A Duplicated Copy of the Meiotic Gene ZIP4 Preserves up to 50% Pollen Viability and Grain Number in Polyploid Wheat
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Pollen Profiling
2.3. Pollen Viability
2.4. Grain Number Per Spike Assessment
2.5. Female Sterility Assessment
2.6. Seed Germination Rate
2.7. Meiotic Analysis
2.8. Image Processing
2.9. TaZIP4 Protein Sequence Analysis
3. Results
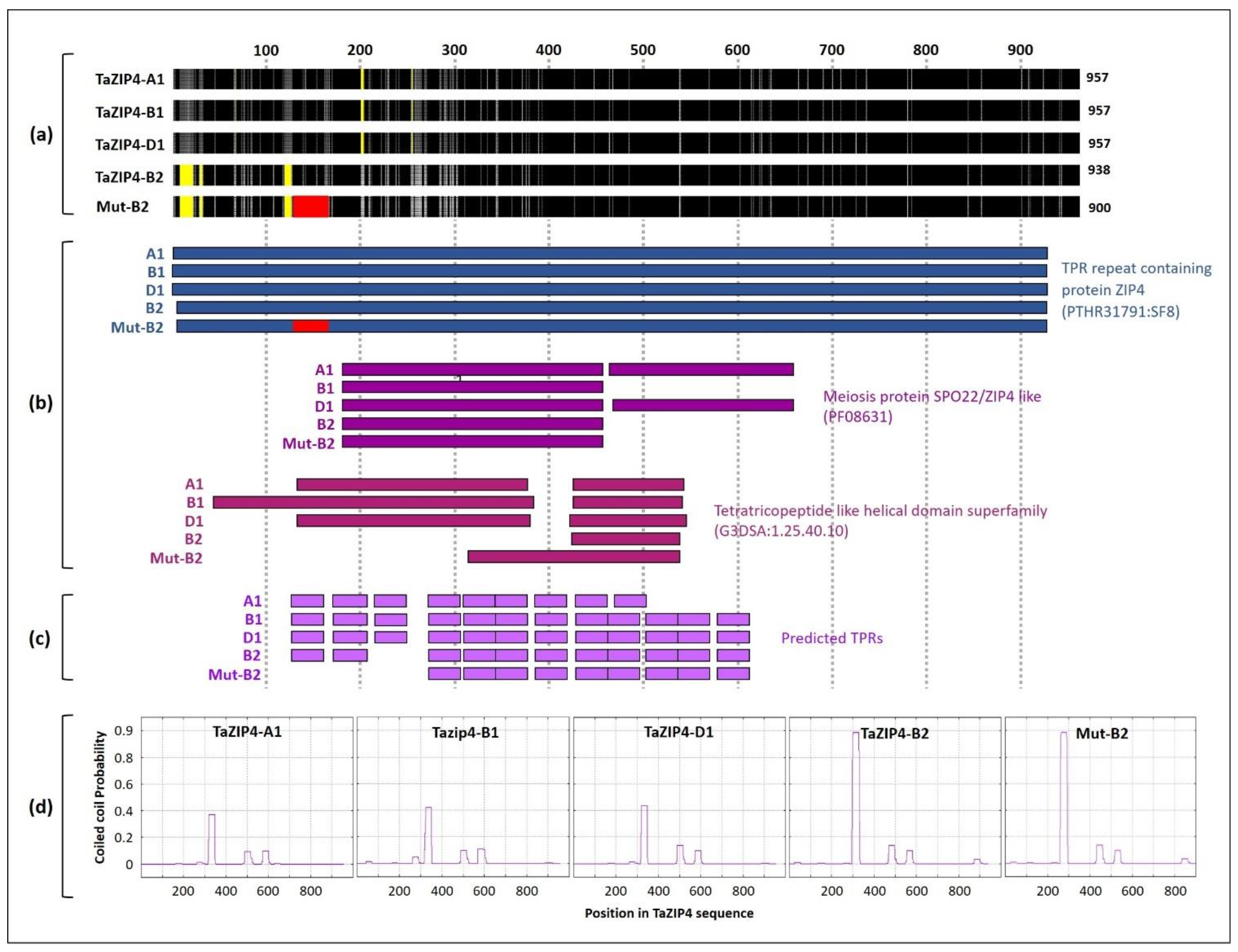
3.1. Divergence and the CRISPR Deletion Occur within the TaZIP4-B2 TPR Domain
3.2. Effect of the TaZIP4-B2 Deletion on Meiotic and Tetrad Stages
3.3. Effect of the Tazip4-B2 Deletion on Wheat Grain Number Per Spike (Grain Setting)
3.4. Pollen Contributes to the Tazip4-B2 Effect on Grain Setting
3.5. A Pollen Profiling Approach Reveals 50% Tazip4-B2 Mutant Pollen Is Small
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 838–846. [Google Scholar] [CrossRef]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelé, A.; Rousseau-Gueutin, M.; Chevre, A.-M. Speciation Success of polyploid plants closely relates to regulation of meiotic recombination. Front. Plant Sci. 2018, 9, 907. [Google Scholar] [CrossRef] [PubMed]
- Feliner, G.N.; Casacuberta, J.; Wendel, J.C. Genomics of Evolutionary Novelty in Hybrids and Polyploids. Front. Genet. 2020, 11, 792. [Google Scholar] [CrossRef]
- Osborn, T.C.; Pires, J.C.; Birchler, J.A.; Donald, L.; Auger, D.L.; Chen, Z.J.; Lee, H.-S.; Comai, L.; Madlung, A.; Doerge, R.W.; et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003, 19, 141–147. [Google Scholar] [CrossRef]
- Adams, K.L.; Wendel, J.F. Novel patterns of gene expression in polyploid plants. Trends Gen. 2005, 21, 539–543. [Google Scholar] [CrossRef]
- Mason, A.S.; Wendel, J.F. Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 2020, 11, 1014. [Google Scholar] [CrossRef]
- Ramírez-González, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, e6089. [Google Scholar] [CrossRef] [Green Version]
- Alabdullah, A.K.; Borrill, P.; Martin, A.C.; Ramirez-Gonzalez, R.H.; Uauy, C.; Shaw, P.; Moore, G. A co-expression network in hexaploid wheat reveals mostly balanced expression and lack of significant gene loss of homeologous meiotic genes upon polyploidization. Front. Plant Sci. 2019, 10, 1325. [Google Scholar] [CrossRef]
- Riley, R.; Chapman, V. Genetic control of the cytological diploid behaviour of hexaploid wheat. Nature 1958, 182, 713–715. [Google Scholar] [CrossRef]
- Sears, E.R.; Okamoto, M. Intergenomic chromosome relationships in hexaploid wheat. Theor. Appl. Genet. 1991, 82, 441–449. [Google Scholar]
- Wall, A.M.; Riley, R.; Gale, M.D. The position of a locus of chromosome 5B in Triticum aestivum affecting homoeologous meiotic pairing. Genet. Res. 1971, 18, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Sears, E.R. Induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 1977, 19, 585–593. [Google Scholar] [CrossRef]
