Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress
Abstract
Simple Summary
Abstract
1. Introduction
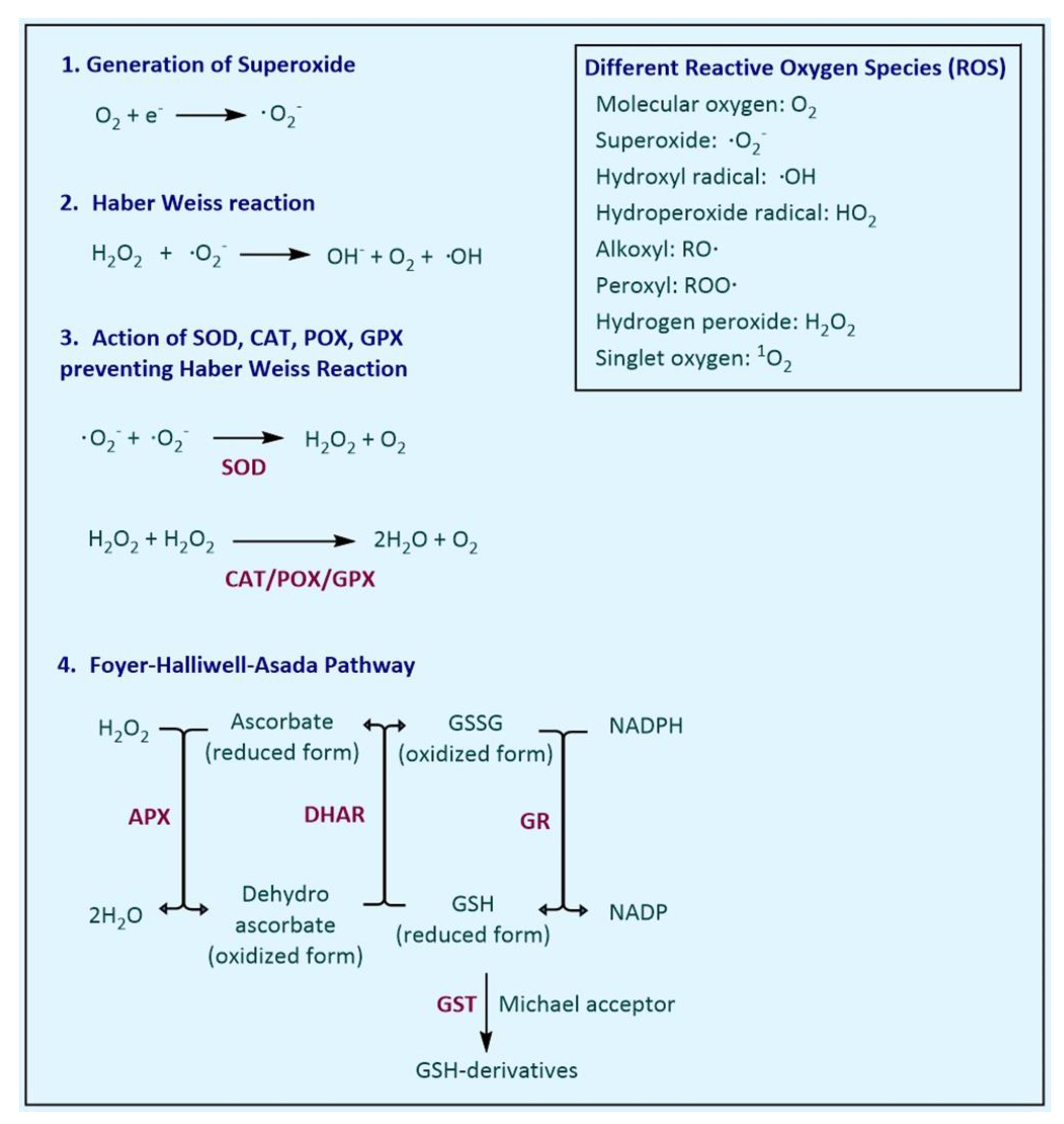
2. Enzymatic Antioxidant Defence Systems in Plants
2.1. Superoxide Dismutase
2.2. Catalase
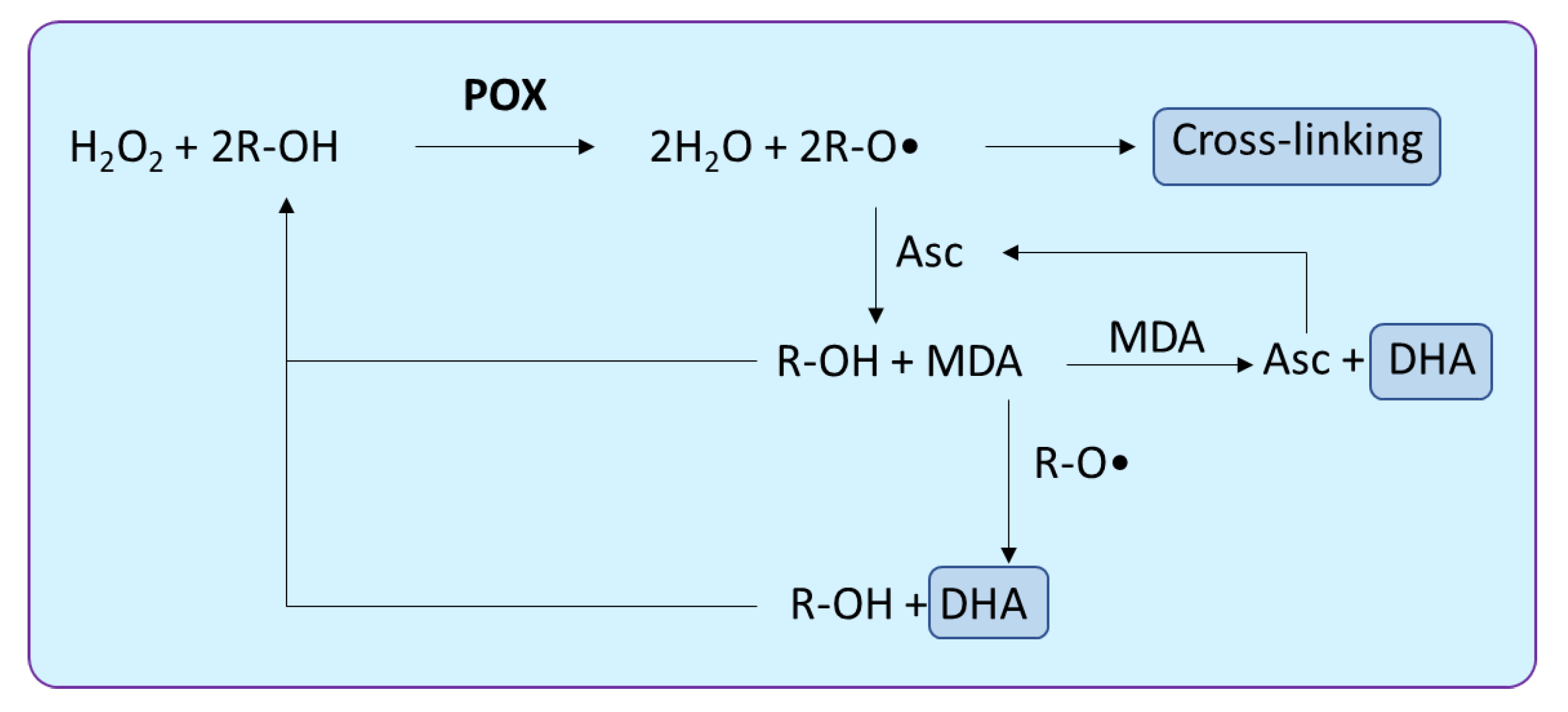
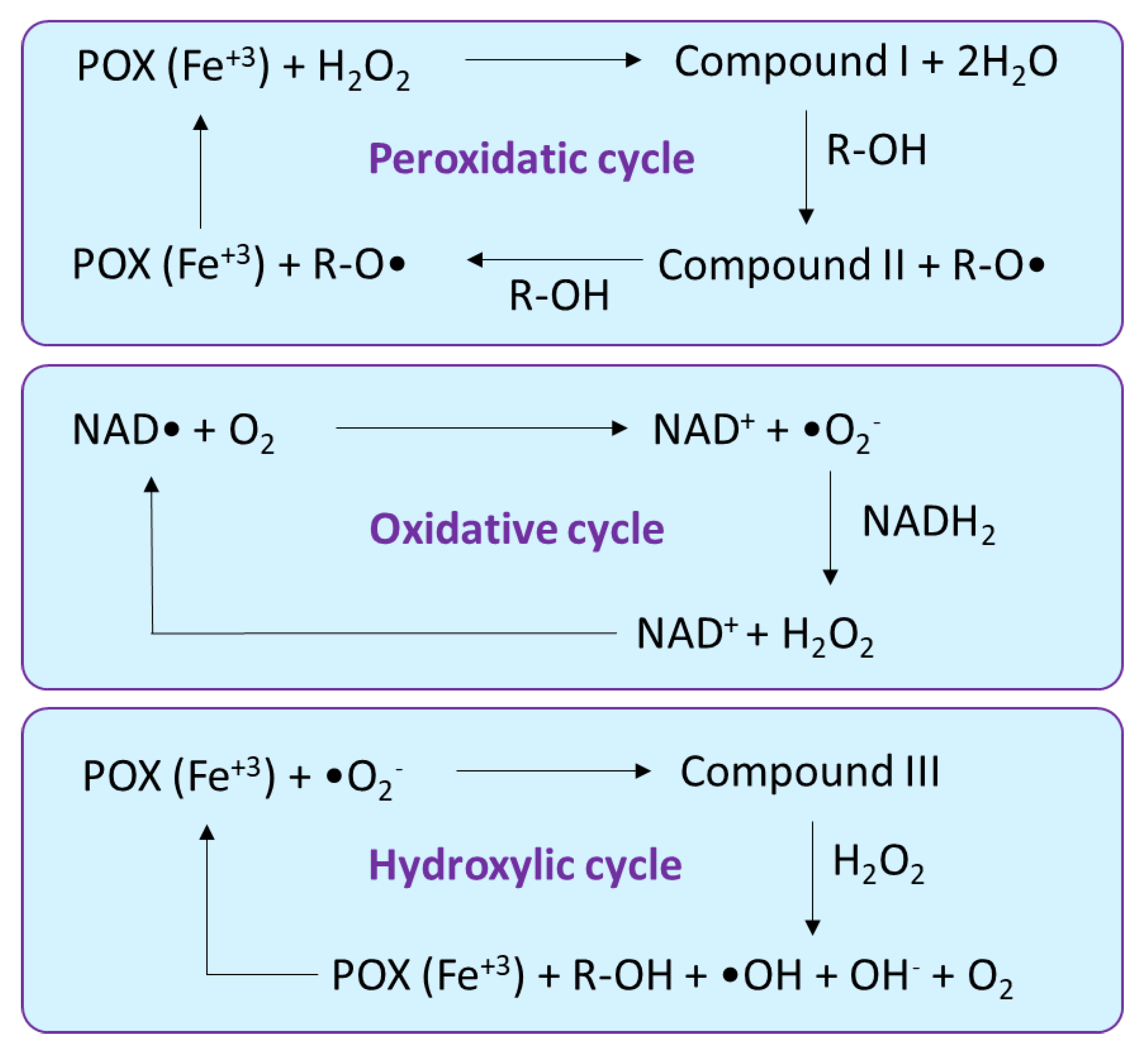
2.3. Peroxidase
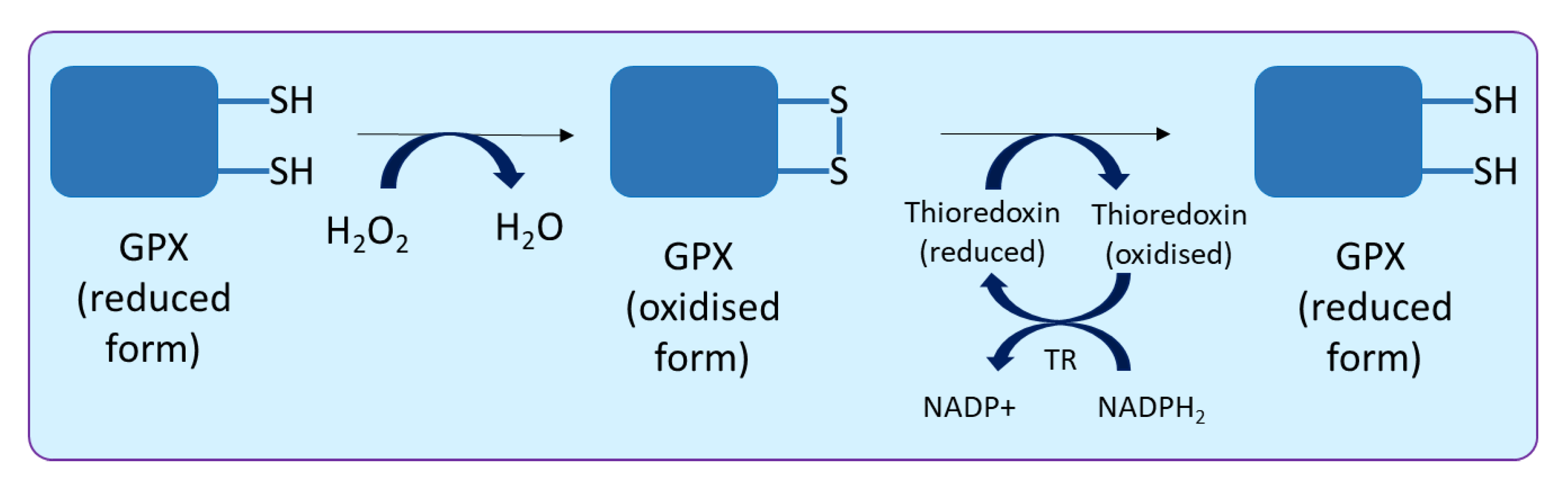
2.4. Glutathione Peroxidase
2.5. Glutathione Reductase
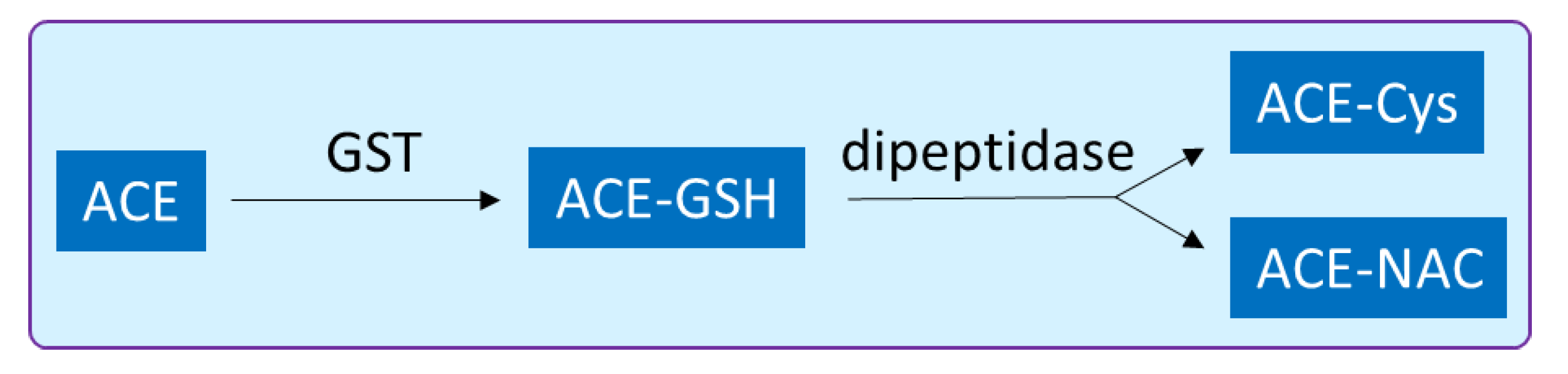
2.6. Glutathione-S-Transferase
2.7. Ascorbate Peroxidase
| APX | ||
| Ascorbate + H2O2 | → | Monodehydroascorbate (MDHA) + 2 H2O |
| Spontaneous oxidation | ||
| MDHA | → | Dehydroascorbate (DHA) |
2.8. Monodehydroascorbate Reductase
| NADH or NADHP | ||
| Monodehydroascorbate (MDHA) | → | Ascorbate |
| MDHAR |
2.9. Dehydroascorbate Reductase
3. Applications of Antioxidant Enzymes in Developing Stress-Tolerant Transgenic Plants
| S.No. | Transgenic Plant(s) | Gene(s)/Source | Stress Condition | Significant Finding(s) | Reference |
|---|---|---|---|---|---|
| 1. | Transgenic S. lycopersicum | FeSOD gene from Arabidopsis | Salt stress | Overexpression of antioxidant enzymes significantly mitigates the harmful effects of salt stress on cytoskeleton structural organisation in roots of the transgenic line cells. | [172] |
| 2. | Transgenic S. tuberosum | Cu-ZnSOD (StSOD1 gene overexpressed under CaMV 35S promoter) | Low temperature | Activity of SOD is 1.38-fold higher compared to non-transgenic lines. Furthermore, the activity of POX and CAT were also enhanced in transgenic line, signifying the fact that increasing the activity of one antioxidant enzyme can influence the activity of other defence enzymes via cross-talk. | [173] |
| 3. | Transgenic Citrus sps | CsPIF8 influencing SOD gene expression | Low temperature | Phytochrome-interacting transcription factor CsPIF8 positively regulate CsSOD expression in citrus, highlighting the cross-talk between phytochrome genes and antioxidant enzymes. In this study, it is found that CsPIF8 directly bound to the E-box (CANNTG) of CsSOD promoter and activated the promoter of CsSOD. | [50] |
| 4. | Transgenic Arabidopsis | CmSOD gene (from winter squash; Cucurbita moschata) and AtSOD gene (from Arabidopsis) under a ubiquitin promoter | Low temperature | Increased resistance to chilling and less oxidative injury in transgenic lines than wild type, indicating that the overexpression of AtSOD and CmSOD led to higher SOD activity in Arabidopsis-enhanced chilling tolerance by eliminating •O2−. Furthermore, the activity of SOD in transgenic lines is influenced by ABA, indicating the role of plant hormone in the cross-talk with enzymes of the antioxidant defence system. | [174] |
| 5. | Transgenic Arabidopsis | Cu-Zn SOD gene (SaCu/Zn SOD), from Sedum alfredii | Oxidative stress due to Cadmium | Cadmium stress induces the production of ROS, leading to oxidative stress. Cd-hyperaccumulator plant S. alfredii is used as a source of SOD gene, resulting in enhanced antioxidative defence capacity in transgenic Arabidopsis plants. The SaCu/Zn SOD is implicated as being responsible for conferring Cd tolerance. | [175] |
| 6. | Transgenic tobacco | Cu/Zn-SOD gene, SiCSD from Saussurea involucrata | Drought, cold and oxidative stress | Higher activities of SODs, CAT and APX are reported in transgenic lines, and SOD is found as a positive regulator in drought and cold stress by reducing oxidant injury. | [176] |
| 7. | Transgenic C. grandis | The basic helix-loop-helix (bHLH) family of transcription factors (PtrbHLH) from Poncirus trifoliata | Low temperature | Transgenic plant was found to exhibit lower electrolyte leakage and malondialdehyde content after chilling stress, lower ROS levels and elevated activity of antioxidant enzymes, including CAT, POX and SOD. Interestingly, PtrbHLH was found to bind to the promoter and activate the PtrCAT gene, thereby implicated as regulating the CAT gene activity. | [177] |
| 8. | Manihot esculenta | SOD (MeCu/ZnSOD) and catalase (MeCAT1) | Biotic stress (Mite Tetranychus cinnabarinus) | The transgenic approach led to mite-resistant traits, as survival, reproduction and development of T. cinnabarinus feeding on transgenic cassava is significantly inhibited. Furthermore, the activities of SOD and CAT in transgenic cassava plants damaged by T. cinnabarinus significantly increased. This study highlights the role of antioxidant enzymes in developing pest resistant crops. | [178] |
| 9. | Transgenic Ipomoea batatas | Peroxidase gene swpa4 in I. batatas | Salt stress | Overexpressing the swpa4 gene under CaMV 35S promoter led to 3- to 13-fold higher expression in transgenic sweet potato. Transgenic plants also showed increased tolerance to salinity conditions, with 13–26% less damage than control plants. Furthermore, photosynthetic capacity and total chlorophyll contents were less severely impacted in transgenic plants. | [179] |
| 10. | Transgenic Arabidopsis | Glutathione peroxidase-like 5 gene (AtGPXL5) from Arabidopsis | Salt stress | Constitutive overexpression of AtGPXL5 led to an increase in gene expression by 17–24 times in 6-week-old plants. It results in an increase in GSH pool and more negative redox potential than wild type and increased salt tolerance. | [91] |
| 11. | Transgenic Arabidopsis | AtGR1 encoding glutathione reductase (GR) from Arabidopsis | Aluminium toxicity | The overexpression of AtGR1 led to a higher GSH pool and improved ratio of GSH/GSSG, and increased aluminium tolerance, with better root growth in comparison to the wild type under aluminium stress. Increased GSH levels were found to increase the capacity of RCS detoxification, which indicates that GR overexpression contributes to the mitigating of not only ROS, but also RCS. | [180] |
| 12. | Transgenic O. sativa | OsGSTU5 (a tau class GST in O. sativa) | Biotic stress | Overexpression of OsGSTU5 provided tolerance against sheath blight disease, caused by Rhizoctonia solani. | [181] |
| 13. | Transgenic Arabidopsis | Glutathione S-transferase from Thermosynechococcus elongatus BP-1 (TeGST) | Thiocyanate (SCN−) stress | Overexpression of TeGST in transgenic plant increased the tolerance to thiocyanate (SCN-) up to 5 mmol L−1. This approach was found to be potentially effective to enhance the phytoremediation of environmental thiocyanates. | [182] |
| 14. | Transgenic Arabidopsis | Ascorbate peroxidase (AgAPX1) from Apium graveolens | Drought tolerance | Overexpression of the AgAPX1 gene enhanced ascorbate content, antioxidant capacity and drought resistance. Furthermore, increased antioxidant capacity does not affect the growth parameters of the plant much, as a comparatively smaller decrease in the net photosynthetic rate is observed, and a high survival rate of transgenic Arabidopsis lines after drought is reported. | [43] |
| 15. | Transgenic Arabidopsis | Ascorbate peroxidase gene (DaAPX) from Dioscorea alata | Flood/Chilling stress | This study reports the effect of different types of stress on the expression of DaAPX. Yam variety Minghuai 1 (MH1), when exposed to a flood situation, showed an increase in the expression of DaAPX; however, chilling stress did not influence the expression profile of DaAPX, thereby making this variety sensitive to chilling stress. However, overexpression of DaAPX in Arabidopsis led to increased tolerance towards several abiotic stress, including flooding and chilling. | [152] |
