Antibiotics Modulate Intestinal Regeneration
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibiotic Effect on Intestinal Regeneration
2.1.1. Animal Care
2.1.2. Immunohistochemistry
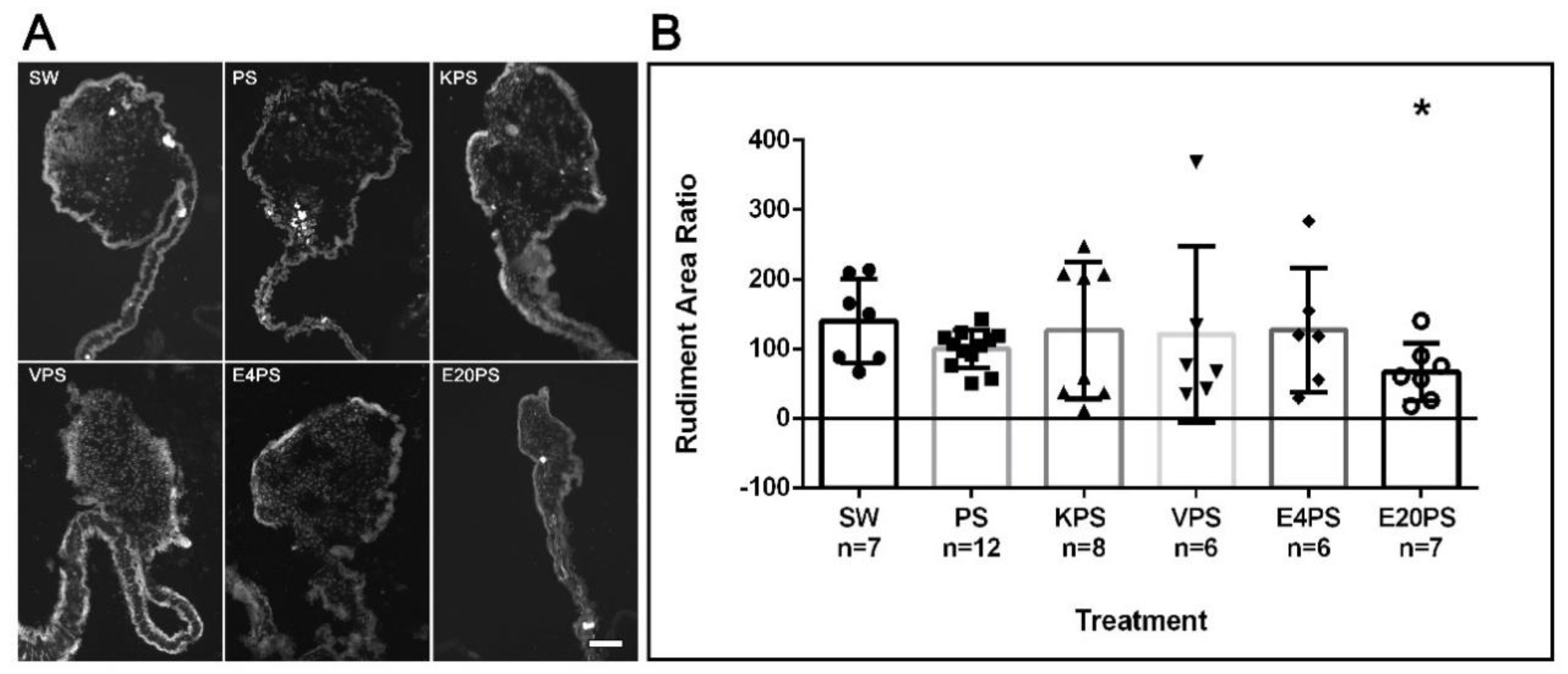
2.1.3. Measurement of Rudiment Area
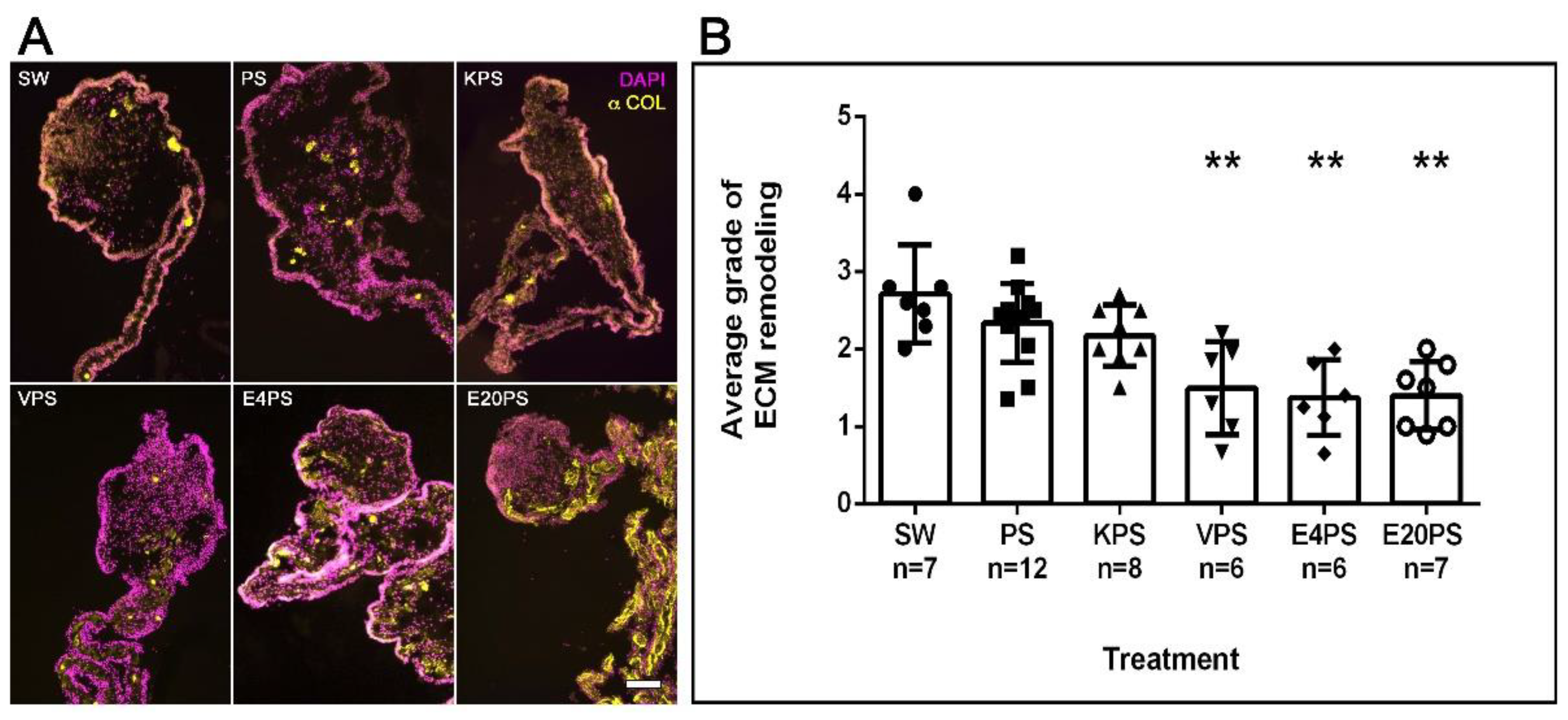
2.1.4. Remodeling of the ECM
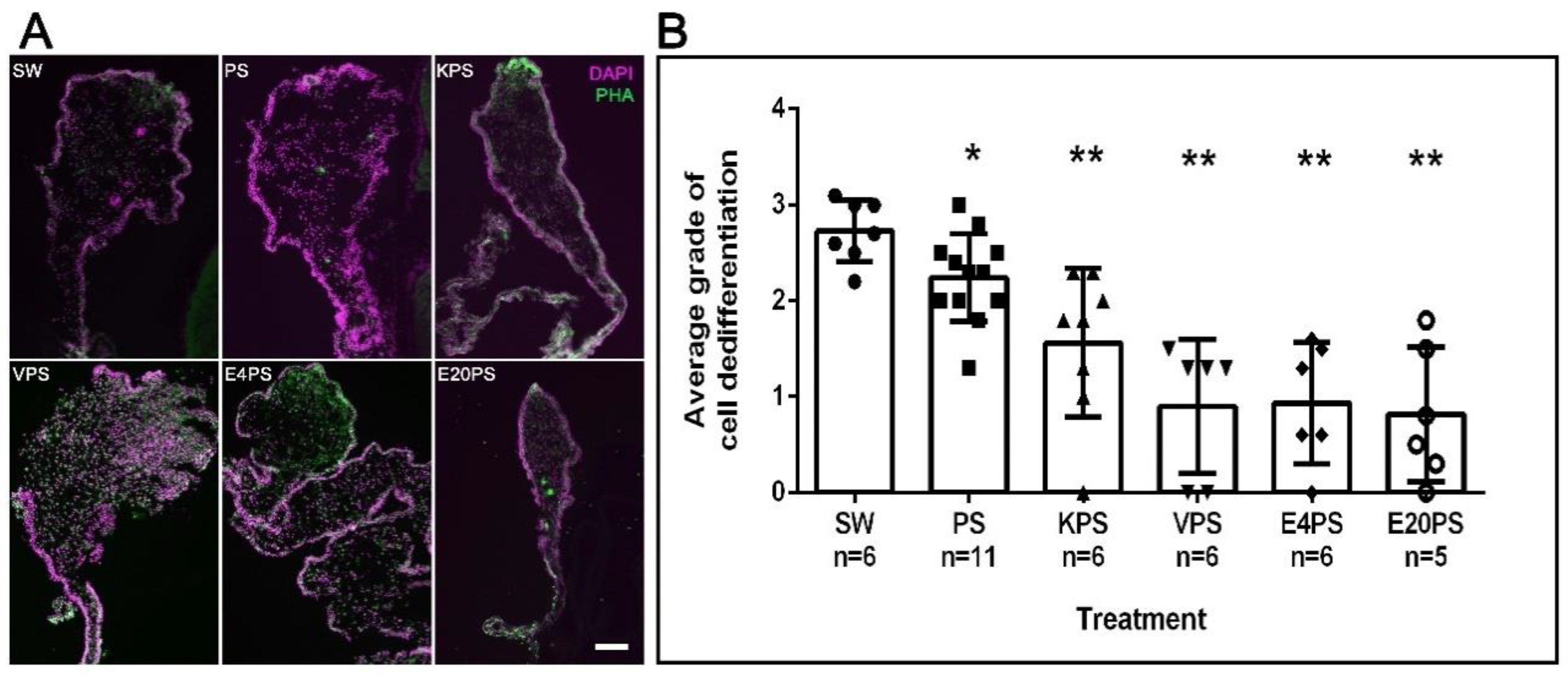
2.1.5. Muscle Dedifferentiation
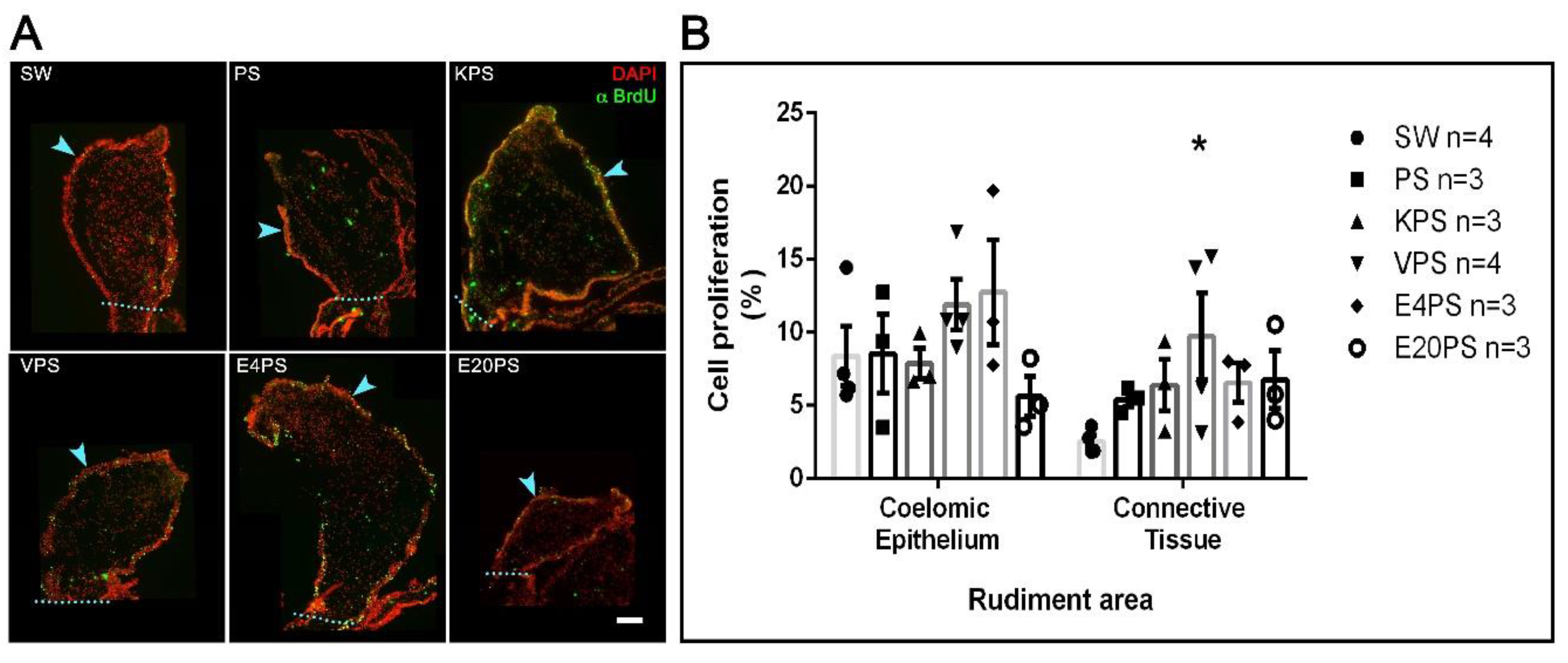
2.1.6. Cellular Proliferation
2.2. Minimum Inhibitory Concentration (MIC)
MIC Determination
2.3. Antibiotic Toxicity
2.3.1. Muscle Dissection for In Vivo and Ex Vivo Assays
2.3.2. Explant Culture and Toxicity Essay
2.3.3. Protein Quantification for MTT Normalization
2.4. Statistical Analyses
3. Results
3.1. Survival Rate in Regenerating Sea Cucumbers during Antibiotic Treatments
3.2. Rudiment Formation Is Perturbed by Erythromycin Exposure
3.3. Intestinal Cellular Dedifferentiation Is Delayed by Antibacterial Treatments
3.4. ECM Remodeling Is Altered by Antibiotics
3.5. Vancomycin PS-Based Treatment Alters the Cell Proliferation Rate in the Connective Tissue
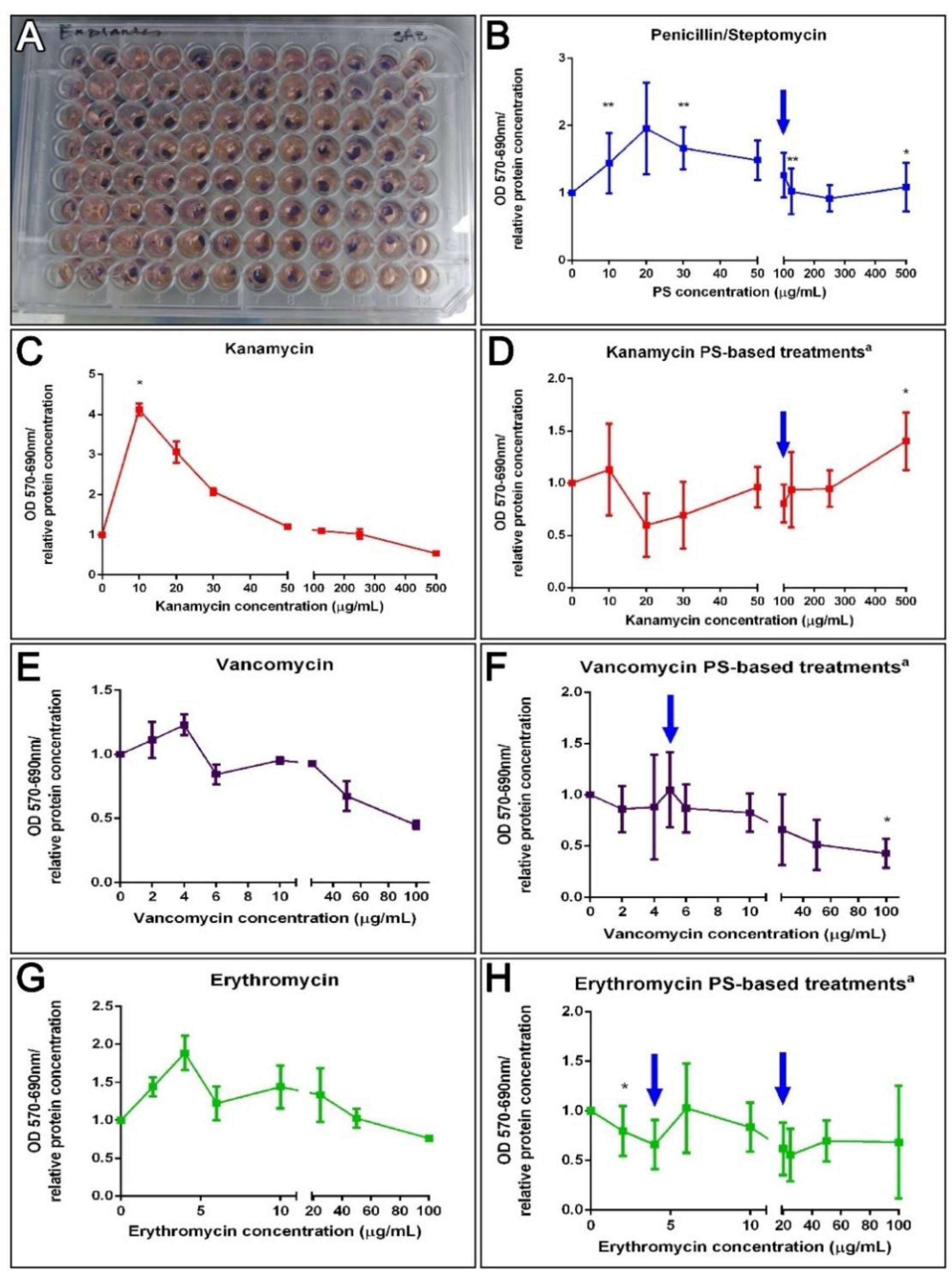
3.6. Antibiotics and Holothurian Cellular Toxicity
3.6.1. Cell Cultures
3.6.2. Explant Cultures
3.7. Holothurians Gut Bacteria Growth Inhibition
4. Discussion
4.1. The Survival Rate of Sea Cucumbers In Vivo Is Maintained after Antibiotic Treatments
4.2. Sea Cucumber Intestinal Regeneration Is Perturbed by Antibiotic Treatments
4.3. The Metabolic Activity Remains Unaffected after Antibiotic Treatments Ex Vivo
4.4. Antibiotics Cocktails Inhibit Gut Bacterial Populations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B.A. A Global Perspective on the Use, Sales, Exposure Pathways, Occurrence, Fate and Effects of Veterinary Antibiotics (VAs) in the Environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Zhu, M.; Zhao, H.; Chen, J.; Xie, H.; Du, J. Investigation of Antibiotics in Sea Cucumbers: Occurrence, Pollution Characteristics, and Human Risk Assessment. Environ. Sci. Pollut. Res. 2018, 25, 32081–32087. [Google Scholar] [CrossRef] [PubMed]
- National Research Council (US). Committee to Study the Human Health Effects of Subtherapeutic Antibiotic Use in Animal Feeds. In The Effects on Human Health of Subtherapeutic Use of Antimicrobials in Animal Feeds; Appendix K, Antibiotics In Animal Feeds; National Academies Press (US): Washington DC, USA, 1980. Available online: https://www.ncbi.nlm.nih.gov/books/NBK216502/ (accessed on 7 January 2021).
- Ok, Y.S.; Kim, S.-C.; Kim, K.-R.; Lee, S.S.; Moon, D.H.; Lim, K.J.; Sung, J.-K.; Hur, S.-O.; Yang, J.E. Monitoring of Selected Veterinary Antibiotics in Environmental Compartments near a Composting Facility in Gangwon Province, Korea. Environ. Monit. Assess. 2011, 174, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Austin, B. Antibiotic Pollution from Fish Farms: Effects on Aquatic Microflora. Microbiol. Sci. 1985, 2, 113–117. [Google Scholar]
- Chen, J.; Sun, R.; Pan, C.; Sun, Y.; Mai, B.; Li, Q.X. Antibiotics and Food Safety in Aquaculture. J. Agric. Food Chem. 2020, 68, 11908–11919. [Google Scholar] [CrossRef]
- Armstrong, S.M.; Hargrave, B.T.; Haya, K. Antibiotic Use in Finfish Aquaculture: Modes of Action, Environmental Fate, and Microbial Resistance. In Environmental Effects of Marine Finfish Aquaculture; Handbook of Environmental, Chemistry; Hargrave, B.T., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 5M, pp. 341–357. [Google Scholar] [CrossRef]
- Arthur, J.R.; Lavilla-Pitogo, C.; Subasinghe, R.P.; Southeast Asian Fisheries Development Center; FAO; Kanada (Eds.) Use of Chemicals in Aquaculture in Asia. In Proceedings of the Meeting on the Use of Chemicals in Aquaculture in Asia, Tigbauan, Iloilo, Philippnes, 20–22 May 1996; Southeast Asian Fisheries Development Center, Aquaculture Department: Tigbauan, Iloilo, Philippines, 2000. [Google Scholar]