- Martín, A.C.; Borrill, P.; Higgins, J.; Alabdullah, A.K.; Ramírez-González, R.H.; Swarbeck, D.; Uauy, C.; Shaw, P.; Moore, G. Overall wheat transcription during early meiosis is independent of synapsis, ploidy level and the Ph1 locus. Front. Plant Sci. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Roberts, M.A.; Reader, S.; Dalgliesh, C.; Miller, T.; Foote, T.; Fish, L.J.; Snape, J.W.; Moore, G. Induction and characterization of Ph1 wheat mutants. Genetics 1999, 153, 1909–1918. [Google Scholar]
- Griffiths, S.; Sharp, R.; Foote, T.N.; Bertin, I.; Wanous, M.; Reader, S.; Colas, I.; Moore, G. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 2006, 439, 749–752. [Google Scholar] [CrossRef]
- Al-Kaff, N.; Knight, E.; Bertin, I.; Foote, T.; Hart, N.; Griffiths, S.; Moore, G. Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: With deletion mutants and expression profiling. Annu. Bot. 2008, 105, 863–872. [Google Scholar] [CrossRef] [Green Version]
- Rey, M.-D.; Martín, A.C.; Higgins, J.; Swarbreck, D.; Uauy, C.; Shaw, P.; Moore, G. Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. 2017, 37, 95. [Google Scholar] [CrossRef]
- Rey, M.-D.; Martín, A.C.; Smedley, M.; Hayta, S.; Harwood, W.; Shaw, P.; Moore, G. Magnesium increases homoeologous crossover frequency during meiosis in Tazip4-B2 (Ph1 gene) mutant wheat-wild relative hybrids. Front. Plant Sci. 2018, 9, 509. [Google Scholar] [CrossRef] [Green Version]
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limiting in wheat research and breeding using a fully annotated reference genome. Science 2018, 362, eaar7191. [Google Scholar]
- Chelysheva, L.; Gendrot, G.; Vezon, D.; Doutriaux, M.-P.; Mercier, R.; Grelon, M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007, 3, e83. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, T.K.; Wang, W.; Huang, J.; Luo, W.; Luo, Q.; Hong, L.H.; Li, M.; Cheng, Z. ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J. Cell Sci. 2012, 125, 2581–2591. [Google Scholar] [CrossRef] [Green Version]
- Pagliarini, M.S. Meiotic behavior of economically important plant species: The relationship between fertility and male sterility. Genet. Mol. Biol. 2000, 23, 997–1002. [Google Scholar] [CrossRef]
- Sheidai, M.; Jafari, S.; Taleban, P.; Keshavarzi, M. Cytomixis and unreduced pollen grain formation in Alopecurus L. and Catbrosa Beauv. (Poaceae). Cytologia 2009, 74, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Sheidai, M.; Jafari, S.; Nourmohammadi, Z.; Beheshti, S. Cytomixis and unreduced pollen grain formation in six Hordeum species. Gene Conserve. 2010, 9, 40–50. [Google Scholar]