| 16. | Transgenic Brassica juncea | Ascorbate peroxidase gene (Apx1) from Arabidopsis | Salt stress | Overexpression of cytosolic AtApx1 gene increased salinity stress tolerance in B. juncea. APX, along with higher activity of other enzymes such as GPX, CAT and POX, maintains the ROS homeostasis and provides tolerance to the cell, greater proline accumulation, increased chlorophyll stability index and lower chlorophyll a/b ratio. | [150] |
| 17. | Transgenic Nicotiana tabacum | Monodehydroascorbate reductase from S. lycopersicum (SlMDHAR) | Salt stress | Overexpression of SlMDHAR in transgenic tobacco is found to increase salt stress tolerance and NO accumulation and the S-nitrosyalted SlMDHAR levels were found to be higher in transgenic tobacco. Results suggested that SlMDHAR confers salt stress tolerance by probably involving the S-nitrosylation (post-translational modification of cysteine thiol by nitric oxide group) of MDHAR. | [183] |
| 18. | Transgenic Arabidopsis | Monodehydroascorbate reductase (BvM14-MDHAR) from B. vulgaris | Salt stress | The MDHAR gene is constitutively expressed in Arabidopsis, resulting in an enhanced salt stress tolerance phenotype, with higher AsA/DHA levels than wild-type. In addition, the overexpression seedlings showed higher activities of MDHAR and DHAR and decreased cell membrane damage. | [184] |
| 19. | Transgenic Arabidopsis | DHAR (AcDHAR1 and AcDHAR2) from Actinidia chinensis (kiwi fruit) | Salt stress | Transgenic overexpression of these two genes (separately) in Arabidopsis plants was found to significantly enhance the ascorbic acid concentration and enhance the tolerance to salinity. | [185] |
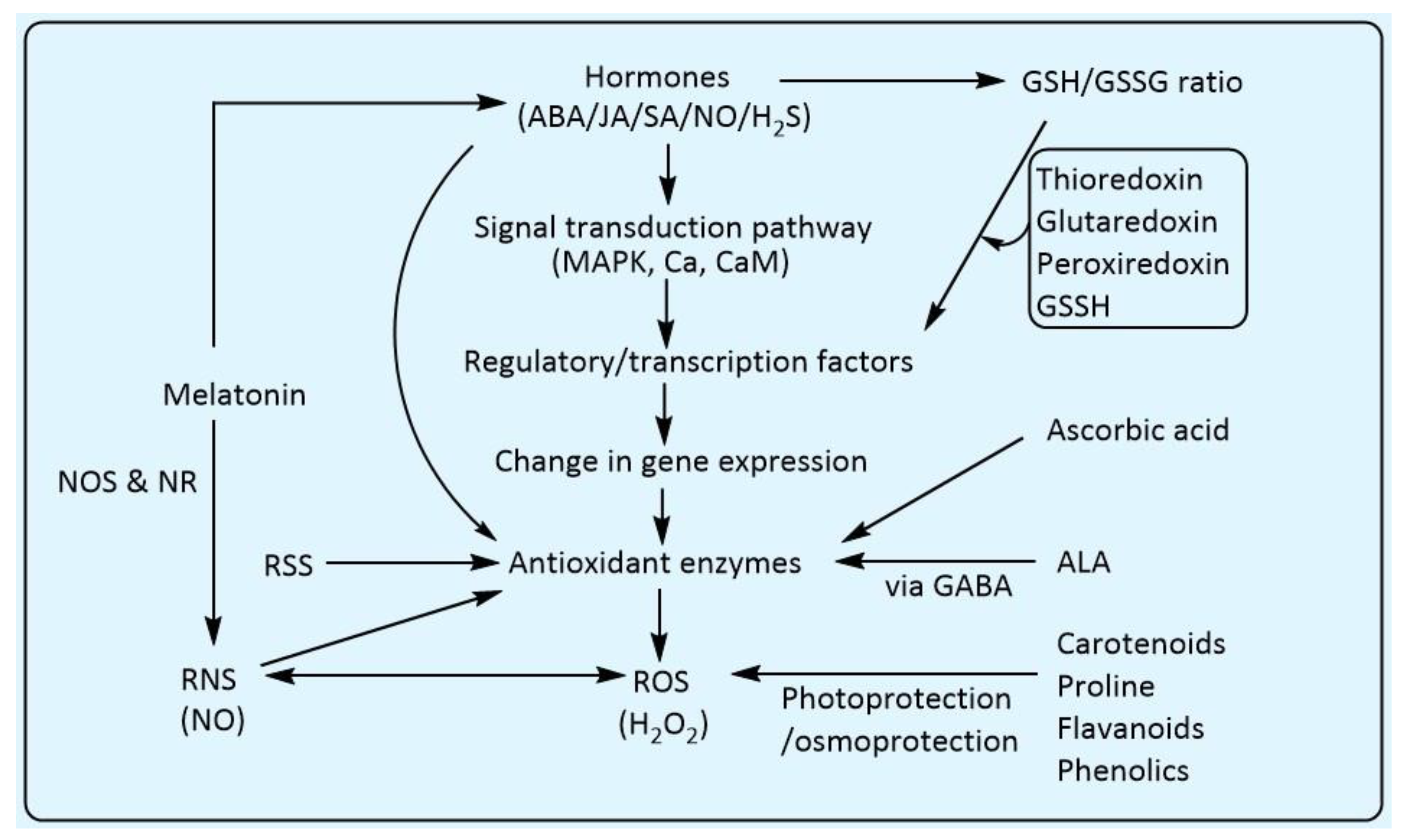
4. Significance of the Cross-Talk of the Antioxidants in Plant Biology
5. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Herbette, S.; Labrouhe, D.T.d.; Drevet, J.R.; Roeckel-Drevet, P. Transgenic tomatoes showing higher glutathione peroxydase antioxidant activity are more resistant to an abiotic stress but more susceptible to biotic stresses. Plant Sci. 2011, 180, 548–553. [Google Scholar] [CrossRef]
- Rajput, V.D.; Minkina, T.; Yaning, C.; Sushkova, S.; Chapligin, V.A.; Mandzhieva, S. A review on salinity adaptation mechanism and characteristics of Populus euphratica, a boon for arid ecosystems. Acta Ecol. Sin. 2016, 36, 497–503. [Google Scholar] [CrossRef]
- Miller, G.A.D.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R.O.N. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and Salt Tolerance in Plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Shibahara, T.; Inanaga, S.; Tanaka, K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, D.M.; Umbach, A.L.; Subbaiah, C.C.; Siedow, J.N. Mitochondrial Reactive Oxygen Species. Contribution to Oxidative Stress and Interorganellar Signaling. Plant Physiol. 2006, 141, 357. [Google Scholar] [CrossRef]
- Noctor, G.; De Paepe, R.; Foyer, C.H. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007, 12, 125–134. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.; Dubey, R.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Ye, S.; Jiang, L.; Liu, S. Genome-wide identification of glutathione peroxidase (GPX) gene family and their response to abiotic stress in cucumber. 3 Biotech 2018, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Hu, L.; Wu, H.; Jiang, L.; Liu, S. Genome-wide identification and transcriptional expression analysis of cucumber superoxide dismutase (SOD) family in response to various abiotic stresses. Int. J. Genom. 2017, 2017, 7243973. [Google Scholar] [CrossRef]
- Rajput, V.D.; Chen, Y.; Ayup, M. Effects of high salinity on physiological and anatomical indices in the early stages of Populus euphratica growth. Russ. J. Plant Physiol. 2015, 62, 229–236. [Google Scholar] [CrossRef]
- Remacle, J.; Michiels, C.; Raes, M. The importance of antioxidant enzymes in cellular aging and degeneration. Free Radic. Aging 1992, 99–108. [Google Scholar] [CrossRef]
- Liu, N.; Lin, Z.; Guan, L.; Gaughan, G.; Lin, G. Antioxidant enzymes regulate reactive oxygen species during pod elongation in Pisum sativum and Brassica chinensis. PLoS ONE 2014, 9, e87588. [Google Scholar] [CrossRef]
- Prochazkova, D.; Wilhelmova, N. Leaf senescence and activities of the antioxidant enzymes. Biol. Plant. 2007, 51, 401–406. [Google Scholar] [CrossRef]
- De Pinto, M.C.; De Gara, L. Changes in the ascorbate metabolism of apoplastic and symplastic spaces are associated with cell differentiation. J. Exp. Bot. 2004, 55, 2559–2569. [Google Scholar] [CrossRef]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef]
- Abdel Latef, A.A.H. Ameliorative effect of calcium chloride on growth, antioxidant enzymes, protein patterns and some metabolic activities of canola (Brassica napus L.) under seawater stress. J. Plant Nutr. 2011, 34, 1303–1320. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Zhao, R.; Yu, W.; Li, R.; Li, Y.; Sheng, J.; Shen, L. Knockout of SlMAPK3 Reduced Disease Resistance to Botrytis cinerea in Tomato Plants. J. Agric. Food Chem. 2018, 66, 8949–8956. [Google Scholar] [CrossRef] [PubMed]
- Sairam, R.K.; Vasanthan, B.; Arora, A. Calcium regulates Gladiolus flower senescence by influencing antioxidative enzymes activity. Acta Physiol. Plant. 2011, 33, 1897–1904. [Google Scholar] [CrossRef]
- Schröder, P.; Daubner, D.; Maier, H.; Neustifter, J.; Debus, R. Phytoremediation of organic xenobiotics–Glutathione dependent detoxification in Phragmites plants from European treatment sites. Bioresour. Technol. 2008, 99, 7183–7191. [Google Scholar] [CrossRef] [PubMed]
- Bartha, B.; Huber, C.; Harpaintner, R.; Schröder, P. Effects of acetaminophen in Brassica juncea L. Czern.: Investigation of uptake, translocation, detoxification, and the induced defense pathways. Environ. Sci. Pollut. Res. 2010, 17, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Sofy, A.R.; Dawoud, R.A.; Sofy, M.R.; Mohamed, H.I.; Hmed, A.A.; El-Dougdoug, N.K. Improving regulation of enzymatic and non-enzymatic antioxidants and stress-related gene stimulation in Cucumber mosaic cucumovirus-infected cucumber plants treated with glycine betaine, chitosan and combination. Molecules 2020, 25, 2341. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Lu, W.; Ghergurovich, J.M.; Guo, L.; Blair, I.A.; Rabinowitz, J.D.; Yang, X. Upregulation of antioxidant capacity and nucleotide precursor availability suffices for oncogenic transformation. Cell Metab. 2021, 33, 94–109.e108. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.; Mayo, J.C.; Sainz, R.M.; Antolín, I.; Herrera, F.; Martín, V.; Reiter, R.J. Regulation of antioxidant enzymes: A significant role for melatonin. J. Pineal Res. 2004, 36, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Srivastava, S.; Tripathi, R.; Govindarajan, R.; Kuriakose, S.; Prasad, M. Phytochelatin synthesis and response of antioxidants during cadmium stress in Bacopa monnieri L◊. Plant Physiol. Biochem. 2006, 44, 25–37. [Google Scholar] [CrossRef]
