaouni, S.; Selim, S.; Hassan, A.H.A.; Khamis, G. Elevated CO2 improves glucosinolate metabolism and stimulates anticancer and anti-inflammatory properties of broccoli sprouts. Food Chem. 2020, 328, 127102. [Google Scholar] [CrossRef]
- Williams, S.T. Genus Streptomyces waksman and henrici 1943. Bwergey’s Man. Syst. Bacteriol. 1989, 4, 2452–2492. [Google Scholar]
- Manfio, G.P. Towards minimal standards for the description of Streptomyces species. Biotekhnologiya 1995, 8, 228–237. [Google Scholar]
- Ohnishi, Y.; Seo, J.-W.; Horinouchi, S. Deprogrammed sporulation in Streptomyces. FEMS Microbiol. Lett. 2002, 216, 1–7. [Google Scholar] [CrossRef]
- Mukherjee, G.; Sen, S.K. Characterization and identification of chitinase producing Streptomyces venezuelae P 10. Indian J. Exp. Biol. 2004, 42, 541–544. [Google Scholar]
- Ser, H.-L.; Law, J.W.-F.; Chaiyakunapruk, N.; Jacob, S.A.; Palanisamy, U.D.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Fermentation conditions that affect clavulanic acid production in Streptomyces clavuligerus: A systematic review. Front. Microbiol. 2016, 7, 522. [Google Scholar] [CrossRef]
- Boroujeni, M.E.; Arijit, D.; Prashanthi, K.; Sandeep, S.; Sourav, B. Enzymatic screening and random amplified polymorphic DNA fingerprinting of soil streptomycetes isolated from Wayanad district in Kerala, India. J. Biol. Sci. 2012, 12, 43–50. [Google Scholar]
- Thirumurugan, D.; Vijayakumar, R.; Vadivalagan, C.; Karthika, P.; Khan, M.K.A. Isolation, structure elucidation and antibacterial activity of methyl-4, 8-dimethylundecanate from the marine actinobacterium Streptomyces albogriseolus ECR64. Microb. Pathog. 2018, 121, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Risdian, C.; Primahana, G.; Mozef, T.; Dewi, R.T.; Ratnakomala, S.; Lisdiyanti, P.; Wink, J. Screening of antimicrobial producing Actinobacteria from Enggano Island, Indonesia. In Proceedings of the AIP Conference Proceedings; AIP Publishing LLC: New York, NY, USA, 7 November 2018; Volume 2024, p. 20039. [Google Scholar]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Xu, C.; Wang, J.; Lu, F.; Bie, X.; Lu, Z. Identification and characterization of Streptomyces flavogriseus NJ-4 as a novel producer of actinomycin D and holomycin. PeerJ 2017, 5, e3601. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhao, G.; Chen, H.; Wang, H.; Qin, S.; Zhu, W.; Xu, L.; Jiang, C.; Li, W. Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Lett. Appl. Microbiol. 2008, 47, 574–580. [Google Scholar] [CrossRef]
- Akintobi, O.A.; Agunbiade, S.O.; Okonko, I.O.; Ojo, O.V. Antimicrobial evaluation and phytochemical analysis of leaf extracts of Mirabilis jalapa against some human pathogenic bacteria. Nat. Sci. 2011, 9, 45–53. [Google Scholar]
- Verma, V.C.; Gond, S.K.; Kumar, A.; Mishra, A.; Kharwar, R.N.; Gange, A.C. Endophytic actinomycetes from Azadirachta indica A. Juss.: Isolation, diversity, and anti-microbial activity. Microb. Ecol. 2009, 57, 749–756. [Google Scholar] [CrossRef]
- Kumar, P.S.; Al-Dhabi, N.A.; Duraipandiyan, V.; Balachandran, C.; Kumar, P.P.; Ignacimuthu, S. In vitro antimicrobial, antioxidant and cytotoxic properties of Streptomyces lavendulae strain SCA5. BMC Microbiol. 2014, 14, 291. [Google Scholar]
- Hozzein, W.N.; Abuelsoud, W.; Wadaan, M.A.M.; Shuikan, A.M.; Selim, S.; Al Jaouni, S.; AbdElgawad, H. Exploring the potential of actinomycetes in improving soil fertility and grain quality of economically important cereals. Sci. Total Environ. 2019, 651, 2787–2798. [Google Scholar] [CrossRef]