- Giorgi, B. Origin, behaviour and utilization of a Ph1 mutant of durum wheat, Triticum turgidum (L.) var. durum. In Proceedings of the 6th International Wheat Genetetics Symposium, Kyoto, Japan, 28 November–3 December 1983; pp. 1033–1040. [Google Scholar]
- Alexander, M.P. Differential staining of aborted and nonaborted pollen. Stain Technol. 1969, 44, 117–122. [Google Scholar] [CrossRef]
- Martín, A.C.; Rey, M.-D.; Shaw, P.; Moore, G. Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 2017, 126, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.K.; Sharma, A. Chromosome Techniques: Theory and Practice; Butterworth-Heinemann: Oxford, UK, 2014. [Google Scholar]
- Rey, M.D.; Moore, G.; Martín, A.C. Identification and comparison of individual chromosomes of three accessions of Hordeum chilense, Hordeum vulgare, and Triticum aestivum by FISH. Genome 2018, 61, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Higgins, D.G.; Sharp, P.M. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 1988, 73, 237–244. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro: Improving coverage.; classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [Green Version]
- Blatch, G.L.; Lassle, M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 1999, 21, 932–939. [Google Scholar] [CrossRef]
- D’Andrea, L.D.; Regan, L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662. [Google Scholar] [CrossRef]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Delorenzi, M.; Speed, T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 2002, 18, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.C.; Shaw, P.; Phillips, D.; Reader, S.; Moore, G. Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 2014, 5, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Li, W.; Fellers, J.; Friebe, B.; Gill, B.S. BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 2004, 112, 288–299. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and charcacterisation of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 11, 1869–1885. [Google Scholar] [CrossRef]
- Morrison, J.W. Chromosome behaviour in wheat monosomics. Heredity 1953, 7, 203–217. [Google Scholar] [CrossRef]
- Lamborn, E.; Cresswell, J.E.; Macnair, M.R. The potential for adaptive evolution of pollen grain size in Mimulus guttatus. New Phytol. 2005, 16, 289–296. [Google Scholar] [CrossRef] [PubMed]
- De Storme, N.; Zamariola, L.; Mau, M.; Sharbel, T.F.; Geelen, D. Volume-based pollen size analysis: An advanced method to assess somatic and gametophytic ploidy in flowering plants. Plant Reprod. 2013, 26, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Cetl, I. The size of pollen grain of the genus Triticum L. Biol. Plantarum 1960, 2, 287–291. [Google Scholar] [CrossRef]
- Saps. Pollen Images in Size Order. Available online: https://saps.org.uk/pollen/index2.htm (accessed on 20 January 2021).