- Mullineaux, P.M.; Rausch, T. Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression. Photosynth. Res. 2005, 86, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Dumont, S.; Rivoal, J. Consequences of Oxidative Stress on Plant Glycolytic and Respiratory Metabolism. Front. Plant Sci. 2019, 10, 166. [Google Scholar] [CrossRef]
- Alscher, R.G.; Donahue, J.L.; Cramer, C.L. Reactive oxygen species and antioxidants: Relationships in green cells. Physiol. Plant. 1997, 100, 224–233. [Google Scholar] [CrossRef]
- Pan, S.-M.; Yau, Y.-Y. Characterization of superoxide dismutase in Arabidopsis. Taiwania 1992, 37, 58–66. [Google Scholar]
- Fink, R.C.; Scandalios, J.G. Molecular Evolution and Structure–Function Relationships of the Superoxide Dismutase Gene Families in Angiosperms and Their Relationship to Other Eukaryotic and Prokaryotic Superoxide Dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef]
- Wuerges, J.; Lee, J.-W.; Yim, Y.-I.; Yim, H.-S.; Kang, S.-O.; Carugo, K.D. Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proc. Natl. Acad. Sci. USA 2004, 101, 8569–8574. [Google Scholar] [CrossRef]
- Stephenie, S.; Chang, Y.P.; Gnanasekaran, A.; Esa, N.M.; Gnanaraj, C. An insight on superoxide dismutase (SOD) from plants for mammalian health enhancement. J. Funct. Foods 2020, 68, 103917. [Google Scholar] [CrossRef]
- Perry, J.J.P.; Shin, D.S.; Getzoff, E.D.; Tainer, J.A. The structural biochemistry of the superoxide dismutases. Biochim. Et Biophys. Acta -Proteins Proteom. 2010, 1804, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Dehury, B.; Sarma, K.; Sarmah, R.; Sahu, J.; Sahoo, S.; Sahu, M.; Sen, P.; Modi, M.K.; Sharma, G.D.; Choudhury, M.D.; et al. In silico analyses of superoxide dismutases (SODs) of rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 2013, 22, 150–156. [Google Scholar] [CrossRef]
- Leitch, J.M.; Li, C.X.; Baron, J.A.; Matthews, L.M.; Cao, X.; Hart, P.J.; Culotta, V.C. Post-translational modification of Cu/Zn superoxide dismutase under anaerobic conditions. Biochemistry 2012, 51, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.M.; López-Huertas, E.; Corpas, F.J.; Sandalio, L.M.; Gómez, M.; Del Río, L.A. Peroxisomal manganese superoxide dismutase: Purification and properties of the isozyme from pea leaves. Physiol. Plant. 1998, 104, 720–726. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Pilon, M.; Ravet, K.; Tapken, W. The biogenesis and physiological function of chloroplast superoxide dismutases. Biochim. Et Biophys. Acta -Bioenerg. 2011, 1807, 989–998. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Gill, R.; Yadav, S.; Hasanuzzaman, M.; Fujita, M.; Mishra, P.; Sabat, S.C.; Tuteja, N. Superoxide dismutase--mentor of abiotic stress tolerance in crop plants. Environ. Sci. Pollut. Res. Int. 2015, 22, 10375–10394. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhan, J.; Li, J.; Lu, X.; Liu, J.; Wang, Y.; Zhao, Q.; Ye, G. Genome-wide Association Study (GWAS) for Mesocotyl Elongation in Rice (Oryza sativa L.) under Multiple Culture Conditions. Genes 2019, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-X.; Feng, K.; Duan, A.-Q.; Li, H.; Yang, Q.-Q.; Xu, Z.-S.; Xiong, A.-S. Isolation, purification and characterization of an ascorbate peroxidase from celery and overexpression of the AgAPX1 gene enhanced ascorbate content and drought tolerance in Arabidopsis. Bmc Plant Biol. 2019, 19, 488. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, R.; Cui, D.; Cao, Q.; Shan, Z.; Jiao, Z. Physio-biochemical and molecular mechanism underlying the enhanced heavy metal tolerance in highland barley seedlings pre-treated with low-dose gamma irradiation. Sci. Rep. 2017, 7, 14233. [Google Scholar] [CrossRef]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Genome-wide analysis of superoxide dismutase Gene Family in Gossypium raimondii and G. arboreum. Plant Gene 2016, 6. [Google Scholar] [CrossRef]
- Zhou, C.; Zhu, C.; Fu, H.; Li, X.; Chen, L.; Lin, Y.; Lai, Z.; Guo, Y. Genome-wide investigation of superoxide dismutase (SOD) gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant (Camellia sinensis). PLoS ONE 2019, 14, e0223609. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Li, Z.; Tian, H.; Du, X.; Liu, Z.; Huang, H.; Wang, P.; Ye, Z.; Zhang, X.; Tu, L. Cytosolic Ascorbate Peroxidases Plays a Critical Role in Photosynthesis by Modulating Reactive Oxygen Species Level in Stomatal Guard Cell. Front. Plant Sci. 2020, 11, 446. [Google Scholar] [CrossRef]
- Pilati, S.; Perazzolli, M.; Malossini, A.; Cestaro, A.; Demattè, L.; Fontana, P.; Dal Ri, A.; Viola, R.; Velasco, R.; Moser, C. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at vèraison. BMC Genom. 2007, 8, 428. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.; Zhang, Z.; Ma, M.; Wang, R.; Qian, M.; Zhang, S. Genome-wide identification and comparative analysis of the superoxide dismutase gene family in pear and their functions during fruit ripening. Postharvest Biol. Technol. 2018, 143, 68–77. [Google Scholar] [CrossRef]
- He, Z.; Zhao, T.; Yin, Z.; Liu, J.; Cheng, Y.; Xu, J. The phytochrome-interacting transcription factor CsPIF8 contributes to cold tolerance in citrus by regulating superoxide dismutase expression. Plant Sci. 2020, 298, 110584. [Google Scholar] [CrossRef]
- Fei, X.; Li, J.; Kong, L.; Hu, H.; Tian, J.; Liu, Y.; Wei, A. miRNAs and their target genes regulate the antioxidant system of Zanthoxylum bungeanum under drought stress. Plant Physiol. Biochem. 2020, 150, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Zlatev, Z.S.; Lidon, F.C.; Ramalho, J.C.; Yordanov, I.T. Comparison of resistance to drought of three bean cultivars. Biol. Plant. 2006, 50, 389–394. [Google Scholar] [CrossRef]
- Sharma, P.; Dubey, R.S. Drought Induces Oxidative Stress and Enhances the Activities of Antioxidant Enzymes in Growing Rice Seedlings. Plant Growth Regul. 2005, 46, 209–221. [Google Scholar] [CrossRef]
- Sharma, P.; Shanker Dubey, R. Modulation of nitrate reductase activity in rice seedlings under aluminium toxicity and water stress: Role of osmolytes as enzyme protectant. J. Plant Physiol. 2005, 162, 854–864. [Google Scholar] [CrossRef]
- Chang-Quan, W.; Rui-Chang, L. Enhancement of superoxide dismutase activity in the leaves of white clover (Trifolium repens L.) in response to polyethylene glycol-induced water stress. Acta Physiol. Plant. 2008, 30, 841. [Google Scholar] [CrossRef]
- Kukreja, S.; Nandwal, A.S.; Kumar, N.; Sharma, S.K.; Sharma, S.K.; Unvi, V.; Sharma, P.K. Plant water status, H2O2 scavenging enzymes, ethylene evolution and membrane integrity of Cicer arietinum roots as affected by salinity. Biol. Plant. 2005, 49, 305–308. [Google Scholar] [CrossRef]
- Gapińska, M.; Skłodowska, M.; Gabara, B. Effect of short- and long-term salinity on the activities of antioxidative enzymes and lipid peroxidation in tomato roots. Acta Physiol. Plant. 2008, 30, 11–18. [Google Scholar] [CrossRef]
- Eyidogan, F.; Oz, T. Effect of salinity on antioxidant responses of chickpea seedlings. Acta Physiol. Plant. 2007, 29, 485–493. [Google Scholar] [CrossRef]
- Wang, Y.; Ying, Y.; Chen, J.; Wang, X. Transgenic Arabidopsis overexpressing Mn-SOD enhanced salt-tolerance. Plant Sci. 2004, 167, 671–677. [Google Scholar] [CrossRef]
- Agrawal, S.; Singh, S.; Agrawal, M. Chapter 3 Ultraviolet-B Induced Changes in Gene Expression and Antioxidants in Plants. Adv. Bot. Res. 2009, 52, 47–86. [Google Scholar]