- Krügel, H.; Krubasik, P.; Weber, K.; Saluz, H.P.; Sandmann, G. Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 1999, 1439, 57–64. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Y.; Chen, M.; Xie, B.; Yang, J.; Chen, J.; Sun, Z. Isorenieratene interaction with human serum albumin: Multi-spectroscopic analyses and docking simulation. Food Chem. 2018, 258, 393–399. [Google Scholar] [CrossRef]
- Sweetline, C.; Usha, R.; Palaniswamy, M. Antibacterial activity of actinomycetes from Pichavaram Mangrove of Tamil Nadu. Appl. J. Hyg. 2012, 1, 15–18. [Google Scholar]
- Lee, D.-R.; Lee, S.-K.; Choi, B.-K.; Cheng, J.; Lee, Y.-S.; Yang, S.H.; Suh, J.-W. Antioxidant activity and free radical scavenging activities of Streptomyces sp. strain MJM 10778. Asian Pac. J. Trop. Med. 2014, 7, 962–967. [Google Scholar] [CrossRef]
- Wadetwar, R.N.; Patil, A.T. Isolation and characterization of bioactive actinomycetes from soil in and around Nagpur. Int. J. Pharm. Sci. Res. 2013, 4, 1428. [Google Scholar]
- Oskay, A.M.; Üsame, T.; Cem, A. Antibacterial activity of some actinomycetes isolated from farming soils of Turkey. Afr. J. Biotechnol. 2004, 3, 441–446. [Google Scholar]
- Arasu, M.V.; Duraipandiyan, V.; Agastian, P.; Ignacimuthu, S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. J. Mycol. Med. 2008, 18, 147–153. [Google Scholar] [CrossRef]
- Alharbi, S.A.; Arunachalam, C.; Murugan, A.M.; Wainwright, M. Antibacterial activity of actinomycetes isolated from terrestrial soil of Saudi Arabia. J. Food Agric. Environ. 2012, 10, 1093–1097. [Google Scholar]
- Dalin, H.; Guifeng, Y.; Yajuan, X.; Naixiang, L.; Jianhong, C. Jiang lianxiu L, Senzhou, C. Isolation and identification of Actinomycetes sp. BH0954 from the mangrove forest soil of Guangxi Beihai. Chin. Agric. Sci. Bull. 2010, 26, 406–409. [Google Scholar]
- Castillo, U.F.; Strobel, G.A.; Ford, E.J.; Hess, W.M.; Porter, H.; Jensen, J.B.; Albert, H.; Robison, R.; Condron, M.A.M.; Teplow, D.B. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscansaaThe GenBank accession number for the sequence determined in this work is AY127079. Microbiology 2002, 148, 2675–2685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.T.-H.; Chan, K.-G.; Khan, T.M.; Bukhari, S.I.; Saokaew, S.; Duangjai, A.; Pusparajah, P.; Lee, L.-H.; Goh, B.-H. Streptomyces sp. MUM212 as a source of antioxidants with radical scavenging and metal chelating properties. Front. Pharmacol. 2017, 8, 276. [Google Scholar] [CrossRef]
- Tseng, M.; Su, Y.; Cheng, M.; Liu, T.; Chen, I.; Wu, M.; Chang, H.; Yuan, G. Chemical constituents from a soil-derived actinomycete, actinomadura miaoliensis BCRC 16873, and their inhibitory activities on lipopolysaccharide-induced tumor necrosis factor production. Chem. Biodivers. 2013, 10, 303–312. [Google Scholar] [CrossRef]
- Hatsu, M.; Sasaki, T.; Gomi, S.; Kodama, Y.; Sezaki, M.; Inouye, S.; Kondo, S. A new tetracycline antibiotic with antitumor activity. J. Antibiot. 1992, 45, 325–330. [Google Scholar] [CrossRef]
- Rather, S.A.; Kumar, S.; Rah, B.; Arif, M.; Ali, A.; Qazi, P. A potent cytotoxic metabolite from terrestrial actinomycete, Streptomyces collinus. Med. Chem. Res. 2014, 23, 382–387. [Google Scholar] [CrossRef]
- Graf, E.; Schneider, K.; Nicholson, G.; Ströbele, M.; Jones, A.L.; Goodfellow, M.; Beil, W.; DS, R.; Fiedler, H.-P. Elloxazinones A and B, new aminophenoxazinones from Streptomyces griseus Acta 2871. J. Antibiot. 2007, 60, 277–284. [Google Scholar] [CrossRef]