- De Vries, A. Some aspects of cross-pollination in wheat (Triticum aestivum L.). 3. Anther length and number of pollen grains per anther. Euphytica 1974, 23, 11–19. [Google Scholar] [CrossRef]
- Yant, Y.; Hollister, J.D.; Wright, K.M.; Arnold, B.J.; Higgins, J.D.; Franklin, F.C.H.; Bomblies, K. Meiotic adaption to genome duplication in Arabidopsis arenosa. Curr. Biol. 2013, 23, 2151–2156. [Google Scholar] [CrossRef] [Green Version]
- Morgan, C.; Zhang, H.; Henry, C.E.; Franklin, C.F.H.; Bomblies, K. Derived alleles of two axis proteins affect meiotic traits in autotetraploid Arabidopsis arenosa. Proc. Natl. Acad. Sci. USA 2020, 117, 8980–8988. [Google Scholar] [CrossRef] [Green Version]
- Seear, P.J.; France, M.J.; Gregory, C.L.; Heavens, D.; Schmickl, R.; Yant, L.; Higgins, J.D. A novel allele of ASY3 is associated with greater meiotic stability in autotetraploid Arabidopsis lyrata. PLoS Genet. 2020, 16, e1008900. [Google Scholar] [CrossRef]
- Jenczewski, E.; Eber, F.; Grimaud, A.; Huet, S.; Lucas, M.O.; Monod, H.; Chevre, A.M. PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 2003, 164, 645–653. [Google Scholar]
- Liu, Z.; Adamczyk, K.; Manzanares-Dauleux, M.; Eber, F.; Lucas, M.O.; Delourme, R.; Chevre, A.M.; Jenczewski, E. Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 2006, 174, 1583–1596. [Google Scholar] [CrossRef] [Green Version]
- Higgins, E.E.; Howell, E.C.; Armstrong, S.J.; Parkin, I.A.P. A major quantitative trait locus on chromosome A9, BnaPh1 controls homoeologous recombination in Brassica napus. New Phytol. 2020, 229, 3041–3043. [Google Scholar]
- Cruden, R.W.; Miller-Ward, S. Pollen-ovule ratio, pollen size, and the ratio of stigmatic area to the pollen-bearing area of the pollinator: An hypothesis. Evolution 1981, 35, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Németh, M.B.; Smith-Huerta, N.L. Pollen deposition, pollen tube growth, seed production, and seedling performance in natural populations of Clarkia Unguiculata (Onagraceae). Int. J. Plant Sci. 2003, 164, 153–164. [Google Scholar] [CrossRef]
- Benjahya, F.; Naduad, I.; Da Innes, O.; Rimbert, H.; White, C.; Sourdille, P. SPOII.2 is essential for programmes double strand break formation during meiosis in bread wheat (Triticum aestivum L.). Plant J. 2020, 104, 30–43. [Google Scholar] [CrossRef]