- Oshino, N.; Jamieson, D.; Sugano, T.; Chance, B. Optical measurement of the catalase-hydrogen peroxide intermediate (Compound I) in the liver of anaesthetized rats and its implication to hydrogen peroxide production in situ. Biochem. J. 1975, 146, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef]
- Del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.A.; Singh, H.P.; Khan, M.I.; Masood, A.; Per, T.S.; Negi, A.; Batish, D.R.; Khan, N.A.; Duarte, A.C.; Pereira, E.; et al. Too much is bad--an appraisal of phytotoxicity of elevated plant-beneficial heavy metal ions. Environ. Sci. Pollut. Res. Int. 2015, 22, 3361–3382. [Google Scholar] [CrossRef]
- Palma, J.M.; Mateos, R.M.; López-Jaramillo, J.; Rodríguez-Ruiz, M.; González-Gordo, S.; Lechuga-Sancho, A.M.; Corpas, F.J. Plant catalases as NO and H2S targets. Redox Biol. 2020, 34, 101525. [Google Scholar] [CrossRef]
- Anjum, N.A.; Sharma, P.; Gill, S.S.; Hasanuzzaman, M.; Khan, E.A.; Kachhap, K.; Mohamed, A.A.; Thangavel, P.; Devi, G.D.; Vasudhevan, P.; et al. Catalase and ascorbate peroxidase—representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. Res. 2016, 23, 19002–19029. [Google Scholar] [CrossRef]
- Niewiadomska, E.; Miszalski, Z. Partial characterization and expression of leaf catalase in the CAM-inducible halophyte Mesembryanthemum crystallinum L. Plant Physiol. Biochem. 2008, 46, 421–427. [Google Scholar] [CrossRef]
- Rodríguez-Ruiz, M.; González-Gordo, S.; Cañas, A.; Campos, M.J.; Paradela, A.; Corpas, F.J.; Palma, J.M. Sweet Pepper (Capsicum annuum L.) Fruits Contain an Atypical Peroxisomal Catalase That is Modulated by Reactive Oxygen and Nitrogen Species. Antioxidants 2019, 8, 374. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, Y.; Chen, D.; Liu, D.; Hu, M.; Dong, J.; Zhang, X.; Song, L.; Shen, F. The catalase gene family in cotton: Genome-wide characterization and bioinformatics analysis. Cells 2019, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.-Q.; Fu, J.-Y.; Rong, L.-J.; Zhang, P.-Y.; Guo, C.-J.; Xiao, K. TaMIR1119, a miRNA family member of wheat (Triticum aestivum), is essential in the regulation of plant drought tolerance. J. Integr. Agric. 2018, 17, 2369–2378. [Google Scholar] [CrossRef]
- Simova-Stoilova, L.; Vaseva, I.; Grigorova, B.; Demirevska, K.; Feller, U. Proteolytic activity and cysteine protease expression in wheat leaves under severe soil drought and recovery. Plant Physiol. Biochem. 2010, 48, 200–206. [Google Scholar] [CrossRef]
- Zafar, S.A.; Hameed, A.; Ashraf, M.; Khan, A.S.; Qamar, Z.-U.; Li, X.; Siddique, K.H.M. Agronomic, physiological and molecular characterisation of rice mutants revealed the key role of reactive oxygen species and catalase in high-temperature stress tolerance. Funct. Plant Biol. 2020, 47, 440–453. [Google Scholar] [CrossRef]
- Abuelsoud, W.; Cortleven, A.; Schmülling, T. Photoperiod stress alters the cellular redox status and is associated with an increased peroxidase and decreased catalase activity. bioRxiv 2020, bioRxiv:2020.2003.2005.978270. [Google Scholar] [CrossRef]
- Dai, L.; Li, J.; Harmens, H.; Zheng, X.; Zhang, C. Melatonin enhances drought resistance by regulating leaf stomatal behaviour, root growth and catalase activity in two contrasting rapeseed (Brassica napus L.) genotypes. Plant Physiol. Biochem. 2020, 149, 86–95. [Google Scholar] [CrossRef]
- Hiraga, S.; Sasaki, K.; Ito, H.; Ohashi, Y.; Matsui, H. A Large Family of Class III Plant Peroxidases. Plant Cell Physiol. 2001, 42, 462–468. [Google Scholar] [CrossRef]
- Mujer, C.V.; Mendoza, E.M.T.; Ramirez, D.A. Coconut peroxidase isoenzymes: Isolation, partial purification and physicochemical properties. Phytochemistry 1983, 22, 1335–1340. [Google Scholar] [CrossRef]
- Regalado, C.; Arvizu, O.P.; GarcÍA-Almendarez, B.E.; Whitaker, J.R. Purification and properties of two acid peroxidases from brussels sprouts (Brassica oleraceae L.). J. Food Biochem. 1999, 23, 435–450. [Google Scholar] [CrossRef]
- Pandey, V.; Dwivedi, U. Purification and characterization of peroxidase from Leucaena leucocephala, a tree legume. J. Mol. Catal. B Enzym. 2011, 68, 168–173. [Google Scholar] [CrossRef]
- Suzuki, T.; Honda, Y.; Mukasa, Y.; Kim, S.-j. Characterization of peroxidase in buckwheat seed. Phytochemistry 2006, 67, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Lüthje, S. Properties of Guaiacol Peroxidase Activities Isolated from Corn Root Plasma Membranes. Plant Physiol. 2003, 132, 1489–1498. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative damage, lipid peroxidation and antioxidant protection in chloroplasts. Chem. Phys. Lipids 1987, 44, 327–340. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Glušac, J.; Isaschar-Ovdat, S.; Fishman, A.; Kukavica, B. Partial characterization of bean and maize root peroxidases and their ability to crosslink potato protein. Arch. Biol. Sci. 2019, 71, 293–303. [Google Scholar] [CrossRef]
- Coelho, D.G.; Marinato, C.S.; de Matos, L.P.; de Andrade, H.M.; da Silva, V.M.; Santos-Neves, P.H.; Araújo, S.C.; Oliveira, J.A. Is arsenite more toxic than arsenate in plants? Ecotoxicology 2020, 29, 196–202. [Google Scholar] [CrossRef]
- Lukić, N.; Kukavica, B.; Davidović-Plavšić, B.; Hasanagić, D.; Walter, J. Plant stress memory is linked to high levels of anti-oxidative enzymes over several weeks. Environ. Exp. Bot. 2020, 178, 104166. [Google Scholar] [CrossRef]
- Xiao, H.; Wang, C.; Khan, N.; Chen, M.; Fu, W.; Guan, L.; Leng, X. Genome-wide identification of the class III POD gene family and their expression profiling in grapevine (Vitis vinifera L). BMC Genom. 2020, 21, 444. [Google Scholar] [CrossRef]
- Jovanović, S.V.; Kukavica, B.; Vidović, M.; Morina, F.; Menckhoff, L. Class III peroxidases: Functions, localization and redox regulation of isoenzymes. In Antioxidants and Antioxidant Enzymes in Higher Plants; Springer: Cham, Switzerland, 2018; pp. 269–300. [Google Scholar]
- Passaia, G.; Margis-Pinheiro, M. Glutathione peroxidases as redox sensor proteins in plant cells. Plant Sci. 2015, 234, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Sabeh, F.; Wright, T.; Norton, S.J. Purification and Characterization of a Glutathione Peroxidase from the Aloe vera Plant. Enzym. Protein 1993, 47, 92–98. [Google Scholar] [CrossRef]
- Dixon, D.P.; Cummins, I.; Cole, D.J.; Edwards, R. Glutathione-mediated detoxification systems in plants. Curr. Opin. Plant Biol. 1998, 1, 258–266. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Bela, K.; Horváth, E.; Rigó, G.; Gallé, Á.; Szabados, L.; Fehér, A.; Csiszár, J. Overexpression of the Arabidopsis glutathione peroxidase-like 5 gene (AtGPXL5) resulted in altered plant development and redox status. Environ. Exp. Bot. 2019, 167, 103849. [Google Scholar] [CrossRef]
- Wang, Z.; Tang, S.-F.; Hou, X. Glutathione peroxidase 6 from Arabidopsis thaliana as potential biomarker for plants exposure assessment to di-(2-ethylhexyl) phthalate. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117955. [Google Scholar] [CrossRef]
- Hassan, N.M.; Nemat Alla, M.M. Kinetics of inhibition of isoproturon to glutathione-associated enzymes in wheat. Physiol. Mol. Biol. Plants 2020, 26, 1505–1518. [Google Scholar] [CrossRef]