- Takahashi, I.; Takahashi, K.-I.; Asano, K.; Kawamoto, I.; Yasuzawa, T.; Ashizawat, T.; Tomita, F.; Nakano, H. DC92-B, a new antitumor antibiotic from Actinomadura. J. Antibiot. 1988, 41, 1151–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sletta, H.; Degnes, K.F.; Herfindal, L.; Klinkenberg, G.; Fjærvik, E.; Zahlsen, K.; Brunsvik, A.; Nygaard, G.; Aachmann, F.L.; Ellingsen, T.E. Anti-microbial and cytotoxic 1, 6-dihydroxyphenazine-5, 10-dioxide (iodinin) produced by Streptosporangium sp. DSM 45942 isolated from the fjord sediment. Appl. Microbiol. Biotechnol. 2014, 98, 603–610. [Google Scholar] [CrossRef]
- Sun, C.-H.; Wang, Y.; Wang, Z.; Zhou, J.-Q.; Jin, W.-Z.; You, X.-F.; Gao, H.; Zhao, L.-X.; Si, S.-Y.; Li, X. Chemomicin A, a new angucyclinone antibiotic produced by Nocardia mediterranei subsp. kanglensis 1747-64. J. Antibiot. 2007, 60, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.M.; Wani, A.; Qazi, P.H.; Rehman, S.; Mushtaq, S.; Ali, S.A.; Hussain, A.; Shah, A.; Qazi, A.K.; Makhdoomi, U.S. Isolation and characterization of alborixin from Streptomyces scabrisporus: A potent cytotoxic agent against human colon (HCT-116) cancer cells. Chem. Biol. Interact. 2016, 256, 198–208. [Google Scholar] [CrossRef]
- Jakubowicz-Gil, J.; Langner, E.; Wertel, I.; Piersiak, T.; Rzeski, W. Temozolomide, quercetin and cell death in the MOGGCCM astrocytoma cell line. Chem. Biol. Interact. 2010, 188, 190–203. [Google Scholar] [CrossRef]
- Xiao, D.; Gu, Z.L.; Zhu, S.P. Quercetin down-regulated bcl-2 gene expression in human leukemia HL-60 cells. Zhongguo Yao Li Xue Bao Acta Pharmacol. Sin. 1998, 19, 551–553. [Google Scholar]
- König, A.; Schwartz, G.K.; Mohammad, R.M.; Al-Katib, A.; Gabrilove, J.L. The novel cyclin-dependent kinase inhibitor flavopiridol downregulates Bcl-2 and induces growth arrest and apoptosis in chronic B-cell leukemia lines. Blood J. Am. Soc. Hematol. 1997, 90, 4307–4312. [Google Scholar]
- Lin, J.-P.; Yang, J.-S.; Lu, C.-C.; Chiang, J.-H.; Wu, C.-L.; Lin, J.-J.; Lin, H.-L.; Yang, M.-D.; Liu, K.-C.; Chiu, T.-H. Rutin inhibits the proliferation of murine leukemia WEHI-3 cells in vivo and promotes immune response in vivo. Leuk. Res. 2009, 33, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Miao, Q.; Geng, M.; Liu, J.; Hu, Y.; Tian, L.; Pan, J.; Yang, Y. Anti-tumor effect of rutin on human neuroblastoma cell lines through inducing G2/M cell cycle arrest and promoting apoptosis. Sci. World J. 2013, 2013, 269165. [Google Scholar] [CrossRef] [Green Version]
- Nile, S.H.; Park, S.W. Edible berries: Bioactive components and their effect on human health. Nutrition 2014, 30, 134–144. [Google Scholar] [CrossRef]
- Hoensch, H.; Oertel, R. Anti-inflammatory effects of tea-flavonoids. Dtsch. Med. Wochenschr. 2012, 137, 2738–2740. [Google Scholar]
- Liu, X.; Ye, F.; Wu, J.; How, B.; Li, W.; Zhang, D.Y. Signaling proteins and pathways affected by flavonoids in leukemia cells. Nutr. Cancer 2015, 67, 238–249. [Google Scholar] [CrossRef]
- Valipour, B.; Mohammadi, S.M.; Abedelahi, A.; Maragheh, B.F.A.; Naderali, E.; Dehnad, A.; Charoudeh, H.N. Culture filtrate ether extracted metabolites from Streptomyces levis ABRIINW111 increased apoptosis and reduced proliferation in acute lymphoblastic leukemia. Biomed. Pharm. 2018, 108, 216–223. [Google Scholar] [CrossRef]
- Basha, N.S.; Rekha, R.; Komala, M.; Ruby, S. Production of extracellular anti-leukaemic enzyme lasparaginase from marine actinomycetes by solidstate and submerged fermentation: Purification and characterisation. Trop. J. Pharm. Res. 2009, 8. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; El-Shweihy, N.M. Bioprocess development for L-asparaginase production by Streptomyces rochei, purification and in-vitro efficacy against various human carcinoma cell lines. Sci. Rep. 2020, 10, 1–21. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; El-Ewasy, S.M.; El-Shweihy, N.M. of Acute Lymphoblastic Leukemia: The Pros and Cons. Int. J. Pharmacol. 2014, 10, 182–199. [Google Scholar] [CrossRef]
