Anchored Phylogenomics, Evolution and Systematics of Elateridae: Are All Bioluminescent Elateroidea Derived Click Beetles?
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. ElaterBaits Probe Design
2.3. Sample Preparation and DNA Extraction
2.4. Library Preparation, Hybridization and Sequencing
2.5. Read Assembly and Orthology Assessment Pipeline
2.6. Alignment, Trimming and Manual Inspection
2.7. Phylogenetic Analyses
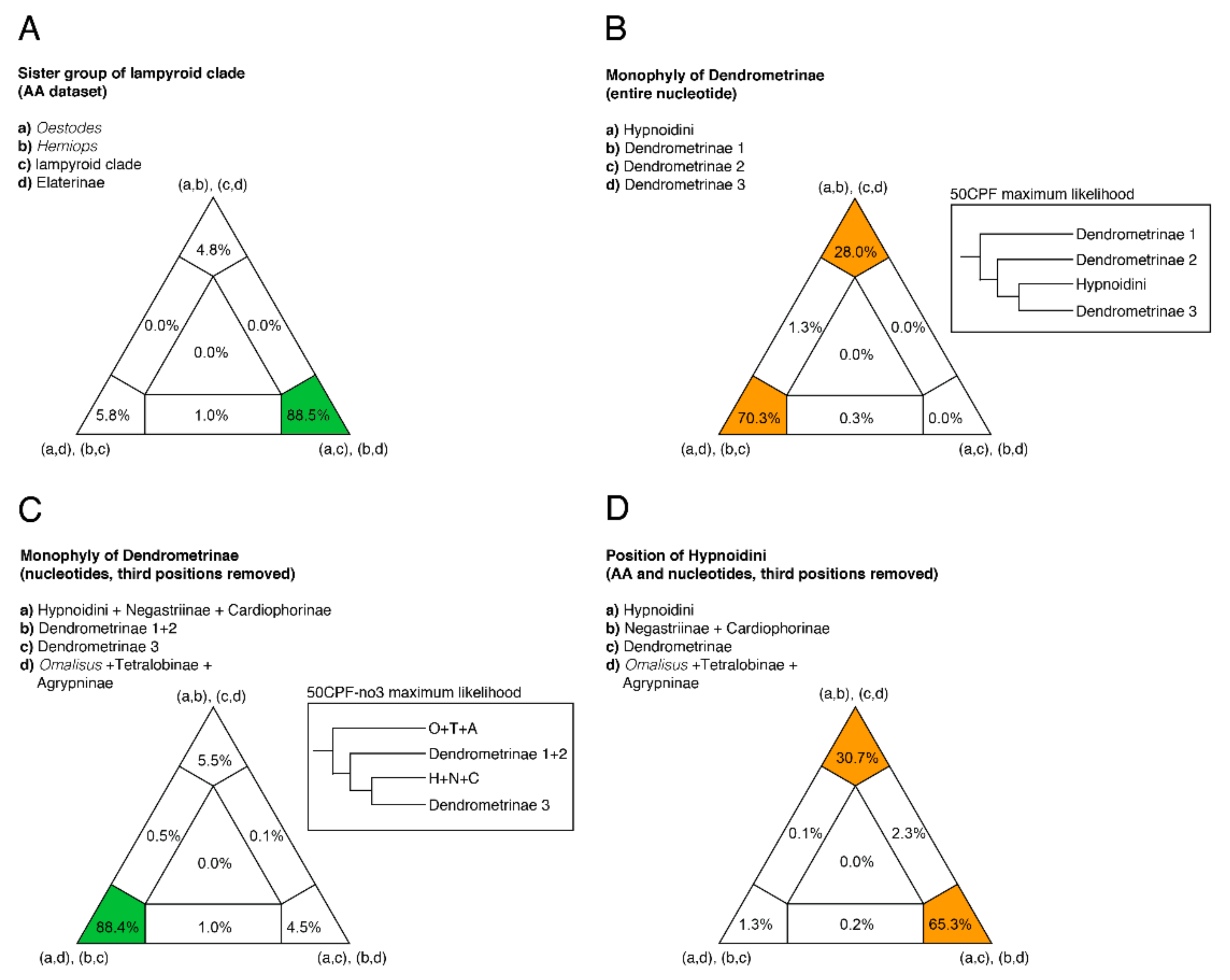
2.8. Four-cluster Likelihood Mapping
3. Results
3.1. Dataset and Target Capture
3.2. Phylogenetic Analyses
4. Discussion
4.1. Monophyly of the Elateridae
4.2. Major Divisions of the Elateridae and Their Morphology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, C.; Lawrence, J.F.; Rosa, S.P. Elateridae Leach, 1815. In Handbook of Zoology, Arthropoda: Insecta: Coleoptera Beetles; Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformia partim); Leschen, R.A.B., Beutel, R.G., Lawrence, J.F., Eds.; Walter de Gruyter GmbH & Co.: Berlin, Germany, 2010; Volume 2, pp. 75–103. [Google Scholar]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef] [Green Version]
- McKenna, D.D.; Wild, A.L.; Kanda, K.; Bellamy, C.L.; Beutel, R.G.; Caterino, M.S.; Farnum, C.W.; Hawks, D.C.; Ivie, M.A.; Jameson, M.L.; et al. The beetle tree of life reveals that Coleoptera survived end-Permian mass extinction to diversify during the Cretaceous terrestrial revolution. Syst. Entomol. 2015, 40, 835–880. [Google Scholar] [CrossRef] [Green Version]
- Kundrata, R.; Packova, G.; Hoffmannova, J. Fossil genera in Elateridae (Insecta, Coleoptera): A Triassic origin and Jurassic diversification. Insects 2020, 11, 394. [Google Scholar] [CrossRef]
- Kundrata, R.; Packova, G.; Prosvirov, A.; Hoffmannova, J. The fossil record of Elateridae (Coleoptera: Elateroidea): Described species, current problems and future prospects. Insects 2021, 12, 286. [Google Scholar] [CrossRef]
- Kusy, D.; He, J.-W.; Bybee, S.M.; Motyka, M.; Bi, W.-X.; Podsiadlowski, L.; Li, X.-Y.; Bocak, L. Phylogenomic relationships of bio-luminescent elateroids define the ‘lampyroid’ clade with clicking Sinopyrophoridae as its earliest member. Syst. Entomol. 2021, 46, 111–123. [Google Scholar] [CrossRef]
- Bouchard, P.; Smith, A.B.T.; Douglas, H.B.; Gimmel, M.L.; Brunke, A.J.; Kanda, K. Biodiversity of Coleoptera. In Insect Biodiversity: Science and Society, 2nd ed.; Foottit, R.G., Adler, P.H., Eds.; John Wiley and Sons Ltd.: West Sussex, UK, 2017; pp. 337–417. [Google Scholar]
- Calder, A.A.; Lawrence, J.F.; Trueman, J.W.H. Austrelater, gen. nov. (Coleoptera: Elateridae), with a description of the larva and comments on elaterid relationships. Invertebr. Taxon. 1993, 7, 1349–1394. [Google Scholar] [CrossRef]