- Asche, F.; Roll, K.H.; Tveterås, S. Future Trends in Aquaculture: Productivity Growth and Increased Production. In Aquaculture in the Ecosystem; Holmer, M., Black, K., Duarte, C.M., Marbà, N., Karakassis, I., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 271–292. [Google Scholar] [CrossRef]
- Asche, F. Farming the sea. Mar. Resour. Econ. 2008, 23, 527–547. [Google Scholar] [CrossRef]
- Asche, F.; Hansen, H.; Tveteras, R.; Tveterås, S. The Salmon Disease Crisis in Chile. Mar. Resour. Econ. 2009, 24, 405–411. [Google Scholar] [CrossRef]
- Buschmann, A.H.; Cabello, F.; Young, K.; Carvajal, J.; Varela, D.A.; Henríquez, L. Salmon Aquaculture and Coastal Ecosystem Health in Chile: Analysis of Regulations, Environmental Impacts and Bioremediation Systems. Ocean Coast. Manag. 2009, 52, 243–249. [Google Scholar] [CrossRef]
- Burridge, L.; Weis, J.S.; Cabello, F.; Pizarro, J.; Bostick, K. Chemical Use in Salmon Aquaculture: A Review of Current Practices and Possible Environmental Effects. Aquaculture 2010, 306, 7–23. [Google Scholar] [CrossRef]
- Cabello, F.C. Heavy Use of Prophylactic Antibiotics in Aquaculture: A Growing Problem for Human and Animal Health and for the Environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.W.; Cole, R.; Gaydos, S.J.; Gray, J.; Hyland, G.; Jacques, M.L.; Powell-Dunford, N.; Sawhney, C.; Au, W.W. Aquaculture: Environmental, Toxicological, and Health Issues. Int. J. Hyg. Environ. Health 2009, 212, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Costa-Pierce, B.A. Use of Ecosystems Science in Ecological Aquaculture. Bull. Aquac. Assoc. Can. 2003, 2, 32–40. [Google Scholar]
- Costa-Pierce, B.A. Sustainable Ecological Aquaculture Systems: The Need for a New Social Contract for Aquaculture Development. Mar. Technol. Soc. J. 2010, 44, 88–112. [Google Scholar] [CrossRef]
- Diana, J.S. Aquaculture Production and Biodiversity Conservation. BioScience 2009, 59, 27–38. [Google Scholar] [CrossRef]
- Haya, K.; Burridge, L.E.; Chang, B.D. Environmental Impact of Chemical Wastes Produced by the Salmon Aquaculture Industry. ICES J. Mar. Sci. 2001, 58, 492–496. [Google Scholar] [CrossRef] [Green Version]
- Millanao, B.A.; Barrientos, H.M.; Gómez, C.C.; Tomova, A.; Buschmann, A.; Dölz, H.; Cabello, F.C. Uso Inadecuado y Excesivo de Antibióticos: Salud Pública y Salmonicultura En Chile. Rev. Méd. Chile 2011, 139, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Naylor, R.; Burke, M. Aquaculture and Ocean Resourses: Raising Tigers of the Sea. Annu. Rev. Environ. Resour. 2005, 30, 185–218. [Google Scholar] [CrossRef] [Green Version]
- Samanidou, V.F.; Evaggelopoulou, E.N. Analytical Strategies to Determine Antibiotic Residues in Fish. J. Sep. Sci. 2007, 30, 2549–2569. [Google Scholar] [CrossRef]
- Liu, X.; Steele, J.C.; Meng, X.-Z. Usage, Residue, and Human Health Risk of Antibiotics in Chinese Aquaculture: A Review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Chung, J.K. The Research of the Antibiotics Reduction in Aquarium and the Environmental Influence of Parasiticide; Ministry of Food, Agriculture, Forestry and Fisheries & Pukyong National University Press: Busan, Korea, 2008.
- Li, Z.; Zhang, J.; Li, X.; Wang, X.; Cao, Z.; Wang, L.; Xu, Y. Efficiency of a Bacteriophage in Controlling Vibrio Infection in the Juvenile Sea Cucumber Apostichopus Japonicus. Aquaculture 2016, 451, 345–352. [Google Scholar] [CrossRef]
- Santos, L.; Soares, B.; Rosa, J.; Freitas, A.; Leston, S.; Barbosa, J.; Ramos, F. Detection and Quantification of 41 Antibiotic Residues in Gilthead Sea Bream (Sparus aurata) from Aquaculture Origin, Using a Multiclass and Multi-Residue UHPLC-MS/MS Method. Food Anal. Methods 2016, 9, 2749–2753. [Google Scholar] [CrossRef]