- Verma, N.; Kumar, K.; Kaur, G.; Anand, S. L-asparaginase: A promising chemotherapeutic agent. Crit. Rev. Biotechnol. 2007, 27, 45–62. [Google Scholar] [CrossRef]
- Sudhir, A.P.; Dave, B.R.; Trivedi, K.A.; Subramanian, R.B. Production and amplification of an l-asparaginase gene from actinomycete isolate Streptomyces ABR2. Ann. Microbiol. 2012, 62, 1609–1614. [Google Scholar] [CrossRef]
- Deshpande, N.; Choubey, P.; Agashe, M. Studies on optimization of growth parameters for L-asparaginase production by Streptomyces ginsengisoli. Sci. World J. 2014, 2014, 895167. [Google Scholar] [CrossRef]
- Narta, U.K.; Kanwar, S.S.; Azmi, W. Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia. Crit. Rev. Oncol. Hematol. 2007, 61, 208–221. [Google Scholar] [CrossRef]
- Kamble, V.P.; Rao, R.S.; Borkar, P.S. Purification of L-asparaginase from a bacteria Erwinia carotovora and effect of a dihydropyrimidine derivative on some of its kinetic parameters. Indian J. Biochem. Bbiophys 2006, 43, 391–394. [Google Scholar]
- Song, P.; Ye, L.; Fan, J.; Li, Y.; Zeng, X.; Wang, Z.; Wang, S.; Zhang, G.; Yang, P.; Cao, Z. Asparaginase induces apoptosis and cytoprotective autophagy in chronic myeloid leukemia cells. Oncotarget 2015, 6, 3861. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, D.; Nissen, S.E.; Topol, E.J. Risk of cardiovascular events associated with selective COX-2 inhibitors. JAMA 2001, 286, 954–959. [Google Scholar] [CrossRef]
- Ondua, M.; Adebayo, S.A.; Shai, L.J.; Lebelo, S.L. The anti-inflammatory and anti-nociceptive activities of some medicinal plant species used to treat inflammatory pain conditions in southern Africa. Int. J. Pharmacogn. Phytochem. Res. 2016, 8, 1571–1575. [Google Scholar]
- Kimata, M.; Shichijo, M.; Miura, T.; Serizawa, I.; Inagaki, N.; Nagai, H. Effects of luteolin, quercetin and baicalein on immunoglobulin E-mediated mediator release from human cultured mast cells. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2000, 30, 501–508. [Google Scholar] [CrossRef]
- Islam, V.I.H.; Saravanan, S.; Ignacimuthu, S. Microbicidal and anti-inflammatory effects of Actinomadura spadix (EHA-2) active metabolites from Himalayan soils, India. World J. Microbiol. Biotechnol. 2014, 30, 9–18. [Google Scholar] [CrossRef]
- Jayaprakashvel, M. Therapeutically active biomolecules from marine actinomycetes. J. Mod. Biotechnol. 2012, 1, 1–7. [Google Scholar]

| Isolate | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Colony | Aerial mycelium | + | + | + | + | − | + | + | + | + | − | + | − | + | + | − | + | + | − | + | + | + |
| Pigmentation | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Spore chain | Spiral | + | + | − | + | + | − | − | + | − | + | + | + | − | + | − | + | + | + | + | + | + |
| Rectiflexibels | − | − | + | − | − | + | − | − | − | − | + | − | − | − | − | − | + | − | − | − | − | |
| Verticillate | − | − | − | − | − | − | + | − | + | − | + | − | + | − | + | − | + | − | − | − | − | |
| Spore color | Yellow | + | + | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | + | − | − | − |
| Orange | − | − | + | + | − | + | − | + | − | + | − | + | − | − | − | + | + | − | + | − | + | |
| Red | − | − | − | − | + | − | + | − | + | − | + | − | − | + | − | − | − | − | − | + | − | |