- Douglas, H. Phylogenetic relationships of Elateridae inferred from adult morphology, with special reference to the position of Cardiophorinae. Zootaxa 2011, 2900, 1–45. [Google Scholar] [CrossRef]
- Kundrata, R.; Bocak, L. The phylogeny and limits of Elateridae (Insecta, Coleoptera): Is there a common tendency of click beetles to soft-bodiedness and neoteny? Zool. Scr. 2011, 40, 364–378. [Google Scholar] [CrossRef]
- Kundrata, R.; Bocakova, M.; Bocak, L. The comprehensive phylogeny of the superfamily Elateroidea (Coleoptera: Elateriformia). Mol. Phylogenet. Evol. 2014, 76, 162–171. [Google Scholar] [CrossRef]
- Kundrata, R.; Gunter, N.L.; Douglas, H.; Bocak, L. Next step toward a molecular phylogeny of click-beetles (Coleoptera: Elateridae): Redefinition of Pityobiinae, with a description of a new subfamily Parablacinae from the Australasian Region. Austral. Entomol. 2016, 55, 291–302. [Google Scholar] [CrossRef]
- Kundrata, R.; Gunter, N.L.; Janosikova, D.; Bocak, L. Molecular evidence for the subfamilial status of Tetralobinae (Coleoptera: Elateridae), with comments on parallel evolution of some phenotypic characters. Arthropod. Syst. Phyl. 2018, 76, 137–145. [Google Scholar]
- Bocak, L.; Motyka, M.; Bocek, M.; Bocakova, M. Incomplete sclerotization and phylogeny: The phylogenetic classification of Plastocerus (Coleoptera: Elateroidea). PLoS ONE 2018, 13, e0194026. [Google Scholar] [CrossRef]
- Bi, W.-X.; He, J.-W.; Chen, C.-C.; Kundrata, R.; Li, X.-Y. Sinopyrophorinae, a new subfamily of Elateridae (Coleoptera, Elateroidea) with the first record of a luminous click beetle in Asia and evidence for multiple origins of bioluminescence in Elateridae. ZooKeys 2019, 864, 79–97. [Google Scholar] [CrossRef] [Green Version]
- Kusy, D.; Motyka, M.; Bocek, M.; Vogler, A.P.; Bocak, L. Genome sequences identify three families of Coleoptera as morphologically derived click beetles (Elateridae). Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.J.; Stanger-Hall, K.F.; Branham, M.A.; Da Silveira, L.F.L.; Lower, S.E.; Hall, D.W.; Li, X.-Y.; Lemmon, A.R.; Lemmon, E.M.; Bybee, S.M. Higher-level phylogeny and reclassification of Lampyridae (Coleoptera: Elateroidea). Insect Syst. Divers. 2019, 3, 1–15. [Google Scholar] [CrossRef]
- Douglas, H.B. World reclassification of the Cardiophorinae (Coleoptera, Elateridae), based on phylogenetic analyses of morpho-logical characters. ZooKeys 2017, 655, 1–130. [Google Scholar] [CrossRef] [Green Version]
- Kusy, D.; Motyka, M.; Bocak, L. Click Beetle mitogenomics with the definition of a new subfamily Hapatesinae from Australasia (Coleoptera: Elateridae). Insects 2021, 12, 17. [Google Scholar] [CrossRef]
- Sagegami-Oba, R.; Oba, Y.; Ôhira, H. Phylogenetic relationships of click beetles (Coleoptera: Elateridae) inferred from 28S ribosomal DNA: Insights into the evolution of bioluminescence in Elateridae. Mol. Phylogenet. Evol. 2007, 42, 410–421. [Google Scholar] [CrossRef]