- Heuer, O.E.; Kruse, H.; Grave, K.; Collignon, P.; Karunasagar, I.; Angulo, F.J. Human Health Consequences of Use of Antimicrobial Agents in Aquaculture. Clin. Infect Dis. 2009, 49, 1248–1253. [Google Scholar] [CrossRef]
- Liu, Y.-B.; Tabashnik, B.E.; Moar, W.J.; Smith, R.A. Synergism between Bacillus Thuringiensis Spores and Toxins against Resistant and Susceptible Diamondback Moths (Plutella xylostella). Appl. Environ. Microbiol. 1998, 64, 1385–1389. [Google Scholar] [CrossRef] [Green Version]
- Thakur, A.; Dhammi, P.; Saini, H.S.; Kaur, S. Effect of Antibiotic on Survival and Development of Spodoptera litura (Lepidoptera: Noctuidae) and Its Gut Microbial Diversity. Bull. Entomol. Res. 2016, 106, 387–394. [Google Scholar] [CrossRef]
- Desbois, A.P.; Coote, P.J. Wax Moth Larva (Galleria mellonella): An in Vivo Model for Assessing the Efficacy of Antistaphylococcal Agents. J. Antimicrob. Chemother. 2011, 66, 1785–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buyukguzel, K.; Yazgan, S. Effects of antimicrobial agents on the survival and development of larvae of Pimpla turionellae L. (Hymenoptera: Ichneumonidae) reared on an artificial diet. Turk. J. Zool. 2002, 26, 111–119. [Google Scholar]
- Lin, X.-L.; Kang, Z.-W.; Pan, Q.-J.; Liu, T.-X. Evaluation of Five Antibiotics on Larval Gut Bacterial Diversity of Plutella xylostella (Lepidoptera: Plutellidae): Gut Bacterial Diversity in Plutella xylostella Larvae. Insect Sci. 2015, 22, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Büyükgüzel, E.; Kalender, Y. Galleria mellonella (L.) Survivorship, Development and Protein Content in Response to Dietary Antibiotics. J. Entomol. Sci. 2008, 43, 27–40. [Google Scholar] [CrossRef]
- Ji, K.; Kim, S.; Han, S.; Seo, J.; Lee, S.; Park, Y.; Choi, K.; Kho, Y.-L.; Kim, P.-G.; Park, J.; et al. Risk Assessment of Chlortetracycline, Oxytetracycline, Sulfamethazine, Sulfathiazole, and Erythromycin in Aquatic Environment: Are the Current Environmental Concentrations Safe? Ecotoxicology 2012, 21, 2031–2050. [Google Scholar] [CrossRef] [PubMed]
- Catnach, S.M.; Fairclough, P.D.; Trembath, R.C.; O’donnell, L.J.D.; Mclean, A.M.; Law, P.A.; Wickham, J.E.A. Effect of Oral Erythromycin on Gallbladder Motility in Normal Subjects and Subjects with Gallstones. Gastroenterology 1992, 102, 2071–2076. [Google Scholar] [CrossRef]
- Knoop, K.A.; Gustafsson, J.K.; McDonald, K.G.; Kulkarni, D.H.; Kassel, R.; Newberry, R.D. Antibiotics Promote the Sampling of Luminal Antigens and Bacteria via Colonic Goblet Cell Associated Antigen Passages. Gut Microbes 2017, 8, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Million, M.; Hugon, P.; Armougom, F.; Raoult, D. Human Gut Microbiota: Repertoire and Variations. Front. Cell. Inf. Microbio. 2012, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, K.; Buerger, M.; Stallmach, A.; Bruns, T. Effects of Antibiotics on Gut Microbiota. Dig. Dis. 2016, 34, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Tulstrup, M.V.-L.; Christensen, E.G.; Carvalho, V.; Linninge, C.; Ahrné, S.; Højberg, O.; Licht, T.R.; Bahl, M.I. Antibiotic Treatment Affects Intestinal Permeability and Gut Microbial Composition in Wistar Rats Dependent on Antibiotic Class. PLoS ONE 2015, 10, e0144854. [Google Scholar] [CrossRef]
- Aguilera, M.; Cerdà-Cuéllar, M.; Martínez, V. Antibiotic-Induced Dysbiosis Alters Host-Bacterial Interactions and Leads to Colonic Sensory and Motor Changes in Mice. Gut Microbes 2015, 6, 10–23. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Zheng, Y.; Wen, R.; Deng, X.; Zhou, L.; Liao, H. Effects of Ceftriaxone Induced Intestinal Dysbacteriosis on Lymphocytes in Different Tissues in Mice. Immunobiology 2016, 221, 994–1000. [Google Scholar] [CrossRef]
- Lichtman, J.S.; Ferreyra, J.A.; Ng, K.M.; Smits, S.A.; Sonnenburg, J.L.; Elias, J.E. Host-Microbiota Interactions in the Pathogenesis of Antibiotic-Associated Diseases. Cell Rep. 2016, 14, 1049–1061. [Google Scholar] [CrossRef] [Green Version]