- Islam, T.; Manna, M.; Kaul, T.; Pandey, S.; Reddy, C.S.; Reddy, M. Genome-wide dissection of Arabidopsis and rice for the identification and expression analysis of glutathione peroxidases reveals their stress-specific and overlapping response patterns. Plant Mol. Biol. Rep. 2015, 33, 1413–1427. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Jang, M.-G.; Noh, H.-Y.; Lee, H.-J.; Sukweenadhi, J.; Kim, J.-H.; Kim, S.-Y.; Kwon, W.-S.; Yang, D.-C. Molecular characterization of two glutathione peroxidase genes of Panax ginseng and their expression analysis against environmental stresses. Gene 2014, 535, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.-P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Passaia, G.; Fonini, L.S.; Caverzan, A.; Jardim-Messeder, D.; Christoff, A.P.; Gaeta, M.L.; de Araujo Mariath, J.E.; Margis, R.; Margis-Pinheiro, M. The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci. 2013, 208, 93–101. [Google Scholar] [CrossRef]
- Diao, Y.; Xu, H.; Li, G.; Yu, A.; Yu, X.; Hu, W.; Zheng, X.; Li, S.; Wang, Y.; Hu, Z. Cloning a glutathione peroxidase gene from Nelumbo nucifera and enhanced salt tolerance by overexpressing in rice. Mol. Biol. Rep. 2014, 41, 4919–4927. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, A.; Basu, S.; Sengupta, D.N. Antioxidants and stress-related metabolites in the seedlings of two indica rice varieties exposed to cadmium chloride toxicity. Acta Physiol. Plant. 2012, 34, 835–847. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, M.; Teng, Y.; Jia, S.; Yu, D.; Wei, T.; Chen, C.; Song, W. Overexpression of the Glutathione Peroxidase 5 (RcGPX5) Gene From Rhodiola crenulata Increases Drought Tolerance in Salvia miltiorrhiza. Front. Plant Sci. 2019, 9, 1950. [Google Scholar] [CrossRef]
- Herbette, S.; Lenne, C.; De Labrouhe, D.T.; Drevet, J.R.; Roeckel-Drevet, P. Transcripts of sunflower antioxidant scavengers of the SOD and GPX families accumulate differentially in response to downy mildew infection, phytohormones, reactive oxygen species, nitric oxide, protein kinase and phosphatase inhibitors. Physiol. Plant. 2003, 119, 418–428. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Rakwal, R.; Jwa, N.-S.; Agrawal, V.P. Effects of signaling molecules, protein phosphatase inhibitors and blast pathogen (Magnaporthe grisea) on the mRNA level of a rice (Oryza sativa L.) phospholipid hydroperoxide glutathione peroxidase (OsPHGPX) gene in seedling leaves. Gene 2002, 283, 227–236. [Google Scholar] [CrossRef]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Govrin, E.M.; Levine, A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef]
- Orozco-Cárdenas, M.L.; Narváez-Vásquez, J.; Ryan, C.A. Hydrogen Peroxide Acts as a Second Messenger for the Induction of Defense Genes in Tomato Plants in Response to Wounding, Systemin, and Methyl Jasmonate. Plant Cell 2001, 13, 179. [Google Scholar] [CrossRef]
- Bolwell, G.P.; Bindschedler, L.V.; Blee, K.A.; Butt, V.S.; Davies, D.R.; Gardner, S.L.; Gerrish, C.; Minibayeva, F. The apoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 2002, 53, 1367–1376. [Google Scholar] [CrossRef]
- De Jaegher, G.; Boyer, N.; Bon, M.-C.; Gaspar, T. Thigmomorphogenesis in Bryonia dioica: Early Events in Ethylene Biosynthesis Pathway. Biochem. Und Physiol. Der Pflanz. 1987, 182, 49–56. [Google Scholar] [CrossRef]
- De Jaegher, G.; Boyer, N.; Gaspar, T. Thigmomorphogenesis inBryonia dioica: Changes in soluble and wall peroxidases, phenylalanine ammonia-lyase activity, cellulose, lignin content and monomeric constituents. Plant Growth Regul. 1985, 3, 133–148. [Google Scholar] [CrossRef]
- Depège, N.; Drevet, J.; Boyer, N. Molecular cloning and characterization of tomato cDNAs encoding glutathione peroxidase-like proteins. Eur. J. Biochem. 1998, 253, 445–451. [Google Scholar] [CrossRef]
- Bolwell, G.P.; Wojtaszek, P. Mechanisms for the generation of reactive oxygen species in plant defence–a broad perspective. Physiol. Mol. Plant Pathol. 1997, 51, 347–366. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef]
- Schulz, G.; Schirmer, R.H.; Sachsenheimer, W.; Pai, E.F. The structure of the flavoenzyme glutathione reductase. Nature 1978, 273, 120–124. [Google Scholar] [CrossRef]
- Mahan, J.R.; Burke, J.J. Purification and characterization of glutathione reductase from corn mesophyll chloroplasts. Physiol. Plant. 1987, 71, 352–358. [Google Scholar] [CrossRef]
- Hausladen, A.; Alscher, R.G. Purification and characterization of glutathione reductase isozymes specific for the state of cold hardiness of red spruce. Plant Physiol. 1994, 105, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Connell, J.P.; Mullet, J.E. Pea chloroplast glutathione reductase: Purification and characterization. Plant Physiol. 1986, 82, 351–356. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Hasanuzzaman, M.; Gill, R.; Trivedi, D.K.; Ahmad, I.; Pereira, E.; Tuteja, N. Glutathione and glutathione reductase: A boon in disguise for plant abiotic stress defense operations. Plant Physiol. Biochem. 2013, 70, 204–212. [Google Scholar] [CrossRef]
- Cohen, A.; Hacham, Y.; Welfe, Y.; Khatib, S.; Avice, J.-C.; Amir, R. Evidence of a significant role of glutathione reductase in the sulfur assimilation pathway. Plant J. 2020, 102, 246–261. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Gill, S.S.; Yadav, S.; Tuteja, N. Genome-wide analysis of glutathione reductase (GR) genes from rice and Arabidopsis. Plant Signal. Behav. 2013, 8, e23021. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Jiang, R.; Lu, Q.; Wen, X.; Lu, C. Glutathione reductase 2 maintains the function of photosystem II in Arabidopsis under excess light. Biochim. Et Biophys. Acta -Bioenerg. 2016, 1857, 665–677. [Google Scholar] [CrossRef]
- Marty, L.; Bausewein, D.; Müller, C.; Bangash, S.A.K.; Moseler, A.; Schwarzländer, M.; Müller-Schüssele, S.J.; Zechmann, B.; Riondet, C.; Balk, J.; et al. Arabidopsis glutathione reductase 2 is indispensable in plastids, while mitochondrial glutathione is safeguarded by additional reduction and transport systems. New Phytol. 2019, 224, 1569–1584. [Google Scholar] [CrossRef]
- Creissen, G.; Reynolds, H.; Xue, Y.; Mullineaux, P. Simultaneous targeting of pea glutathione reductase and of a bacterial fusion protein to chloroplasts and mitochondria in transgenic tobacco. Plant J. 1995, 8, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Contour-Ansel, D.; Torres-Franklin, M.L.; Cruz De Carvalho, M.H.; D’Arcy-Lameta, A.; Zuily-Fodil, Y. Glutathione reductase in leaves of cowpea: Cloning of two cDNAs, expression and enzymatic activity under progressive drought stress, desiccation and abscisic acid treatment. Ann. Bot. 2006, 98, 1279–1287. [Google Scholar] [CrossRef]
- Torres-Franklin, M.L.; Gigon, A.; De Melo, D.F.; Zuily-Fodil, Y.; Pham-Thi, A.T. Drought stress and rehydration affect the balance between MGDG and DGDG synthesis in cowpea leaves. Physiol. Plant. 2007, 131, 201–210. [Google Scholar] [CrossRef]
- Stevens, R.G.; Creissen, G.P.; Mullineaux, P.M. Cloning and characterisation of a cytosolic glutathione reductase cDNA from pea (Pisum sativum L.) and its expression in response to stress. Plant Mol. Biol. 1997, 35, 641–654. [Google Scholar] [CrossRef]
- Lee, H.; Jo, J.; Son, D. Molecular cloning and characterization of the gene encoding glutathione reductase in Brassica campestris. Biochim. Et Biophys. Acta -Gene Struct. Expr. 1998, 1395, 309–314. [Google Scholar] [CrossRef]