| N source utilization | L-Cysteine | + | − | − | + | + | + | + | + | + | − | − | + | + | − | + | + | − | − | + | − | + |
| L-Phenylalanine | − | + | + | − | − | + | + | − | + | − | + | − | + | + | + | − | + | + | − | + | − | |
| L-Histidine | − | + | − | + | + | − | + | + | − | − | + | − | + | − | − | − | + | + | − | + | − | |
| L-Lysine | + | + | + | − | − | + | + | − | + | − | − | + | + | + | − | + | + | + | + | + | + | |
| L-Asparagine | + | + | − | + | + | − | − | + | − | + | + | + | + | + | − | + | + | − | + | − | + | |
| L-Arginine | + | − | + | − | − | + | + | − | + | − | + | + | − | − | − | + | − | − | + | + | + | |
| L-proline | + | + | − | + | − | − | + | + | + | − | − | + | + | + | + | + | + | − | + | − | + | |
| L-Valine | − | + | − | − | + | + | + | − | − | − | + | − | + | − | − | − | + | + | − | − | −- | |
| Tyrosine | + | + | − | + | − | + | − | + | − | − | + | + | − | + | − | + | − | − | + | − | + | |
| C source utilization | D-fructose | − | − | − | + | + | − | + | − | − | + | − | − | + | − | − | − | + | + | − | + | − |
| D-glucose | + | + | − | + | − | − | − | + | + | − | − | + | − | + | + | + | + | + | + | + | + | |
| Sucrose | − | + | + | − | + | − | + | − | + | + | + | + | + | + | − | + | + | − | + | + | + | |
| Maltose | − | − | − | − | + | + | + | − | + | + | + | − | + | − | − | − | − | − | − | − | − | |
| Raffinose | + | + | + | + | + | + | + | + | + | − | + | + | + | + | − | + | + | + | + | + | + | |
| Lactose | − | − | − | + | − | − | − | + | + | − | + | − | − | − | − | − | + | + | − | − | − | |
| Galactose | + | + | − | + | + | − | + | + | − | − | + | + | − | − | − | + | + | − | + | + | + | |
| Meso-Inositol | + | − | − | − | + | + | + | − | + | + | + | + | − | − | + | + | − | − | + | + | + | |
| Celullose | − | − | − | + | − | − | − | + | + | + | + | − | + | + | + | − | + | + | − | + | − | |
| Xylose | + | − | − | − | + | − | − | − | + | + | − | + | + | + | + | + | + | − | − | − | + | |
| Dextran | + | − | + | − | − | − | + | − | + | − | + | + | − | + | − | + | − | + | + | + | + | |
| Enzymes activity | Catalase | − | − | + | − | + | − | + | − | + | − | + | − | + | − | − | − | + | − | − | + | − |
| Peroxidase | + | − | − | + | + | + | + | + | − | + | − | + | + | + | + | + | − | − | − | + | + | |
| Starch hydrolysis | + | + | + | − | − | − | − | − | + | + | − | + | − | − | − | + | − | + | + | − | + | |
| Gelatin liquefication | + | − | − | + | − | + | − | + | + | + | − | + | + | − | + | + | − | + | + | − | + | |
| Casein hydrolysis | − | − | + | − | + | + | + | − | − | − | − | − | − | − | + | − | − | − | − | + | − | |
| Lipolysis | + | + | + | + | − | + | + | + | + | + | + | + | + | − | − | + | − | + | + | − | + | |
| Citrate utilization | + | + | − | + | + | + | − | + | + | + | + | + | + | + | + | + | − | + | + | − | + | |
| H2S Production | − | + | − | − | + | + | + | − | + | − | + | − | − | + | + | − | + | + | − | + | − | |
| DNase | + | − | − | + | − | − | + | + | − | + | − | + | + | − | − | + | − | − | + | + | + | |
| Nitrate reduction | + | + | + | − | − | − | − | − | + | + | − | + | − | − | + | + | + | + | + | + | + | |
| Urease | + | + | − | + | + | − | + | − | + | + | + | + | − | + | + | + | + | + | + | + | + | |
| L-asparaginase | + | − | + | − | + | + | − | − | − | + | + | + | − | + | − | + | + | − | − | + | + | |
| L-glutaminase | + | − | − | + | + | + | + | + | − | + | − | + | + | + | + | + | + | − | + | + | + |
| Isolates | FRAP | DPPH | ABTS | SOS | XO inh | Anti-Hemolytic | Anti-Peroxi | Anti-Hemolysis | Cytotoxicity | T. acruzi |
|---|---|---|---|---|---|---|---|---|---|---|