- Miki, T.; Goto, R.; Fujimoto, M.; Okada, N.; Hardt, W.-D. The Bactericidal Lectin RegIIIβ Prolongs Gut Colonization and Enteropathy in the Streptomycin Mouse Model for Salmonella Diarrhea. Cell Host Microbe 2017, 21, 195–207. [Google Scholar] [CrossRef] [Green Version]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Bäumler, A.J. Dysbiotic Proteobacteria Expansion: A Microbial Signature of Epithelial Dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wurm, P.; Spindelboeck, W.; Krause, R.; Plank, J.; Fuchs, G.; Bashir, M.; Petritsch, W.; Halwachs, B.; Langner, C.; Högenauer, C.; et al. Antibiotic-Associated Apoptotic Enterocolitis in the Absence of a Defined Pathogen: The Role of Intestinal Microbiota Depletion. Crit. Care Med. 2017, 45, e600–e606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivera-Chávez, F.; Zhang, L.F.; Faber, F.; Lopez, C.A.; Byndloss, M.X.; Olsan, E.E.; Xu, G.; Velazquez, E.M.; Lebrilla, C.B.; Winter, S.E.; et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 2016, 19, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y.; et al. Microbiota-Activated PPAR-γ Signaling Inhibits Dysbiotic Enterobacteriaceae Expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.Y.; Li, M.; Li, S.S.; He, M.; Yu, X.H.; Shi, L.; He, F. Vancomycin and Ceftriaxone Can Damage Intestinal Microbiota and Affect the Development of the Intestinal Tract and Immune System to Different Degrees in Neonatal Mice. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef]
- Hou, Q.; Ye, L.; Huang, L.; Yu, Q. The Research Progress on Intestinal Stem Cells and Its Relationship with Intestinal Microbiota. Front. Immunol. 2017, 8, 599. [Google Scholar] [CrossRef] [Green Version]
- Ghoshal, U.C.; Ghoshal, U. Small Intestinal Bacterial Overgrowth and Other Intestinal Disorders. Gastroenterol. Clin. N. Am. 2017, 46, 103–120. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cervantes, J. Use Your Antibiotics Wisely. Consequences to the Intestinal Microbiome. FEMS Microbiol. Lett. 2016, 363, fnw081. [Google Scholar] [CrossRef] [Green Version]
- Peterson, C.T.; Sharma, V.; Elmén, L.; Peterson, S.N. Immune Homeostasis, Dysbiosis and Therapeutic Modulation of the Gut Microbiota: Gut Microbiota and Immune Homeostasis. Clin. Exp. Immunol. 2015, 179, 363–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassir, N.; Simeoni, U.; La Scola, B. Gut Microbiota and the Pathogenesis of Necrotizing Enterocolitis in Preterm Neonates. Future Microbiol. 2016, 11, 273–292. [Google Scholar] [CrossRef] [PubMed]
- Dik, V.K.; van Oijen, M.G.H.; Smeets, H.M.; Siersema, P.D. Frequent Use of Antibiotics Is Associated with Colorectal Cancer Risk: Results of a Nested Case–Control Study. Dig. Dis. Sci. 2016, 61, 255–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabouridis, P.S.; Pachnis, V. Emerging Roles of Gut Microbiota and the Immune System in the Development of the Enteric Nervous System. J. Clin. Investig. 2015, 125, 956–964. [Google Scholar] [CrossRef] [Green Version]
- Savidge, T.C.; Newman, P.; Pothoulakis, C.; Ruhl, A.; Neunlist, M.; Bourreille, A.; Hurst, R.; Sofroniew, M.V. Enteric Glia Regulate Intestinal Barrier Function and Inflammation Via Release of S-Nitrosoglutathione. Gastroenterology 2007, 132, 1344–1358. [Google Scholar] [CrossRef]
- Cornet, A.; Savidge, T.C.; Cabarrocas, J.; Deng, W.-L.; Colombel, J.-F.; Lassmann, H.; Desreumaux, P.; Liblau, R.S. Enterocolitis Induced by Autoimmune Targeting of Enteric Glial Cells: A Possible Mechanism in Crohn’s Disease? Proc. Natl. Acad. Sci. USA 2001, 98, 13306–13311. [Google Scholar] [CrossRef] [Green Version]