- Lascano, H.; Casano, L.; Melchiorre, M.; Trippi, V. Biochemical and molecular characterisation of wheat chloroplastic glutathione reductase. Biol. Plant. 2001, 44, 509–516. [Google Scholar] [CrossRef]
- Bashir, K.; Nagasaka, S.; Itai, R.N.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Expression and enzyme activity of glutathione reductase is upregulated by Fe-deficiency in graminaceous plants. Plant Mol. Biol. 2007, 65, 277–284. [Google Scholar] [CrossRef]
- Pastori, G.M.; Mullineaux, P.M.; Foyer, C.H. Post-transcriptional regulation prevents accumulation of glutathione reductase protein and activity in the bundle sheath cells of maize. Plant Physiol. 2000, 122, 667–676. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, T.-M.; Lin, W.-R.; Kao, Y.-T.; Hsu, Y.-T.; Yeh, C.-H.; Hong, C.-Y.; Kao, C.H. Identification and characterization of a novel chloroplast/mitochondria co-localized glutathione reductase 3 involved in salt stress response in rice. Plant Mol. Biol. 2013, 83, 379–390. [Google Scholar] [CrossRef]
- Liao, J.; Liu, Z.; Lu, L.; Liu, R.; Yu, W.; Gong, M. The Expression and Enzyme Activity of the Cytosolic Glutathione Reductase was Upregulated by Nitrogen-Deficiency in Wheat (Triticum aestivum L.) Grain. J. Biobased Mater. Bioenergy 2017, 11, 336–342. [Google Scholar] [CrossRef]
- Liu, H.-J.; Wang, X.; Yang, Z.-L.; Ren, L.-L.; Qian, T.-T. Identification and biochemical characterization of the glutathione reductase family from Populus trichocarpa. Plant Sci. 2020, 294, 110459. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A. The functions and regulation of glutathione s-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Vaish, S.; Gupta, D.; Mehrotra, R.; Mehrotra, S.; Basantani, M.K. Glutathione S-transferase: A versatile protein family. 3 Biotech 2020, 10, 321. [Google Scholar] [CrossRef]
- Hernández Estévez, I.; Rodríguez Hernández, M. Plant Glutathione S-transferases: An overview. Plant Gene 2020, 23, 100233. [Google Scholar] [CrossRef]
- Yang, J.; You, S.; Zheng, J. Review in Strengthening Technology for Phytoremediation of Soil Contaminated by Heavy Metals. IOP Conf. Ser. Earth Environ. Sci. 2019, 242, 052003. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, H.; Zhao, Y.; Li, X.; Dai, X.; Zhuang, J.; Zhu, M.; Jiang, X.; Wang, P.; Gao, L.; et al. Three Camellia sinensis glutathione S-transferases are involved in the storage of anthocyanins, flavonols, and proanthocyanidins. Planta 2019, 250, 1163–1175. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Tan, L.; Tang, S.-F. Molecular mechanism study on the interactions of cadmium (II) ions with Arabidopsis thaliana glutathione transferase Phi8. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 216, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wu, L.; Wang, J.; Yu, Q.; Bai, L.; Pan, L. Quizalofop-p-ethyl resistance in Polypogon fugax involves glutathione S-transferases. Pest Manag. Sci. 2020, 76, 3800–3805. [Google Scholar] [CrossRef]
- Sun, C.; Dudley, S.; McGinnis, M.; Trumble, J.; Gan, J. Acetaminophen detoxification in cucumber plants via induction of glutathione S-transferases. Sci. Total Environ. 2019, 649, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef]
- Wang, Y.; Wisniewski, M.; Meilan, R.; Cui, M.; Webb, R.; Fuchigami, L. Overexpression of cytosolic ascorbate peroxidase in tomato confers tolerance to chilling and salt stress. J. Am. Soc. Hortic. Sci. 2005, 130, 167–173. [Google Scholar] [CrossRef]
- Jiménez, A.; Hernández, J.; Barceló, A.R.; Sandalio, L.; Del Río, L.; Sevilla, F. Mitochondrial and peroxisomal ascorbate peroxidase of pea leaves. Physiol. Plant. 1998, 104, 687–692. [Google Scholar] [CrossRef]
- Yoshimura, K.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. Identification of a cis element for tissue-specific alternative splicing of chloroplast ascorbate peroxidase pre-mRNA in higher plants. J. Biol. Chem. 2002, 277, 40623–40632. [Google Scholar] [CrossRef]
- Mullen, R.T.; Trelease, R.N. The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J. Biol. Chem. 2000, 275, 16337–16344. [Google Scholar] [CrossRef]
- Bunkelmann, J.R.; Trelease, R.N. Ascorbate peroxidase (A prominent membrane protein in oilseed glyoxysomes). Plant Physiol. 1996, 110, 589–598. [Google Scholar] [CrossRef]
- Chen, G.-X.; Asada, K. Ascorbate Peroxidase in Tea Leaves: Occurrence of Two Isozymes and the Differences in Their Enzymatic and Molecular Properties. Plant Cell Physiol. 1989, 30, 987–998. [Google Scholar] [CrossRef]
- Madhusudhan, R.; Ishikawa, T.; Sawa, Y.; Shigeoka, S.; Shibata, H. Characterization of an ascorbate peroxidase in plastids of tobacco BY-2 cells. Physiol. Plant. 2003, 117, 550–557. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Mori, H.; Nishimura, M. A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant Cell Physiol. 1995, 36, 1157–1162. [Google Scholar] [CrossRef]
- Liao, G.-L.; Liu, Q.; Li, Y.-Q.; Zhong, M.; Huang, C.-H.; Jia, D.-F.; Xu, X.-B. Identification and expression profiling analysis of ascorbate peroxidase gene family in Actinidia chinensis (Hongyang). J. Plant Res. 2020, 133, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.C.; Salvi, P.; Kamble, N.U.; Joshi, P.K.; Majee, M.; Arora, S. Ectopic overexpression of cytosolic ascorbate peroxidase gene (Apx1) improves salinity stress tolerance in Brassica juncea by strengthening antioxidative defense mechanism. Acta Physiol. Plant. 2020, 42, 45. [Google Scholar] [CrossRef]
- Barros, J.; Escamilla-Trevino, L.; Song, L.; Rao, X.; Serrani-Yarce, J.C.; Palacios, M.D.; Engle, N.; Choudhury, F.K.; Tschaplinski, T.J.; Venables, B.J.; et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019, 10, 1994. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lu, H.-H.; Hua, S.; Lin, K.-H.; Chen, N.; Zhang, Y.; You, Z.; Kuo, Y.-W.; Chen, S.-P. Cloning and overexpression of the ascorbate peroxidase gene from the yam (Dioscorea alata) enhances chilling and flood tolerance in transgenic Arabidopsis. J. Plant Res. 2019, 132, 857–866. [Google Scholar] [CrossRef]
- Rao, M.V.; Paliyath, G.; Ormrod, D.P. Ultraviolet-B- and Ozone-Induced Biochemical Changes in Antioxidant Enzymes of Arabidopsis thaliana. Plant Physiol. 1996, 110, 125. [Google Scholar] [CrossRef]
- Malar, S.; Shivendra Vikram, S.; Jc Favas, P.; Perumal, V. Lead heavy metal toxicity induced changes on growth and antioxidative enzymes level in water hyacinths [Eichhornia crassipes (Mart.)]. Bot. Stud. 2014, 55, 54. [Google Scholar] [CrossRef]
- Bai, Y.; Guo, J.; Reiter, R.J.; Wei, Y.; Shi, H. Melatonin synthesis enzymes interact with ascorbate peroxidase to protect against oxidative stress in cassava. J. Exp. Bot. 2020, 71, 5645–5655. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.; Wang, Y.; Teng, R.-M.; Liu, J.; Lin, S.; Zhuang, J. Cytosolic ascorbate peroxidase 1 modulates ascorbic acid metabolism through cooperating with nitrogen regulatory protein P-II in tea plant under nitrogen deficiency stress. Genomics 2020, 112, 3497–3503. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Lee, C.B. Chilling stress-induced changes of antioxidant enzymes in the leaves of cucumber: In gel enzyme activity assays. Plant Sci 2000, 159, 75–85. [Google Scholar] [CrossRef]