| Act 1 | 42.4 ± 2 | 68 ± 4.4 | 30.5 ± 1.9 | 83 ± 10.2 | 75.2 ± 9.5 | 34.6 ± 2.3 | 17.9 ± 1.2 | 25.7 ± 1.7 | 61.3 ± 9.2 | 4.1 ± 0.3 |
| Act 2 | 67 ± 5.2 | 97 ± 8 | 91.1 ± 8.8 | 83 ± 8.3 | 95.3 ± 9.5 | 61.4 ± 5.1 | 92.2 ± 9.8 | 44.5 ± 3.7 | 71.9 ± 7.8 | 7.1 ± 0.6 |
| Act 3 | 43 ± 1.5 | 58 ± 2.3 | 65.5 ± 2.7 | 81 ± 4.4 | 92.8 ± 5 | 38.9 ± 1.8 | 72.7 ± 3 | 29.3 ± 1.4 | 77.1 ± 4.2 | 4.6 ± 0.2 |
| Act 4 | 28 ± 2.1 | 24 ± 5.7 | 35.1 ± 11 | 42 ± 11.3 | 52.9 ± 11 | 28 ± 0 | 39.3 ± 13 | 30.7 ± 4.7 | 37.6 ± 10.4 | 5.4 ± 1 |
| Act 5 | 25.3 ± 1 | 31.8 ± 1 | 40.5 ± 1.5 | 35.6 ± 1 | 44.8 ± 1.6 | 25.2 ± 1.6 | 41.6 ± 1.3 | 28.4 ± 1.3 | 30.5 ± 0.8 | 5.1 ± 0.2 |
| Act 6 | 28 ± 2.2 | 39 ± 3.1 | 41.9 ± 3.4 | 50 ± 4.4 | 60 ± 5.1 | 30.5 ± 2.4 | 43.9 ± 3.9 | 23.9 ± 1.8 | 46.4 ± 4.1 | 3.8 ± 0.3 |
| Act 7 | 29.3 ± 1 | 53.2 ± 3 | 67.9 ± 5.4 | 28 ± 0.9 | 38.7 ± 1.6 | 35.9 ± 2.6 | 82.5 ± 7 | 27 ± 1.9 | 23.9 ± 0.6 | 4.3 ± 0.3 |
| Act 8 | 43.9 ± 3 | 38 ± 2.1 | 28.7 ± 1.3 | 12.0.9 | 27.5 ± 2.2 | 40.8 ± 3.2 | 61.5 ± 4.1 | 34.3 ± 2.7 | 6.8 ± 0.5 | 5.5 ± 0.4 |
| Act 9 | 41 ± 2.5 | 56 ± 4.7 | 65.3 ± 6 | 50 ± 4.4 | 63.8 ± 5.3 | 39.1 ± 2.7 | 92.5 ± 7.4 | 31.5 ± 2.1 | 45.1 ± 4.1 | 5.1 ± 0.3 |
| Act 10 | 36 ± 2.5 | 36.4 ± 1 | 30.4 ± 0.8 | 66 ± 4.6 | 77.7 ± 5.2 | 24.9 ± 1.5 | 51.6 ± 2.1 | 22.5 ± 1.4 | 63.3 ± 4.4 | 3.6 ± 0.2 |
| Act 11 | 32.2 ± 2 | 47.6 ± 4 | 56.6 ± 5.3 | 55 ± 3.4 | 94.3 ± 6.9 | 30.7 ± 2.4 | 76.6 ± 6.5 | 24.4 ± 1.8 | 80.1 ± 5.9 | 3.9 ± 0.3 |
| Act 12 | 61.4 ± 5 | 124 ± 9 | 115.3 ± 9 | 65 ± 4.9 | 167 ± 13 | 74 ± 6.2 | 123 ± 9.1 | 53.2 ± 4.5 | 139.8 ± 11 | 8.5 ± 0.7 |
| Act 13 | 54 ± 3.2 | 81 ± 3.4 | 68.5 ± 2.7 | 42 ± 2.5 | 86.5 ± 3.1 | 51.7 ± 2.4 | 63.3 ± 2.3 | 39 ± 1.9 | 65.6 ± 2.3 | 6.2 ± 0.3 |
| Act 14 | 37 ± 2.4 | 57.9 ± 4 | 66.9 ± 5.4 | 30 ± 2.3 | 88.4 ± 7.6 | 39.1 ± 2.8 | 75.9 ± 6.5 | 26.5 ± 1.8 | 75.3 ± 6.7 | 4.2 ± 0.3 |
| Act 15 | 58.2 ± 5 | 117 ± 9 | 109.4 ± 8 | 91 ± 7.8 | 129 ± 9.3 | 70.8 ± 5.8 | 137 ± 10.5 | 50.8 ± 4.2 | 103.6 ± 7.1 | 8.1 ± 0.7 |
| Act 16 | 52 ± 3.3 | 84 ± 4 | 77.4 ± 3.9 | 77 ± 4.5 | 90.7 ± 5.1 | 49 ± 2.3 | 86.6 ± 3.9 | 35.9 ± 1.7 | 72 ± 4.2 | 5.7 ± 0.3 |
| Act 17 | 28.2 ± 1 | 33 ± 1.6 | 30.4 ± 1.3 | 27 ± 1.6 | 36.2 ± 2.1 | 28.8 ± 1.8 | 27.1 ± 1.2 | 22.4 ± 1.4 | 23.7 ± 1.4 | 3.6 ± 0.2 |
| Act 18 | 41 ± 3 | 42.5 ± 3 | 30.5 ± 2.4 | 10 ± 1.1 | 21.4 ± 1.7 | 36.5 ± 2.8 | 18.5 ± 1.5 | 27.3 ± 2.1 | 6.5 ± 1 | 4.4 ± 0.3 |
| Act 19 | 84 ± 6.8 | 112 ± 10 | 113.7 ± 11 | 55 ± 6 | 75.9 ± 7.5 | 76.5 ± 6.3 | 115.2 ± 12 | 55.4 ± 4.6 | 46.7 ± 5.3 | 8.9 ± 0.7 |
| Act 20 | 48 ± 1.8 | 72 ± 2.8 | 81.4 ± 3.3 | 70.4.4 | 84 ± 5.2 | 48.3 ± 2.2 | 90.3 ± 3.7 | 36.3 ± 1.7 | 64.4 ± 4.2 | 5.8 ± 0.3 |
| Act 21 | 49 ± 4 | 38 ± 10 | 57.8 ± 21 | 71 ± 2.5 | 88 ± 26.2 | 40.2 ± 0 | 50.6 ± 17.6 | 53.2 ± 8.7 | 62.4 ± 23.4 | 9.3 ± 1.9 |
| Isolates | S. pneumonia | S. aureus | E. coli | B. cereus | E. faecalis | S. typhimurium | P. aeruginosa |
|---|---|---|---|---|---|---|---|
| Act 1 | 9.8 ± 0.6 | 11 ± 0.8 | 13 ± 0.9 | 14.4 ± 1 | 15.7 ± 1 | 17.5 ± 1.1 | 19.1 ± 1.2 |
| Act 2 | 16.3 ± 1.3 | 21 ± 1.8 | 24 ± 2 | 27 ± 2.2 | 21 ± 2.9 | 25.2 ± 0.7 | 24.9 ± 1.6 |
| Act 3 | 11.1 ± 0.6 | 13 ± 0.6 | 14 ± 0.6 | 15 ± 0.6 | 15 ± 0.6 | 15.6 ± 0.5 | 12.5 ± 0.6 |
| Act 4 | 9.9 ± 0.8 | 12 ± 1.1 | 10 ± 1.5 | 12.6 ± 1 | 14 ± 2.8 | 18 ± 6.5 | 7.8 ± 2 |
| Act 5 | 9.2 ± 0.6 | 10 ± 0.5 | 10 ± 0.5 | 7 ± 0.4 | 16 ± 0.9 | 21.5 ± 0.4 | 7.2 ± 0.4 |
| Act 6 | 9.9 ± 0.7 | 10 ± 0.8 | 10 ± 0.8 | 10 ± 0.9 | 17 ± 2.5 | 10.1 ± 0.9 | 10.1 ± 0.9 |
| Act 7 | 10.5 ± 0.7 | 12 ± 0.9 | 13 ± 0.9 | 14 ± 1.0 | 17 ± 2.4 | 15.1 ± 1.3 | 16 ± 1.5 |