- Brun, P.; Giron, M.C.; Qesari, M.; Porzionato, A.; Caputi, V.; Zoppellaro, C.; Banzato, S.; Grillo, A.R.; Spagnol, L.; De Caro, R.; et al. Toll-Like Receptor 2 Regulates Intestinal Inflammation by Controlling Integrity of the Enteric Nervous System. Gastroenterology 2013, 145, 1323–1333. [Google Scholar] [CrossRef] [Green Version]
- Muller, P.A.; Koscsó, B.; Rajani, G.M.; Stevanovic, K.; Berres, M.-L.; Hashimoto, D.; Mortha, A.; Leboeuf, M.; Li, X.-M.; Mucida, D.; et al. Crosstalk between Muscularis Macrophages and Enteric Neurons Regulates Gastrointestinal Motility. Cell 2014, 158, 300–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergstrom, K.; Fu, J.; Johansson, M.E.V.; Liu, X.; Gao, N.; Wu, Q.; Song, J.; McDaniel, J.M.; McGee, S.; Chen, W.; et al. Core 1– and 3–Derived O-Glycans Collectively Maintain the Colonic Mucus Barrier and Protect against Spontaneous Colitis in Mice. Mucosal Immunol. 2017, 10, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Swidsinski, A.; Loening-Baucke, V.; Herber, A. Mucosal Flora in Crohn’s Disease and Ulcerative Colitis—An Overview. J. Physiol. Pharm. 2009, 60 (Suppl. 6), 61–71. [Google Scholar]
- Lee, S.M.; Donaldson, G.P.; Mikulski, Z.; Boyajian, S.; Ley, K.; Mazmanian, S.K. Bacterial Colonization Factors Control Specificity and Stability of the Gut Microbiota. Nature 2013, 501, 426–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Arrarás, J.E.; Lázaro-Peña, M.I.; Díaz-Balzac, C.A. Holothurians as a Model System to Study Regeneration. In Marine Organisms as Model Systems in Biology and Medicine; Kloc, M., Kubiak, J.Z., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 65, pp. 255–283. [Google Scholar] [CrossRef]
- García-Arrarás, J.E.; Estrada-Rodgers, L.; Santiago, R.; Torres, I.I.; Díaz-Miranda, L.; Torres-Avillán, I. Cellular Mechanisms of Intestine Regeneration in the Sea Cucumber, Holothuria Glaberrima Selenka (Holothuroidea: Echinodermata). J. Exp. Zool. 1998, 281, 288–304. [Google Scholar] [CrossRef]
- García-Arrarás, J.E.; Greenberg, M.J. Visceral Regeneration in Holothurians: Holothurian Regeneration. Microsc. Res. Tech. 2001, 55, 438–451. [Google Scholar] [CrossRef]
- García-Arrarás, J.E.; Valentín-Tirado, G.; Flores, J.E.; Rosa, R.J.; Rivera-Cruz, A.; San Miguel-Ruiz, J.E.; Tossas, K. Cell Dedifferentiation and Epithelial to Mesenchymal Transitions during Intestinal Regeneration in H. Glaberrima. BMC Dev. Biol. 2011, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Candelaria, A.G.; Murray, G.; File, S.K.; García-Arrarás, J.E. Contribution of Mesenterial Muscle Dedifferentiation to Intestine Regeneration in the Sea Cucumber Holothuria Glaberrima. Cell Tissue Res. 2006, 325, 55–65. [Google Scholar] [CrossRef] [PubMed]
- García-Arrarás, J.E.; Dolmatov, I.Y. Echinoderms: Potential Model Systems for Studies on Muscle Regeneration. Curr. Pharm. Des. 2010, 16, 942–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashanov, V.S.; García-Arrarás, J.E. Gut Regeneration in Holothurians: A Snapshot of Recent Developments. Biol. Bull. 2011, 221, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, J.L.; Rosa, R.; Ruiz, D.L.; García-Arrarás, J.E. Extracellular Matrix Remodeling and Metalloproteinase Involvement during Intestine Regeneration in the Sea Cucumber Holothuria Glaberrima. Dev. Biol. 2002, 250, 181–197. [Google Scholar] [CrossRef] [Green Version]
- García-Arrarás, J.E.; Schenk, C.; Rodrígues-Ramírez, R.; Torres, I.I.; Valentín, G.; Candelaria, A.G. Spherulocytes in the Echinoderm Holothuria Glaberrima and Their Involvement in Intestinal Regeneration. Dev. Dyn. 2006, 235, 3259–3267. [Google Scholar] [CrossRef]
- Díaz-Balzac, C.A.; Abreu-Arbelo, J.E.; García-Arrarás, J.E. Neuroanatomy of the Tube Feet and Tentacles in Holothuria Glaberrima (Holothuroidea, Echinodermata). Zoomorphology 2010, 129, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Murray, G.; García-Arrarás, J.E. Myogenesis during Holothurian Intestinal Regeneration. Cell Tissue Res. 2004, 318, 515–524. [Google Scholar] [CrossRef]