- Park, A.K.; Kim, I.-S.; Do, H.; Jeon, B.W.; Lee, C.W.; Roh, S.J.; Shin, S.C.; Park, H.; Kim, Y.-S.; Kim, Y.-H.; et al. Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica. Sci. Rep. 2016, 6, 33903. [Google Scholar] [CrossRef]
- Maynard, D.; Kumar, V.; Sproß, J.; Dietz, K.-J. 12-oxophytodienoic acid reductase 3 (OPR3) functions as NADPH-dependent α,β-ketoalkene reductase in detoxification and monodehydroascorbate reductase in redox homeostasis. Plant Cell Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Vanacker, H.; Guichard, M.; Bohrer, A.-S.; Issakidis-Bourguet, E. Redox Regulation of Monodehydroascorbate Reductase by Thioredoxin y in Plastids Revealed in the Context of Water Stress. Antioxidants 2018, 7, 183. [Google Scholar] [CrossRef] [PubMed]
- Shokat, s.; Großkinsky, D.; Roitsch, T.; Liu, F. Higher activity of monodehydroascorbate reductase and lower activities of leaf and spike vacuolar invertase and glutathione S-transferase reveals higher number of grains per spike in spring wheat genotypes grown under well-watered and drought conditions. BMC Plant Biol. 2020. [Google Scholar] [CrossRef]
- Yeh, H.-L.; Lin, T.-H.; Chen, C.-C.; Cheng, T.-X.; Chang, H.-Y.; Lee, T.-M. Monodehydroascorbate Reductase Plays a Role in the Tolerance of Chlamydomonas reinhardtii to Photooxidative Stress. Plant Cell Physiol. 2019, 60, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Bodra, N.; Young, D.; Rosado, L.A.; Pallo, A.; Wahni, K.; De Proft, F.; Huang, J.; Van Breusegem, F.; Messens, J. Erratum: Arabidopsis thaliana dehydroascorbate reductase 2: Conformational flexibility during catalysis. Sci. Rep. 2017, 7, 46896. [Google Scholar] [CrossRef]
- Deutsch, J.C. Dehydroascorbic acid. J. Chromatogr. A 2000, 881, 299–307. [Google Scholar] [CrossRef]
- Do, H.; Kim, I.-S.; Jeon, B.W.; Lee, C.W.; Park, A.K.; Wi, A.R.; Shin, S.C.; Park, H.; Kim, Y.-S.; Yoon, H.-S.; et al. Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica. Sci. Rep. 2016, 6, 19498. [Google Scholar] [CrossRef] [PubMed]
- Krishna Das, B.; Kumar, A.; Maindola, P.; Mahanty, S.; Jain, S.K.; Reddy, M.K.; Arockiasamy, A. Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase. Biochem. Biophys. Res. Commun. 2016, 473, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Lallement, P.-A.; Roret, T.; Tsan, P.; Gualberto, J.M.; Girardet, J.-M.; Didierjean, C.; Rouhier, N.; Hecker, A. Insights into ascorbate regeneration in plants: Investigating the redox and structural properties of dehydroascorbate reductases from Populus trichocarpa. Biochem. J. 2016, 473, 717–731. [Google Scholar] [CrossRef]
- Ding, H.; Wang, B.; Han, Y.; Li, S. The pivotal function of dehydroascorbate reductase in glutathione homeostasis in plants. J. Exp. Bot. 2020, 71, 3405–3416. [Google Scholar] [CrossRef]
- Hao, Z.; Wang, X.; Zong, Y.; Wen, S.; Cheng, Y.; Li, H. Enzymatic activity and functional analysis under multiple abiotic stress conditions of a dehydroascorbate reductase gene derived from Liriodendron Chinense. Environ. Exp. Bot. 2019, 167, 103850. [Google Scholar] [CrossRef]
- Terai, Y.; Ueno, H.; Ogawa, T.; Sawa, Y.; Miyagi, A.; Kawai-Yamada, M.; Ishikawa, T.; Maruta, T. Dehydroascorbate Reductases and Glutathione Set a Threshold for High-Light–Induced Ascorbate Accumulation. Plant Physiol. 2020, 183, 112. [Google Scholar] [CrossRef]
- Loi, M.; De Leonardis, S.; Mulè, G.; Logrieco, A.F.; Paciolla, C. A Novel and Potentially Multifaceted Dehydroascorbate Reductase Increasing the Antioxidant Systems is Induced by Beauvericin in Tomato. Antioxidants 2020, 9, 435. [Google Scholar] [CrossRef]
- Bogoutdinova, L.R.; Lazareva, E.M.; Chaban, I.A.; Kononenko, N.V.; Dilovarova, T.; Khaliluev, M.R.; Kurenina, L.V.; Gulevich, A.A.; Smirnova, E.A.; Baranova, E.N. Salt stress-induced structural changes are mitigated in transgenic tomato plants over-expressing superoxide dismutase. Biology 2020, 9, 297. [Google Scholar] [CrossRef]
- Che, Y.; Zhang, N.; Zhu, X.; Li, S.; Wang, S.; Si, H. Enhanced tolerance of the transgenic potato plants overexpressing Cu/Zn superoxide dismutase to low temperature. Sci. Hortic. 2020, 261, 108949. [Google Scholar] [CrossRef]
- Lin, K.-H.; Sei, S.-C.; Su, Y.-H.; Chiang, C.-M. Overexpression of the Arabidopsis and winter squash superoxide dismutase genes enhances chilling tolerance via ABA-sensitive transcriptional regulation in transgenic Arabidopsis. Plant Signal. Behav. 2019, 14, 1685728. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Han, X.; Song, X.; Zhang, Y.; Jiang, J.; Han, Q.; Liu, M.; Qiao, G.; Zhuo, R. Overexpressing the Sedum alfredii Cu/Zn superoxide dismutase increased resistance to oxidative stress in transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 1010. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, L.; Zhang, L.; Qiu, H.; Liu, C.; Wang, A.; Deng, F.; Zhu, J. A Cu/Zn superoxide dismutase gene from Saussurea involucrata Kar. & Kir., SiCSD, enhances drought, cold, and oxidative stress in transgenic tobacco. Can. J. Plant Sci. 2017, 97, 816–826. [Google Scholar]
- Geng, J.; Wei, T.; Wang, Y.; Huang, X.; Liu, J.-H. Overexpression of PtrbHLH, a basic helix-loop-helix transcription factor from Poncirus trifoliata, confers enhanced cold tolerance in pummelo (Citrus grandis) by modulation of H2O2 level via regulating a CAT gene. Tree Physiol. 2019, 39, 2045–2054. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Liang, X.; Lu, H.; Li, Q.; Chen, Q.; Zhang, P.; Liu, G.; Yan, W.; Song, J.; Duan, C. Overproduction of superoxide dismutase and catalase confers cassava resistance to Tetranychus cinnabarinus. Sci. Rep. 2017, 7, 40179. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Kim, H.S.; Park, S.-C.; Ji, C.Y.; Yang, J.W.; Lee, H.U.; Kwak, S.-S. Overexpression of swpa4 peroxidase enhances tolerance to hydrogen peroxide and high salinity-mediated oxidative stress in transgenic sweetpotato plants. Plant Biotechnol. Rep. 2020, 14, 301–307. [Google Scholar] [CrossRef]
- Yin, L.; Mano, J.; Tanaka, K.; Wang, S.; Zhang, M.; Deng, X.; Zhang, S. High level of reduced glutathione contributes to detoxification of lipid peroxide-derived reactive carbonyl species in transgenic Arabidopsis overexpressing glutathione reductase under aluminum stress. Physiol. Plant. 2017, 161, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Srivastava, S.; Singh, P.C.; Mishra, A.K.; Chakrabarty, D. Functional characterization of tau class glutathione-S-transferase in rice to provide tolerance against sheath blight disease. 3 Biotech 2020, 10, 84. [Google Scholar] [CrossRef]
- Gao, J.-J.; Zhang, L.; Peng, R.-H.; Wang, B.; Feng, H.-J.; Li, Z.-J.; Yao, Q.-H. Recombinant expression of Thermosynechococcus elongatus BP-1 glutathione S-transferase in Arabidopsis thaliana: An efficient tool for phytoremediation of thiocyanate. Biotechnol. Biotechnol. Equip. 2020, 34, 494–505. [Google Scholar] [CrossRef]
- Qi, Q.; Yanyan, D.; Yuanlin, L.; Kunzhi, L.; Huini, X.; Xudong, S. Overexpression of SlMDHAR in transgenic tobacco increased salt stress tolerance involving S-nitrosylation regulation. Plant Sci. 2020, 299, 110609. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Yang, N.; Jiang, S.; Ma, C.; Li, H. Overexpression of a Monodehydroascorbate Reductase Gene from Sugar Beet M14 Increased Salt Stress Tolerance. Sugar Tech 2020, 23, 45–56. [Google Scholar] [CrossRef]