| Act 8 | 15.2 ± 1.2 | 13.6 ± 1 | 12.1 ± 1 | 10 ± 0.8 | 14 ± 1.4 | 7.7 ± 0.5 | 23.3 ± 5.2 |
| Act 9 | 13.6 ± 0.9 | 12 ± 0.9 | 12 ± 0.9 | 12 ± 0.9 | 8 ± 1.2 | 10.8 ± 0.9 | 16.6 ± 3.4 |
| Act 10 | 10.8 ± 0.7 | 8.1 ± 0.4 | 6 ± 0.4 | 11.1 ± 1 | 13 ± 2.4 | 11.9 ± 1.2 | 14.2 ± 1.6 |
| Act 11 | 10.3 ± 0.7 | 10 ± 0.8 | 10 ± 0.9 | 10 ± 0.9 | 5.8 ± 1.8 | 12.7 ± 1.0 | 8.7 ± 1.3 |
| Act 12 | 19.4 ± 1.6 | 20.2 ± 1 | 24 ± 0.5 | 29 ± 2.7 | 21 ± 4.5 | 22.50 ± 4 | 20.0 ± 2.5 |
| Act 13 | 14.8 ± 0.8 | 18 ± 0.8 | 19 ± 0.8 | 15 ± 0.8 | 25 ± 1.2 | 19.8 ± 0.8 | 22.5 ± 1.8 |
| Act 14 | 8.9 ± 0.5 | 13.4 ± 1 | 16 ± 1.3 | 17 ± 0.3 | 11 ± 3.7 | 15.6 ± 0.9 | 9.7 ± 3.7 |
| Act 15 | 18.2 ± 1.5 | 24 ± 2.1 | 15 ± 5.1 | 29 ± 0.9 | 26 ± 4.1 | 27.2 ± 3.2 | 26.3 ± 3.7 |
| Act 16 | 13 ± 0.6 | 17 ± 0.8 | 18 ± 0.9 | 21.4 ± 1 | 23 ± 1.1 | 22.8 ± 1.1 | 27.3 ± 1.7 |
| Act 17 | 8.9 ± 0.6 | 10 ± 0.6 | 10 ± 0.6 | 10 ± 0.6 | 14 ± 1.1 | 9.70 ± 0.7 | 15.1 ± 1.4 |
| Act 18 | 10.5 ± 0.8 | 12.5 ± 1 | 13 ± 1.1 | 14.5 ± 1 | 15 ± 0.8 | 17.1 ± 1.4 | 14 ± 0.8 |
| Act 19 | 20.4 ± 1.4 | 26 ± 2.3 | 24 ± 0.6 | 28 ± 0.2 | 29 ± 2.8 | 26.0 ± 5.5 | 19.2 ± 2 |
| Act 20 | 15.3 ± 0.7 | 16 ± 0.7 | 17 ± 0.7 | 18.0.7 | 14 ± 0.7 | 25.6 ± 0.6 | 12.8 ± 0.7 |
| Act 21 | 16.9 ± 1.9 | 18 ± 0.7 | 20 ± 0.6 | 24.2 ± 1 | 22 ± 2.6 | 21.7 ± 1.1 | 23.5 ± 1.1 |
| Isolates | Total Flavonoids | Total Phenols | Tocopherols | Lutine | Alpha-Carotene | β-Cryptoxanthin | β-Carotene | Gamma-Carotene | Lycopene | Carotenoids |
|---|---|---|---|---|---|---|---|---|---|---|
| Act 1 | 4.6 ± 0.3 | 35.3 ± 2.4 | 0.3 ± 0 | 0.3 ± 0.02 | 0.42 ± 0.03 | 0.24 ± 0.02 | 0.27 ± 0.02 | 0.06 ± 0 | 0.08 ± 0.01 | 0.93 ± 0.09 |
| Act 2 | 8 ± 0.6 | 48.6 ± 3.6 | 0.5 ± 0 | 0.5 ± 0.04 | 0.63 ± 0.05 | 0.28 ± 0.02 | 0.28 ± 0.02 | 0.1 ± 0.01 | 0.13 ± 0.01 | 1.3 ± 0.12 |
| Act 3 | 6.7 ± 0.4 | 31.4 ± 1.7 | ND ± ND | 0.3 ± 0.01 | 0.34 ± 0.01 | 0.3 ± 0.02 | 0.31 ± 0.02 | 0.06 ± 0 | 0.07 ± 0 | 1.07 ± 0.05 |
| Act 4 | 5.4 ± 0.3 | 35.6 ± 3.3 | ND ± ND | 0.2 ± 0.02 | 0.23 ± 0.03 | 0.38 ± 0.06 | 0.42 ± 0.06 | 0.1 ± 0.02 | 0.1 ± 0.02 | 0.59 ± 0.15 |
| Act 5 | 5.8 ± 0.4 | 29.9 ± 1.6 | 0.1 ± 0 | 0.1 ± 0 | 0.14 ± 0 | 0.27 ± 0.01 | 0.29 ± 0.01 | 0.29 ± 0.02 | 0.23 ± 0.02 | 0.61 ± 0.02 |
| Act 6 | 6.9 ± 0.5 | 29.6 ± 2.2 | 0.3 ± 0 | 0.1 ± 0.01 | 0.18 ± 0.02 | 0.35 ± 0.03 | 0.47 ± 0.05 | 0.14 ± 0.01 | 0.12 ± 0.01 | 0.77 ± 0.06 |
| Act 7 | 7.4 ± 0.6 | 31.1 ± 2 | ND ± ND | 0.1 ± 0 | 0.12 ± 0.01 | 0.21 ± 0.01 | 0.23 ± 0.01 | 0.21 ± 0.02 | 0.38 ± 0.04 | 0.8 ± 0.05 |
| Act 8 | 9.5 ± 0.8 | 49.4 ± 4.1 | ND ± ND | 0.2 ± 0.02 | 0.23 ± 0.02 | 0.17 ± 0.03 | 0.18 ± 0.01 | 0.09 ± 0 | 0.16 ± 0.01 | 0.48 ± 0.03 |
| Act 9 | 11.2 ± 0.9 | 33.6 ± 1.7 | 0.4 ± 0 | 0.2 ± 0.01 | 0.27 ± 0.02 | 0.14 ± 0.01 | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.61 ± 0.07 | 0.97 ± 0.08 |
| Act 10 | 10.4 ± 0.7 | 20.7 ± 1.3 | ND ± ND | 0.2 ± 0.01 | 0.17 ± 0.01 | 0.2 ± 0.02 | 0.27 ± 0.04 | 0.09 ± 0.01 | 0.22 ± 0.01 | 0.82 ± 0.04 |
| Act 11 | 8.8 ± 0.6 | 24.5 ± 1.8 | 0.3 ± 0 | 0.2 ± 0.01 | 0.21 ± 0.02 | 0.23 ± 0.02 | 0.19 ± 0.02 | 0.04 ± 0 | 0.04 ± 0.01 | 0.99 ± 0.08 |
| Act 12 | 11.5 ± 0.9 | 48.2 ± 3.9 | 0.5 ± 0 | 0.2 ± 0.02 | 0.33 ± 0.02 | 0.86 ± 0.08 | 0.85 ± 0.09 | 0.1 ± 0.01 | 0.08 ± 0.01 | 1.67 ± 0.13 |
| Act 13 | 9.7 ± 0.6 | 39.3 ± 2.1 | 0.2 ± 0 | 0.3 ± 0.03 | 0.44 ± 0.03 | 0.5 ± 0.02 | 0.56 ± 0.02 | 0.08 ± 0 | 0.1 ± 0.01 | 1.06 ± 0.05 |
| Act 14 | 3.9 ± 0.1 | 35.3 ± 2.5 | ND ± ND | 0.2 ± 0.01 | 0.35 ± 0.02 | 0.14 ± 0 | 0.58 ± 0 | 0.24 ± 0.02 | 0.19 ± 0.03 | 1 ± 0.08 |
| Act 15 | 9.9 ± 0.9 | 48.5 ± 3.6 | 0.5 ± 0 | 0.3 ± 0.03 | 0.45 ± 0.04 | 0.17 ± 0.01 | 0.38 ± 0.02 | 0.19 ± 0.01 | 0.19 ± 0.01 | 1.58 ± 0.12 |