- Pasten, C.; Rosa, R.; Ortiz, S.; González, S.; García-Arrarás, J.E. Characterization of proteolytic activities during intestinal regeneration of the sea cucumber, Holothuria glaberrima. Int. J. Dev. Biol. 2012, 56, 681–691. [Google Scholar] [CrossRef] [Green Version]
- Bello, S.A.; Abreu-Irizarry, R.J.; García-Arrarás, J.E. Primary Cell Cultures of Regenerating Holothurian Tissues. In Tissue Morphogenesis; Nelson, C.M., Ed.; Springer: New York, NY, USA, 2015; Volume 1189, pp. 283–297. [Google Scholar] [CrossRef] [Green Version]
- Nicol, M.R.; Emerson, C.W.; Prince, H.M.; Nelson, J.A.; Fedoriw, Y.; Sykes, C.; Geller, E.J.; Patterson, K.B.; Cohen, M.S.; Kashuba, A.D. Models for predicting effective HIV chemoprevention in women. J. Acquir. Immune Defic. Syndr. 2015, 68, 369–376. [Google Scholar] [CrossRef] [Green Version]
- In Vitro Toxicology Assay Kit MTT Based. Available online: http://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Bulletin/tox1bul.pdf (accessed on 25 January 2021).
- Vistica, D.T.; Skehan, P.; Scudiero, D.; Monks, A.; Pittman, A.; Boyd, M.R. Tetrazolium-Based Assays for Cellular Viability: A Critical Examination of Selected Parameters Affecting Formazan Production. Cancer Res. 1991, 51, 2515–2520. [Google Scholar] [PubMed]
- Maehara, Y.; Anai, H.; Tamada, R.; Sugimachi, K. The ATP Assay Is More Sensitive than the Succinate Dehydrogenase Inhibition Test for Predicting Cell Viability. Eur. J. Cancer Clin. Oncol. 1987, 23, 273–276. [Google Scholar] [CrossRef]
- Berridge, M.V.; Tan, A.S. Characterization of the Cellular Reduction of 3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT): Subcellular Localization, Substrate Dependence, and Involvement of Mitochondrial Electron Transport in MTT Reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Q.; Liu, H.; Li, B.; Zhang, H. High-Throughput Sequencing of 16S rRNA Amplicons Characterizes Gut Microbiota Shift of Juvenile Sea Cucumber Apostichopus japonicus Feeding with Three Antibiotics. J. Oceanol. Limnol. 2019, 37, 1714–1725. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Q.; Zhao, J.; Liu, S.; Zhang, L.; Zhao, Y.; Yang, H.; Sun, L. Quantitative Microbiome Profiling Links Microbial Community Variation to the Intestine Regeneration Rate of the Sea Cucumber Apostichopus Japonicus. Genomics 2020, 112, 5012–5020. [Google Scholar] [CrossRef]
- Limbu, S.M.; Zhou, L.; Sun, S.-X.; Zhang, M.-L.; Du, Z.-Y. Chronic Exposure to Low Environmental Concentrations and Legal Aquaculture Doses of Antibiotics Cause Systemic Adverse Effects in Nile Tilapia and Provoke Differential Human Health Risk. Environ. Int. 2018, 115, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Limbu, S.M.; Shen, M.; Zhai, W.; Qiao, F.; He, A.; Du, Z.-Y.; Zhang, M. Environmental Concentrations of Antibiotics Impair Zebrafish Gut Health. Environ. Pollut. 2018, 235, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.; Zhang, Q.; Zeng, W.; Liu, G.; Shao, H. The Effect of Antibiotic Cocktails on Host Immune Status Is Dynamic and Does Not Always Correspond to Changes in Gut Microbiota. Appl. Microbiol. Biotechnol. 2020, 104, 4995–5009. [Google Scholar] [CrossRef]
- Chuah, L.O.; Effarizah, M.E.; Goni, A.M.; Rusul, G. Antibiotic Application and Emergence of Multiple Antibiotic Resistance (MAR) in Global Catfish Aquaculture. Curr. Environ. Health Rep. 2016, 3, 118–127. [Google Scholar] [CrossRef]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General Principles of Antimicrobial Therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef] [Green Version]
- Quality Control Considerations in Cell Culture. ECACC Laboratory Handbook 4th Edition. Available online: https://www.sigmaaldrich.com/technical-documents/protocols/biology/quality-control-considerations.html (accessed on 25 January 2021).