- Desjardins, S.D.; Ogle, D.E.; Ayoub, M.A.; Heckman, S.; Henderson, I.R.; Edwards, K.J.; Higgins, J.D. MutS homologue 4 and MutS homologue 5 maintain the obligate crossover despite stepwise gene loss following polyploidisation. Plant Physiol. 2020, 183, 1545–1558. [Google Scholar] [CrossRef]
- Serra, H.; Svačina, R.; Baumann, U.; Whitford, R.; Sutton, T.; Bartoš, J.; Sourdille, P. Ph2 encodes the mismatch repair protein MSH7–3D that inhibits wheat homoeologous recombination. Nat. Commun. 2021, 12, 801. [Google Scholar] [CrossRef]
- Tsubouchi, T.; Zhao, H.; Roeder, G.S. The meiosis-specific ZIP4 protein regulates crossover distribution by promoting synaptonemal complex formation together with ZIP2. Dev. Cell 2006, 10, 809–819. [Google Scholar] [CrossRef] [Green Version]
- Dubois, E.; Muyt, A.D.; Soyer, J.L.; Budin, K.; Legras, M.; Piolot, T.; Debuchy, R.; Kleckner, N.; Zickler, D.; Espagne, E. Building bridges to move recombination complexes. Proc. Natl. Acad. Sci. USA 2019, 116, 12400–12409. [Google Scholar] [CrossRef] [Green Version]
- Pyatnitskaya, A.; Borde, V.; Muyt, A.D. Crossing and zipping: Molecular duties of ZMM proteins in meiosis. Chromosoma 2019, 128, 181–198. [Google Scholar] [CrossRef] [Green Version]
- Boden, S.A.; Langridge, P.; Spangenberg, G.; Able, J. TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J. 2009, 57, 487–497. [Google Scholar] [CrossRef]
- Prieto, P.; Shaw, P.; Moore, G. Homologue recognition is associated with a change in chromatin conformation. Nat. Cell Biol. 2004, 6, 906–908. [Google Scholar] [CrossRef] [Green Version]
- Colas, I.; Shaw, P.; Prieto, P.; Wanous, M.; Spielmeyer, W.; Mago, R.; Moore, G. Effective chromosome pairing requires chromatin remodeling at the onset of meiosis. Proc. Natl. Acad. Sci. USA 2008, 105, 6075–6080. [Google Scholar] [CrossRef] [Green Version]
- Golubovskaya, I.N.; Hamant, O.; Timofejeva, L.; Wang, C.-J.R.; Braun, D.; Meeley, R.; Cande, Z.W. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J. Cell Sci. 2006, 119, 3306–3315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, D.-Q.; Matsuda, A.; Okamasa, K.; Nagahama, Y.; Haraguchi, T.; Hiraoka, Y. Meiotic cohesion-based chromosome structure is essential for homologous pairing in Schizosaccharomyces pombe. Chromosoma 2016, 125, 205–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jauhar, P.P.; Almousiem, A.B.; Peterson, T.S.; Joppa, L.R. Inter- and intragenomic chromosome pairing in Haploids of Durum wheat. J. Hered. 