| Act 16 | 8.7 ± 0.5 | 33.4 ± 1.7 | 0.3 ± 0 | 0.2 ± 0.01 | 0.32 ± 0.02 | 0.4 ± 0.03 | 0.44 ± 0.04 | 0.09 ± 0 | 0.09 ± 0.01 | 1.24 ± 0.07 |
| Act 17 | 4.8 ± 0.3 | 31.9 ± 2.4 | ND ± ND | 0.1 ± 0.01 | 0.16 ± 0.01 | 0.36 ± 0.03 | 0.43 ± 0.02 | 0.05 ± 0 | 0.05 ± 0 | 0.53 ± 0.02 |
| Act 18 | 5.1 ± 0.4 | 40.1 ± 3 | 0.2 ± 0 | 0.2 ± 0.02 | 0.32 ± 0.03 | 0.18 ± 0.01 | 0.2 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.02 | 0.49 ± 0.04 |
| Act 19 | 9.8 ± 0.8 | 59.8 ± 4.5 | 0.5 ± 0 | 0.2 ± 0.01 | 0.23 ± 0.01 | 0.27 ± 0 | 0.82 ± 0 | 0.14 ± 0.01 | 0.08 ± 0.01 | 1.29 ± 0.12 |
| Act 20 | 8.3 ± 0.5 | 38.6 ± 2.1 | ND ± ND | 0.1 ± 0 | 0.08 ± 0.01 | 0.17 ± 0.01 | 0.44 ± 0.01 | 0.15 ± 0.01 | 0.38 ± 0.03 | 1.14 ± 0.06 |
| Act 21 | 7.7 ± 0.5 | 51.7 ± 5 | 0.4 ± 0.1 | 0.1 ± 0.02 | 0.2 ± 0.03 | 0.15 ± 0.03 | 0.17 ± 0.02 | 0.29 ± 0.09 | 0.15 ± 0.01 | 0.84 ± 0.25 |
| Phenolic Compounds | Act 2 | Act 12 | Act 15 | Act 19 | Act 21 |
|---|---|---|---|---|---|
| Caffeic acid | 0.79 ± 0.05 | 0.75 ± 0.19 | 0.77 ± 0.06 | 1.25 ± 0.09 | 1.05 ± 0.22 |
| Ferulic ACID | 5.5 ± 0.23 | 5.55 ± 0.23 | 5.15 ± 0.38 | 5.43 ± 0.34 | 6.68 ± 0.32 |
| Protocatechuic acid | 10.2 ± 0.41 | 20 ± 2.49 | 9.26 ± 0.69 | 16.12 ± 0.65 | 21.09 ± 1.61 |
| Catechin | 4.4 ± 0.18 | 6.67 ± 0.13 | 4.01 ± 0.3 | 7.02 ± 0.28 | 8.78 ± 0.13 |
| Gallic acid | 14.4 ± 0.58 | 21.8 ± 1.24 | 13.11 ± 0.97 | 23 ± 0.93 | 24.55 ± 1.92 |
| p-Coumaric acid | 12.1 ± 0.49 | 9.04 ± 0.37 | 11.02 ± 0.82 | 9.15 ± 0.7 | 20.24 ± 1.58 |
| Resorcinol | ND | 0.09 ± 0 | 0.11 ± 0.01 | ND | 0.21 ± 0.02 |
| Chlorogenic acid | 0.74 ± 0.03 | 1.53 ± 0.34 | ND | 1.18 ± 0.05 | 1.76 ± 0.2 |
| Sinapic acid | ND | 8.53 ± 0.21 | ND | 2.3 ± 0.09 | 11.95 ± 0.94 |
| Quercetin | 10.3 ± 0.42 | 7.72 ± 0.31 | 9.41 ± 0.7 | ND | 7.31 ± 0.09 |
| Quercetrin | 1.15 ± 0.05 | 0.86 ± 0.03 | 1.05 ± 0.08 | 1.83 ± 0.07 | 3.32 ± 0.26 |
| Luteolin | 0.34 ± 0.01 | 0.92 ± 0.41 | ND | 0.85 ± 0.03 | ND |
| Apigenin | ND | 1.3 ± 0.2 | ND | 1.89 ± 0.27 | ND |
| Isoquercetrin | ND | 1.2 ± 0.05 | 1.47 ± 0.11 | 1.35 ± 0.12 | 3.27 ± 0.07 |
| Rutin | 4.98 ± 0.36 | 5.41 ± 0.22 | 6.59 ± 0.49 | 6.46 ± 1.02 | 18.57 ± 1.45 |
| Ellagic acid | 2.89 ± 0.12 | 2.16 ± 0.09 | 2.63 ± 0.19 | 3 ± 0.14 | 7.45 ± 0.58 |
| Velutin | ND | 2.61 ± 0.11 | ND | ND | 4.1 ± 0.06 |
| Naringenin | ND | 0.04 ± 0.0 | 0.04 ± 0.003 | 0.11 ± 0.004 | 0.11 ± 0.009 |
| Genistein | ND | 0.03 ± 0.0 | 0.03 ± 0.002 | 0.08 ± 0.003 | 0.09 ± 0.007 |
| Daidzein | ND | 0.02 ± 0.03 | 0.02 ± 0.001 | 0.06 ± 0.002 | ND |
| Fisetin | 0.012 ± 0 | ND | ND | ND | 0.031 ± 0.001 |
| O-hydroxydaidzein | ND | 0.01 ± 0 | 0.06 ± 0.005 | 0.03 ± 0.001 | 0.06 ± 0.003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almuhayawi, M.S.; Mohamed, M.S.M.; Abdel-Mawgoud, M.; Selim, S.; Al Jaouni, S.K.; AbdElgawad, H. Bioactive Potential of Several Actinobacteria Isolated from Microbiologically Barely Explored Desert Habitat, Saudi Arabia. Biology 2021, 10, 235. https://doi.org/10.3390/biology10030235
Almuhayawi MS, Mohamed MSM, Abdel-Mawgoud M, Selim S, Al Jaouni SK, AbdElgawad H. Bioactive Potential of Several Actinobacteria Isolated from Microbiologically Barely Explored Desert Habitat, Saudi Arabia. Biology. 2021; 10(3):235. https://doi.org/10.3390/biology10030235
Chicago/Turabian StyleAlmuhayawi, Mohammed S., Mahmoud S. M. Mohamed, Mohamed Abdel-Mawgoud, Samy Selim, Soad K. Al Jaouni, and Hamada AbdElgawad. 2021. "Bioactive Potential of Several Actinobacteria Isolated from Microbiologically Barely Explored Desert Habitat, Saudi Arabia" Biology 10, no. 3: 235. https://doi.org/10.3390/biology10030235