- Eliopoulos, G.M.; Moellering, R.C. Antibiotic Synergism and Antimicrobial Combinations in Clinical Infections George. Clin. Infect. Dis. 1982, 4, 282–293. [Google Scholar] [CrossRef]
- Acar, J.F. Antibiotic Synergy and Antagonism. Med. Clin. N. Am. 2000, 84, 1391–1406. [Google Scholar] [CrossRef]
- Barza, M.; Cuchural, G. General Principles of Antibiotic Tissue Penetration. J. Antimicrob. Chemother. 1985, 15, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Bergan, T. Pharmacokinetics of Tissue Penetration of Antibiotics. Clin. Infect. Dis. 1981, 3, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Drusano, G.L.; Ryan, P.A.; Standiford, H.C.; Moody, M.R.; Schimpff, S.C. Integration of Selected Pharmacologic and Microbiologic Properties of Three New-Lactam Antibiotics: A Hypothesis for Rational Comparison. Clin. Infect. Dis. 1984, 6, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Schentag, J.J.; Swanson, D.J.; Smith, I.L. Dual Individualization: Antibiotic Dosage Calculation from the Integration of in-Vitro Pharmacodynamics and in-Vivo Pharmacokinetics. J. Antimicrob. Chemother. 1985, 15, 47–57. [Google Scholar] [CrossRef]
- Nix, D.E.; Goodwin, S.D.; Peloquin, C.A.; Rotella, D.L.; Schentag, J.J. Antibiotic Tissue Penetration and Its Relevance: Models of Tissue Penetration and Their Meaning. Antimicrob. Agents Chemother. 1991, 35, 1947–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Derendorf, H. Antimicrobial Tissue Concentrations. Infect. Dis. Clin. N. Am. 2003, 17, 599–613. [Google Scholar] [CrossRef]
- Murray, J.C. Pyoderma gangrenosum with IgA gammopathy. Cutis 1983, 32, 477–480. [Google Scholar]
- Alekshun, M.N.; Levy, S.B. Molecular Mechanisms of Antibacterial Multidrug Resistance. Cell 2007, 128, 1037–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moellering, R.C. Emergence of Enterococcus as a Significant Pathogen. Clin. Infect. Dis. 1992, 14, 1173–1178. [Google Scholar] [CrossRef]
- Washington, J.A.; Wilson, W.R. Erythromycin: A Microbial and Clinical Perspective After 30 Years of Clinical Use (First of Two Parts). Mayo Clin. Proc. 1985, 60, 189–203. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Zhou, Z.; Liu, Y.; Cao, Y.; Meng, K.; Shi, P.; Yao, B.; Ringø, E. Effects of the Antibiotic Growth Promoters Flavomycin and Florfenicol on the Autochthonous Intestinal Microbiota of Hybrid Tilapia (Oreochromis Niloticus ♀ × O. Aureus ♂). Arch. Microbiol. 2010, 192, 985–994. [Google Scholar] [CrossRef]
- He, S.; Zhou, Z.; Liu, Y.; Cao, Y.; Meng, K.; Shi, P.; Yao, B.; Ringø, E. Do Dietary Betaine and the Antibiotic Florfenicol Influence the Intestinal Autochthonous Bacterial Community in Hybrid Tilapia (Oreochromis Niloticus ♀ × O. Aureus ♂)? World J. Microbiol. Biotechnol. 2012, 28, 785–791. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhou, Z.; Meng, K.; Zhao, H.; Yao, B.; Ringø, E.; Yoon, I. Effects of Dietary Antibiotic Growth Promoter and Saccharomyces Cerevisiae Fermentation Product on Production, Intestinal Bacterial Community, and Nonspecific Immunity of Hybrid Tilapia (Oreochromis niloticus Female × Oreochromis aureus Male). J. Anim. Sci. 2011, 89, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, P.; Mardones, P.; Opazo, R.; Espejo, R.; Romero, J. Oxytetracycline Treatment Reduces Bacterial Diversity of Intestinal Microbiota of Atlantic Salmon. J. Aquat. Anim. Health 2008, 20, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Limbu, S.M.; Qiao, F.; Du, Z.-Y.; Zhang, M. Influence of long-term feeding antibiotics on the gut health of zebrafish. Zebrafish 2017, 15, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Pagán-Jiménez, M.; Ruiz-Calderón, J.F.; Dominguez-Bello, M.G.; García-Arrarás, J.E. Characterization of the Intestinal Microbiota of the Sea Cucumber Holothuria Glaberrima. PLoS ONE 2019, 14, e0208011. [Google Scholar] [CrossRef]
- Gao, F.; Li, F.; Tan, J.; Yan, J.; Sun, H. Bacterial Community Composition in the Gut Content and Ambient Sediment of Sea Cucumber Apostichopus japonicus Revealed by 16S RRNA Gene Pyrosequencing. PLoS ONE 2014, 9, e100092. [Google Scholar] [CrossRef]
- Gao, F.; Tan, J.; Sun, H.; Yan, J. Bacterial Diversity of Gut Content in Sea Cucumber (Apostichopus japonicus) and Its Habitat Surface Sediment. J. Ocean Univ. China 2014, 13, 303–310. [Google Scholar] [CrossRef]
- Weigel, B.L. Sea Cucumber Intestinal Regeneration Reveals Deterministic Assembly of the Gut Microbiome. Appl. Environ. Microbiol. 2020, 86, e00489-20. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, X.; Xu, H.; Bao, X.; Liu, X.; Chang, Y.; Ding, J. Characterization of the Bacterial Community in Different Parts of the Gut of Sea Cucumber (Apostichopus japonicus ) and Its Variation during Gut Regeneration. Aquac. Res. 2018, 49, 1987–1996. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Q.; Liu, S.; Huo, D.; Zhao, J.; Zhang, L.; Zhao, Y.; Sun, L.; Yang, H. Genomic and Metagenomic Insights Into the Microbial Community in the Regenerating Intestine of the Sea Cucumber Apostichopus japonicus. Front. Microbiol. 2019, 10, 1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, Y.; Meirelles, P.M.; Mino, S.; Suda, W.; Oshima, K.; Hattori, M.; Thompson, F.L.; Sakai, Y.; Sawabe, T.; Sawabe, T. Individual Apostichopus japonicus Fecal Microbiome Reveals a Link with Polyhydroxybutyrate Producers in Host Growth Gaps. Sci. Rep. 2016, 6, 21631. [Google Scholar] [CrossRef]
- Cox, M.A.; Jackson, J.; Stanton, M.; Rojas-Triana, A.; Bober, L.; Laverty, M.; Yang, X.; Zhu, F.; Liu, J.; Wang, S.; et al. Short-Chain Fatty Acids Act as Antiinflammatory Mediators by Regulating Prostaglandin E2 and Cytokines. WJG 2009, 15, 5549. [Google Scholar] [CrossRef]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. GPR109A Is a G-Protein–Coupled Receptor for the Bacterial Fermentation Product Butyrate and Functions as a Tumor Suppressor in Colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef] [Green Version]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-Sensing Receptors GPR43 and GPR109A Facilitate Dietary Fibre-Induced Gut Homeostasis through Regulation of the Inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borthakur, A.; Priyamvada, S.; Kumar, A.; Natarajan, A.A.; Gill, R.K.; Alrefai, W.A.; Dudeja, P.K. A Novel Nutrient Sensing Mechanism Underlies Substrate-Induced Regulation of Monocarboxylate Transporter-1. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1126–G1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahasrabudhe, N.M.; Beukema, M.; Tian, L.; Troost, B.; Scholte, J.; Bruininx, E.; Bruggeman, G.; van den Berg, M.; Scheurink, A.; Schols, H.A.; et al. Dietary Fiber Pectin Directly Blocks Toll-Like Receptor 2–1 and Prevents Doxorubicin-Induced Ileitis. Front. Immunol. 2018, 9, 383. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The Microbial Metabolite Butyrate Regulates Intestinal Macrophage Function via Histone Deacetylase Inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [Green Version]