1999, 90, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Gonzalo, A.; Lucas, M.-O.; Charpentier, C.; Sandmann, G.; Lloyd, A.; Jenczewski, E. Reducing MSH4 copy number prevents meiotic crossovers between non-homologous chromosomes in Brassica napus. Nat. Commun. 2019, 10, 2354. [Google Scholar] [CrossRef] [PubMed]
- Brinton, J.; Ramirez-Gonzalez, R.H.; Simmonds, J.; Wingen, L.; Orford, S.; Griffiths, S.; 10 Wheat Genome Project; Haberer, G.; Spannagl, M.; Walkowiak, S.; et al. Haplotype-led approach to increase the precision of wheat breeding. Commun. Biol. 2020, 3, 712. [Google Scholar] [CrossRef]
- Wingen, L.U.; West, C.; Leverington-Waite, M.; Collier, S.; Orford, S.; Goram, R.; Yang, C.-Y.; King, J.; Allen, A.M.; Burridge, A.; et al. Wheat landrace genome diversity. Genetics 2017, 205, 1657–1676. [Google Scholar] [CrossRef] [Green Version]

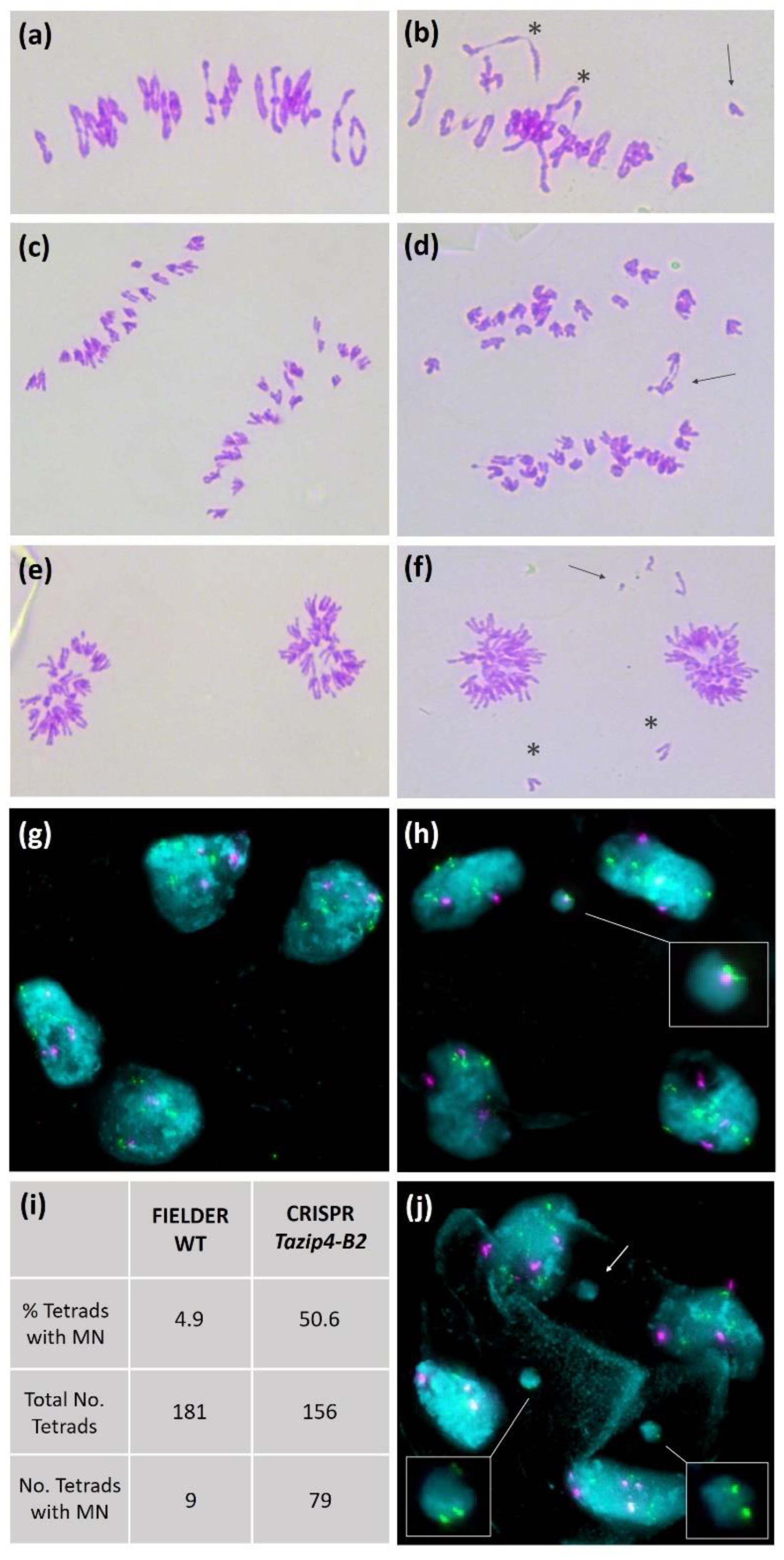




| Genotype | Univalents | Multivalents | Rod Bivalents | Ring Bivalents | Defective Meiocytes * (%) | Reference |
|---|---|---|---|---|---|---|
| Fielder WT ** | 0.16 ± 0.07 | 0 ± 0 | 1.37 ± 0.13 | 19.52 ± 0.14 | 6.70 | [19] |
| CRISPR Tazip4-B2 | 1.16 ± 0.12 | 0.39 ± 0.05 | 4.93 ± 0.15 | 14.84 ± 0.19 | 56.00 | |
| Chinese Spring | 0 ± 0 | 0 ± 0 | 1 ± 0.20 | 20 ± 0.20 | 0.00 | [38] |
| ph1b | 0.80 ± 0.19 | 0.53 ± 0.12 | 4.73 ± 0.26 | 14.83 ± 0.33 | 56.60 |
| Genotype | Controlled Environment Room (CER) | Glasshouse | ||
|---|---|---|---|---|
| N | Normalized Grain Number Per Spike | N | Normalized Grain Number Per Spike | |
| CRISPR Tazip4-B2 | 5 | 50.2 ± 16.7 b,c,d | 15 | 37.7 ± 1.0 d,e |
| cv. Fielder WT | 12 | 78.5 ± 12.9 a | 15 | 67.6 ± 07.9 a,b |
| ph1b | 15 | 46.7 ± 1.0 c,d | 11 | 43.8 ± 08.6 c,d |
| cv. Chinese Spring WT | 20 | 80.9 ± 17.3 a | 15 | 77.3 ± 08.8 a |
| Polyploidy | Variety | N | Pollen Size | Pollen Number Per Anther | ||
|---|---|---|---|---|---|---|
| Mean | Median | Mean | Median | |||
| Hexaploid | Cadenza | 10 | 49.05 ± 1.28 a | 49.65 ± 1.11 a | 2380 ± 320 b | 2407 ± 348 b |
| Hexaploid | Chinese Spring | 12 | 48.58 ± 1.17 a | 49.05 ± 1.15 a | 2807 ± 384 c | 2840 ± 408 c |
| Hexaploid | Fielder | 10 | 48.67 ± 0.98 a | 49.36 ± 1.19 a | 1973 ± 272 a | 1686 ± 250 a |
| Hexaploid | Paragon | 10 | 49.51 ± 1.09 a | 50.25 ± 1.22 a | 3318 ± 236 d | 3293 ± 339 d |