- Liu, F.; Guo, X.; Yao, Y.; Tang, W.; Zhang, W.; Cao, S.; Han, Y.; Liu, Y. Cloning and function characterization of two dehydroascorbate reductases from kiwifruit (Actinidia chinensis L.). Plant Mol. Biol. Rep. 2016, 34, 815–826. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Chaki, M.; Begara-Morales, J.C.; Barroso, J.B. Oxidative Stress in Plants. Antioxidants 2020, 9, 481. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wei, C.-l.; Zhang, W.-r.; Cheng, H.-p.; Liu, J. Cross-talk between calcium and reactive oxygen species signaling. Acta Pharmacol. Sin. 2006, 27, 821–826. [Google Scholar] [CrossRef]
- Chang, C.C.C.; Ślesak, I.; Jordá, L.; Sotnikov, A.; Melzer, M.; Miszalski, Z.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpiński, S. Arabidopsis Chloroplastic Glutathione Peroxidases Play a Role in Cross Talk between Photooxidative Stress and Immune Responses. Plant Physiol. 2009, 150, 670. [Google Scholar] [CrossRef] [PubMed]
- Pastori, G.M.; Foyer, C.H. Common Components, Networks, and Pathways of Cross-Tolerance to Stress. The Central Role of “Redox” and Abscisic Acid-Mediated Controls. Plant Physiol. 2002, 129, 460. [Google Scholar] [CrossRef] [PubMed]
- Sasaki-Sekimoto, Y.; Taki, N.; Obayashi, T.; Aono, M.; Matsumoto, F.; Sakurai, N.; Suzuki, H.; Hirai, M.Y.; Noji, M.; Saito, K.; et al. Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 2005, 44, 653–668. [Google Scholar] [CrossRef]
- Kusumi, K.; Yaeno, T.; Kojo, K.; Hirayama, M.; Hirokawa, D.; Yara, A.; Iba, K. The role of salicylic acid in the glutathione-mediated protection against photooxidative stress in rice. Physiol. Plant. 2006, 128, 651–661. [Google Scholar] [CrossRef]
- Innocenti, G.; Pucciariello, C.; Gleuher, M.; Hopkins, J.; Stefano, M.; Delledonne, M.; Puppo, A.; Baudouin, E.; Frendo, P. Glutathione synthesis is regulated by nitric oxide in Medicago truncatula roots. Planta 2007, 225, 1597–1602. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Bose, S.K.; Howlader, P. Melatonin plays multifunctional role in horticultural crops against environmental stresses: A review. Environ. Exp. Bot. 2020, 176, 104063. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, J.; Fan, X.; Jiang, W. Applications of nitric oxide and melatonin in improving postharvest fruit quality and the separate and crosstalk biochemical mechanisms. Trends Food Sci. Technol. 2020, 99, 531–541. [Google Scholar] [CrossRef]
- Liu, T.; Jiao, X.; Yang, S.; Zhang, Z.; Ye, X.; Li, J.; Qi, H.; Hu, X. Crosstalk between GABA and ALA to improve antioxidation and cell expansion of tomato seedling under cold stress. Environ. Exp. Bot. 2020, 180, 104228. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. Changes in carotenoids, tocopherols and diterpenes during drought and recovery, and the biological significance of chlorophyll loss in Rosmarinus officinalis plants. Planta 2000, 210, 925–931. [Google Scholar] [CrossRef]
- Basu, S.; Roychoudhury, A.; Saha, P.P.; Sengupta, D.N. Comparative analysis of some biochemical responses of three indica rice varieties during polyethylene glycol-mediated water stress exhibits distinct varietal differences. Acta Physiol. Plant. 2010, 32, 551–563. [Google Scholar] [CrossRef]
- Mafakheri, A.; Siosemardeh, A.; Bahramnejad, B.; Struik, P.; Sohrabi, Y. Effect of drought stress on yield, proline and chlorophyll contents in three chickpea cultivars. Aust. J. Crop Sci. 2010, 4, 580–585. [Google Scholar]
- Chutipaijit, S.; Cha-Um, S.; Sompornpailin, K. Differential accumulations of proline and flavonoids in indica rice varieties against salinity. Pak. J. Bot. 2008, 41, 2497–2506. [Google Scholar]
- Malik, S.; Ashraf, M. Exogenous application of ascorbic acid stimulates growth and photosynthesis of wheat (Triticum aestivum L.) under drought. Soil Environ. 2012, 31, 72–77. [Google Scholar]
- Shalata, A.; Neumann, P.M. Exogenous ascorbic acid (vitamin C) increases resistance to salt stress and reduces lipid peroxidation. J. Exp. Bot. 2001, 52, 2207–2211. [Google Scholar] [CrossRef]
- Athar, H.-U.-R.; Khan, A.; Ashraf, M. Exogenously applied ascorbic acid alleviates salt-induced oxidative stress in wheat. Environ. Exp. Bot. 2008, 63, 224–231. [Google Scholar] [CrossRef]
- Vaidyanathan, H.; Sivakumar, P.; Chakrabarty, R.; Thomas, G. Scavenging of reactive oxygen species in NaCl-stressed rice (Oryza sativa L.)—differential response in salt-tolerant and sensitive varieties. Plant Sci. 2003, 165, 1411–1418. [Google Scholar] [CrossRef]
- Roychoudhury, A.; Basu, S. Ascorbate-Glutathione and Plant Tolerance to Various Abiotic Stresses; IK International: New Delhi, India, 2012; pp. 177–258. [Google Scholar]
- Wang, R.; Liu, S.; Zhou, F.; Ding, C. Exogenous Ascorbic Acid and Glutathione Alleviate Oxidative Stress Induced by Salt Stress in the Chloroplasts of Oryza sativa L. Z. Für Nat. C 2014, 69, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Ellouzi, H.; Hamed, K.B.; Cela, J.; Müller, M.; Abdelly, C.; Munné-Bosch, S. Increased sensitivity to salt stress in tocopherol-deficient Arabidopsis mutants growing in a hydroponic system. Plant Signal. Behav. 2013, 8, e23136. [Google Scholar] [CrossRef] [PubMed]
- Jain, K.; Kataria, S.; Guruprasad, K.N. Changes in antioxidant defenses of cucumber cotyledons in response to UV-B and to the free radical generating compound AAPH. Plant Sci. 2003, 165, 551–557. [Google Scholar] [CrossRef]
- Jambunathan, N. Determination and Detection of Reactive Oxygen Species (ROS), Lipid Peroxidation, and Electrolyte Leakage in Plants. In Plant Stress Tolerance: Methods and Protocols; Sunkar, R., Ed.; Humana Press: Totowa, NJ, USA, 2010; pp. 291–297. [Google Scholar]
- Poli, Y.; Nallamothu, V.; Balakrishnan, D.; Ramesh, P.; Desiraju, S.; Mangrauthia, S.K.; Voleti, S.R.; Neelamraju, S. Increased catalase activity and maintenance of Photosystem II distinguishes high-yield mutants from low-yield mutants of rice var. Nagina22 under low-phosphorus stress. Front. Plant Sci. 2018, 9, 1543. [Google Scholar] [CrossRef]
- Seth, K. Current status of potential applications of repurposed Cas9 for structural and functional genomics of plants. Biochem. Biophys. Res. Commun. 2016, 480, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Seth, K. The mysterious circle: Molecular curiosities of RNA mediated gene regulation. Gene Rep. 2017, 9, 13–19. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology 2021, 10, 267. https://doi.org/10.3390/biology10040267
Rajput VD, Harish, Singh RK, Verma KK, Sharma L, Quiroz-Figueroa FR, Meena M, Gour VS, Minkina T, Sushkova S, et al. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology. 2021; 10(4):267. https://doi.org/10.3390/biology10040267
Chicago/Turabian StyleRajput, Vishnu D., Harish, Rupesh Kumar Singh, Krishan K. Verma, Lav Sharma, Francisco Roberto Quiroz-Figueroa, Mukesh Meena, Vinod Singh Gour, Tatiana Minkina, Svetlana Sushkova, and et al. 2021. "Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress" Biology 10, no. 4: 267. https://doi.org/10.3390/biology10040267
APA StyleRajput, V. D., Harish, Singh, R. K., Verma, K. K., Sharma, L., Quiroz-Figueroa, F. R., Meena, M., Gour, V. S., Minkina, T., Sushkova, S., & Mandzhieva, S. (2021). Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology, 10(4), 267. https://doi.org/10.3390/biology10040267