- Riggs, M.G.; Whittaker, R.G.; Neumann, J.R.; Ingram, V.M. N-Butyrate Causes Histone Modification in HeLa and Friend Erythroleukaemia Cells. Nature 1977, 268, 462–464. [Google Scholar] [CrossRef]
- Boffa, L.C.; Vidali, G.; Mann, R.S.; Allfrey, V.G. Suppression of Histone Deacetylation in Vivo and in Vitro by Sodium Butyrate. J. Biol. Chem. 1978, 253, 3364–3366. [Google Scholar] [CrossRef]
- Vidali, G.; Boffa, L.C.; Bradbury, E.M.; Allfrey, V.G. Butyrate Suppression of Histone Deacetylation Leads to Accumulation of Multiacetylated Forms of Histones H3 and H4 and Increased DNase I Sensitivity of the Associated DNA Sequences. Proc. Natl. Acad. Sci. USA 1978, 75, 2239–2243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candido, E.; Reeves, R.; Davie, J.R. Sodium Butyrate Inhibits Histone Deacetylation in Cultured Cells. Cell 1978, 14, 105–113. [Google Scholar] [CrossRef]
- Davie, J.R. Inhibition of Histone Deacetylase Activity by Butyrate. J. Nutr. 2003, 133, 2485S–2493S. [Google Scholar] [CrossRef] [PubMed]
- Shakespear, M.R.; Halili, M.A.; Irvine, K.M.; Fairlie, D.P.; Sweet, M.J. Histone Deacetylases as Regulators of Inflammation and Immunity. Trends Immunol. 2011, 32, 335–343. [Google Scholar] [CrossRef]
- Glauben, R.; Batra, A.; Fedke, I.; Zeitz, M.; Lehr, H.A.; Leoni, F.; Mascagni, P.; Fantuzzi, G.; Dinarello, C.A.; Siegmund, B. Histone Hyperacetylation Is Associated with Amelioration of Experimental Colitis in Mice. J. Immunol. 2006, 176, 5015–5022. [Google Scholar] [CrossRef] [Green Version]
- Thomas, H. Microbiota Promote Gut Healing. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 189. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The Gut Microbiota—Masters of Host Development and Physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Pellegatta, T.; Saler, M.; Bonfanti, V.; Nicoletti, G.; Faga, A. Novel Perspectives on the Role of the Human Microbiota in Regenerative Medicine and Surgery. Biomed. Rep. 2016, 5, 519–524. [Google Scholar] [CrossRef] [Green Version]
- Alam, A.; Leoni, G.; Quiros, M.; Wu, H.; Desai, C.; Nishio, H.; Jones, R.M.; Nusrat, A.; Neish, A.S. The Microenvironment of Injured Murine Gut Elicits a Local Pro-Restitutive Microbiota. Nat. Microbiol. 2016, 1, 15021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaborin, A.; Krezalek, M.; Hyoju, S.; Defazio, J.R.; Setia, N.; Belogortseva, N.; Bindokas, V.P.; Guo, Q.; Zaborina, O.; Alverdy, J.C. Critical Role of Microbiota within Cecal Crypts on the Regenerative Capacity of the Intestinal Epithelium Following Surgical Stress. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G112–G122. [Google Scholar] [CrossRef] [Green Version]
- Rawls, J.F.; Samuel, B.S.; Gordon, J.I. Gnotobiotic Zebrafish Reveal Evolutionarily Conserved Responses to the Gut Microbiota. Proc. Natl. Acad. Sci. USA 2004, 101, 4596–4601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchon, N.; Broderick, N.A.; Poidevin, M.; Pradervand, S.; Lemaitre, B. Drosophila Intestinal Response to Bacterial Infection: Activation of Host Defense and Stem Cell Proliferation. Cell Host Microbe 2009, 5, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Arnold, C.P.; Merryman, M.S.; Harris-Arnold, A.; McKinney, S.A.; Seidel, C.W.; Loethen, S.; Proctor, K.N.; Guo, L.; Sánchez Alvarado, A. Pathogenic Shifts in Endogenous Microbiota Impede Tissue Regeneration via Distinct Activation of TAK1/MKK/P38. eLife 2016, 5, e16793. [Google Scholar] [CrossRef] [PubMed]
- Shavandi, A.; Saeedi, P.; Gérard, P.; Jalalvandi, E.; Cannella, D.; Bekhit, A.E. The Role of Microbiota in Tissue Repair and Regeneration. J. Tissue Eng. Regen. Med. 2020, 14, 539–555. [Google Scholar] [CrossRef]

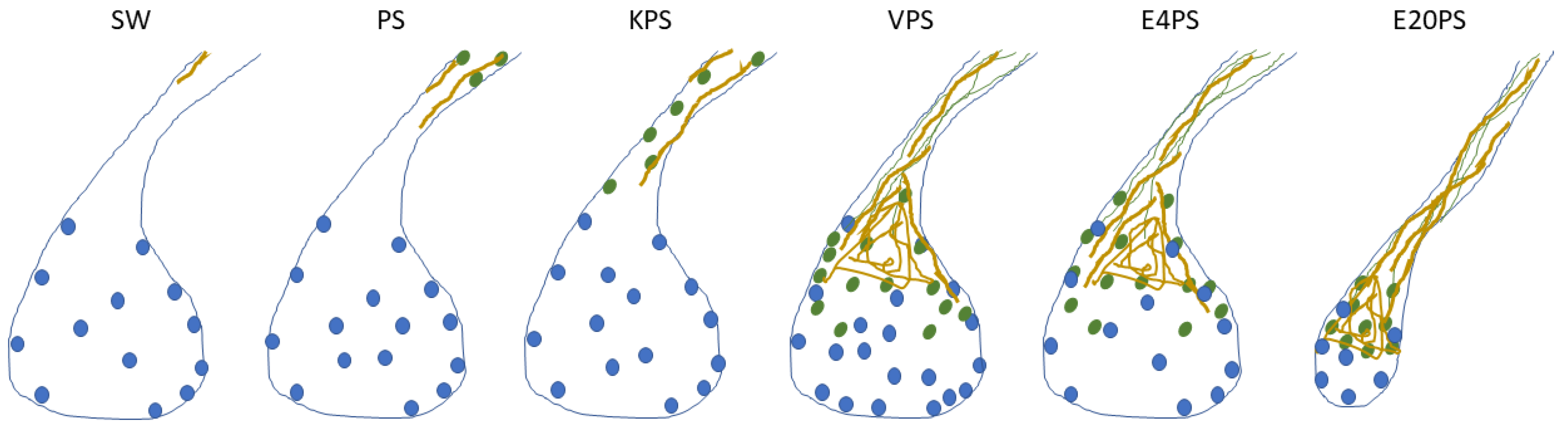
| PS | KPS | V PS | E4PS | E20PS | |
|---|---|---|---|---|---|
| Regeneration-associated processes perturbed | Cell differentiation | Cell differentiation | Cell differentiation ECM remodeling Connective tissue’s cell proliferation | Cell differentiation ECM remodeling | Cell differentiation ECM remodeling Rudiment growth |
| Sea cucumber’s survival rate | No effect | No effect | No effect | No effect | No effect |
| Disassociated cell activity | No effect | No effect | Not measured | No effect | Decreased activity |
| Explant activity | No effect | No effect | No effect | No effect | No effect |
| Bacteria growth | Inhibited | Inhibited | Inhibited | Inhibited | Inhibited |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Díaz, L.M.; Rosario-Meléndez, N.; Rodríguez-Villafañe, A.; Figueroa-Vega, Y.Y.; Pérez-Villafañe, O.A.; Colón-Cruz, A.M.; Rodríguez-Sánchez, P.I.; Cuevas-Cruz, J.M.; Malavez-Cajigas, S.J.; Maldonado-Chaar, S.M.; et al. Antibiotics Modulate Intestinal Regeneration. Biology 2021, 10, 236. https://doi.org/10.3390/biology10030236
Díaz-Díaz LM, Rosario-Meléndez N, Rodríguez-Villafañe A, Figueroa-Vega YY, Pérez-Villafañe OA, Colón-Cruz AM, Rodríguez-Sánchez PI, Cuevas-Cruz JM, Malavez-Cajigas SJ, Maldonado-Chaar SM, et al. Antibiotics Modulate Intestinal Regeneration. Biology. 2021; 10(3):236. https://doi.org/10.3390/biology10030236
Chicago/Turabian StyleDíaz-Díaz, Lymarie M., Natalia Rosario-Meléndez, Andrea Rodríguez-Villafañe, Yariel Y. Figueroa-Vega, Omar A. Pérez-Villafañe, Angela M. Colón-Cruz, Paola I. Rodríguez-Sánchez, Julio M. Cuevas-Cruz, Sonya J. Malavez-Cajigas, Sergio M. Maldonado-Chaar, and et al. 2021. "Antibiotics Modulate Intestinal Regeneration" Biology 10, no. 3: 236. https://doi.org/10.3390/biology10030236
APA StyleDíaz-Díaz, L. M., Rosario-Meléndez, N., Rodríguez-Villafañe, A., Figueroa-Vega, Y. Y., Pérez-Villafañe, O. A., Colón-Cruz, A. M., Rodríguez-Sánchez, P. I., Cuevas-Cruz, J. M., Malavez-Cajigas, S. J., Maldonado-Chaar, S. M., & García-Arrarás, J. E. (2021). Antibiotics Modulate Intestinal Regeneration. Biology, 10(3), 236. https://doi.org/10.3390/biology10030236