| Tetraploid | Cappelli | 10 | 44.77 ± 1.40 b | 44.85 ± 1.48 b | 3515 ± 260 d | 3480 ± 277 d |
| Tetraploid | Kronos | 13 | 44.44 ± 1.39 b | 44.63 ± 1.52 b | 2260 ± 110 b | 2373 ± 256 b |
| All hexaploid wheat varieties | 4 | 48.95 ± 0.43 | 49.58 ± 0.51 | 2533 ± 657 | 2557 ± 684 | |
| All tetraploid wheat varieties | 2 | 44.61 ± 0.23 | 44.74 ± 0.15 | 2915 ± 849 | 2926 ± 783 | |
| Genotype | Total Number of Measured Pollen Grains | N | Pollen Grain Size (µm) | Pollen Number Per Anther | Small Pollen Grains (%) | |||
|---|---|---|---|---|---|---|---|---|
| Mean | Median | Mean | Median | Mean | Median | |||
| ph1b | 10100 | 12 | 42.3 ± 1.3 a,b | 43.2 ± 1.7 a | 2806 ± 426 a | 2817 ± 430 a | 47.2 ± 6.9 a | 45.1 ± 8.3 a |
| cv. Chinese Spring | 11072 | 12 | 47.4 ± 1.3 c | 48.7 ± 1.5 b | 3076 ± 370 a | 3082 ± 364 a | 15.0 ± 5.6 b | 13.2 ± 2.5 b |
| ph1c | 11139 | 10 | 40.9 ± 1.9 b | 42.4 ± 2.6 a | 3713 ± 497 c | 3681 ± 593 c | 34.3 ± 8.2 c | 34.4 ± 8.3 c |
| cv. Cappelli | 11840 | 10 | 43.7 ± 1.5 a | 44.5 ± 1.5 a | 3947 ± 270 c | 3950 ± 248 c | 11.1 ± 3.8 b | 10.7 ± 4.0 b |
| CRISPR Tazip4-B2 | 13904 | 10 | 43.1 ± 2.1 a | 43.4 ± 3 a | 2317 ± 333 b | 2354 ± 379 a,b | 48.5 ± 13.4 a | 48.6 ± 13.4 a |
| cv. Fielder | 11313 | 10 | 47.3 ± 1.0 c | 49.1 ± 1.2 b | 1886 ± 265 b | 1875 ± 231 b | 17.9 ± 3.9 b | 19.1 ± 6.3 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alabdullah, A.K.; Moore, G.; Martín, A.C. A Duplicated Copy of the Meiotic Gene ZIP4 Preserves up to 50% Pollen Viability and Grain Number in Polyploid Wheat. Biology 2021, 10, 290. https://doi.org/10.3390/biology10040290
Alabdullah AK, Moore G, Martín AC. A Duplicated Copy of the Meiotic Gene ZIP4 Preserves up to 50% Pollen Viability and Grain Number in Polyploid Wheat. Biology. 2021; 10(4):290. https://doi.org/10.3390/biology10040290
Chicago/Turabian StyleAlabdullah, Abdul Kader, Graham Moore, and Azahara C. Martín. 2021. "A Duplicated Copy of the Meiotic Gene ZIP4 Preserves up to 50% Pollen Viability and Grain Number in Polyploid Wheat" Biology 10, no. 4: 290. https://doi.org/10.3390/biology10040290
APA StyleAlabdullah, A. K., Moore, G., & Martín, A. C. (2021). A Duplicated Copy of the Meiotic Gene ZIP4 Preserves up to 50% Pollen Viability and Grain Number in Polyploid Wheat. Biology, 10(4), 290. https://doi.org/10.3390/biology10040290