- Hamilton, C.A.; Lemmon, A.R.; Lemmon, E.M.; Bond, J.E. Expanding anchored hybrid enrichment to resolve both deep and shallow relationships within the spider tree of life. BMC Evol. Biol. 2016, 16, 212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, C.H.; Allen, J.M.; Lemmon, A.R.; Lemmon, E.M.; Takiya, D.M.; Evangelista, O.; Walden, K.K.O.; Grady, P.G.S.; Johnson, K.P. Anchored hybrid enrichment-based phylogenomics of leafhoppers and treehoppers (Hemiptera: Cicadomorpha: Membracoidea). Insect Syst. Divers. 2017, 1, 57–72. [Google Scholar] [CrossRef]
- Haddad, S.; Shin, S.; Lemmon, A.R.; Lemmon, E.M.; Svacha, P.; Farrell, B.; Ślipiński, A.; Windsor, D.; McKenna, D.D. Anchored hybrid enrichment provides new insights into the phylogeny and evolution of Longhorned Beetles (Cerambycidae). Syst. Entomol. 2017, 43, 68–89. [Google Scholar] [CrossRef]
- Shin, S.; Clarke, D.J.; Lemmon, A.R.; Lemmon, E.M.; Aitken, A.L.; Haddad, S.; Farrell, B.D.; Marvaldi, A.E.; Oberprieler, R.G.; McKenna, D.D. Phylogenomic data yield new and robust insights into the phylogeny and evolution of Weevils. Mol. Biol. Evol. 2018, 35, 823–836. [Google Scholar] [CrossRef]
- Bouchard, P.; Bousquet, Y.; Davies, A.E.; Alonso-Zarazaga, M.A.; Lawrence, J.F.; Lyal, C.H.C.; Newton, A.F.; Reid, C.A.M.; Schmitt, M.; Ślipiński, S.A.; et al. Family-group names in Coleoptera (Insecta). ZooKeys 2011, 88, 1–972. [Google Scholar] [CrossRef] [Green Version]
- Rosa, S.P. Phylogenetic analysis and taxonomic revision of Physodactylinae (Coleoptera, Elateridae). Pap. Avulsos Zool. 2014, 54, 217–292. [Google Scholar] [CrossRef] [Green Version]
- Petersen, M.; Meusemann, K.; Donath, A.; Dowling, D.; Liu, S.; Peters, R.S.; Podsiadlowski, L.; Vasilikopoulos, A.; Zhou, X.; Misof, B.; et al. Orthograph: A versatile tool for mapping coding nucleotide sequences to clusters of orthologous genes. BMC Bioinform. 2017, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magis, C.; Taly, J.F.; Bussotti, G.; Chang, J.M.; Di Tommaso, P.; Erb, I.; Espinosa-Carrasco, J.; Notredame, C. T-coffee: Tree-based consistency objective function for alignment evaluation. Methods Mol. Biol. 2014, 1079, 117–129. [Google Scholar]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Faircloth, B.C. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 2016, 32, 786–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [Green Version]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate Paired Shotgun Read Merging via Overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L.; et al. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 27, 768–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [Green Version]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: A de novo transcriptome assembler and its application to RNA-Seq data. GigaScience 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedin, M.; Derkarabetian, S.; Ramírez, M.J.; Vink, C.; Bond, J.E. Phylogenomic reclassification of the world’s most venomous spiders (Mygalomorphae, Atracinae), with implications for venom evolution. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. Mafft: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowiec, M.L. AMAS: A fast tool for alignment manipulation and computing of summary statistics. PeerJ 2016, 4, e1660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Espeland, M.; Breinholt, J.; Willmott, K.R.; Warren, A.D.; Vila, R.; Toussaint, E.F.; Maunsell, S.C.; Aduse-Poku, K.; Talavera, G.; Eastwood, R.; et al. A comprehensive and dated phylogenomic analysis of butterflies. Curr. Biol. 2018, 28, 770–778.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gough, H.M.; Allen, J.M.; A Toussaint, E.F.; Storer, C.G.; Kawahara, A.Y. Transcriptomics illuminate the phylogenetic backbone of tiger beetles. Biol. J. Linn. Soc. 2020, 129, 740–751. [Google Scholar] [CrossRef]
- Duchêne, D.; Tong, K.J.; Foster, C.S.P.; Duchêne, S.; Lanfear, R.; Ho, S.Y.W. Linking branch lengths across sets of loci provides the highest statistical support for phylogenetic inference. Mol. Biol. Evol. 2019, 37, 1202–1210. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maxi-mum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 15–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef] [Green Version]
- Strimmer, K.; von Haeseler, A. Likelihood-mapping: A simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 1997, 94, 6815–6819. [Google Scholar] [CrossRef] [Green Version]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef]
- Vasilikopoulos, A.; Balke, M.; Beutel, R.G.; Donath, A.; Podsiadlowski, L.; Pflug, J.M.; Waterhouse, R.; Meusemann, K.; Peters, R.S.; Escalona, H.E.; et al. Phylogenomics of the superfamily Dytiscoidea (Coleoptera: Adephaga) with an evaluation of phylogenetic conflict and systematic error. Mol. Phylogenet. Evol. 2019, 135, 270–285. [Google Scholar] [CrossRef] [Green Version]
- Brunke, A.J.; Hansen, A.K.; Salnitska, M.; Kypke, J.L.; Predeus, A.V.; Escalona, H.; Chapados, J.T.; Eyres, J.; Richter, R.; Smetana, A.; et al. The limits of Quediini at last (Staphylinidae: Staphylininae): A rove beetle mega-radiation resolved by comprehensive sampling and anchored phylogenomics. Syst. Entomol. 2021, 46, 396–421. [Google Scholar] [CrossRef]
- Molloy, E.K.; Warnow, T. To include or not to include: The impact of gene filtering on species tree estimation methods. Syst. Biol. 2017, 67, 285–303. [Google Scholar] [CrossRef]
- Beutel, R.G. Phylogenetic analysis of Elateriformia (Coleoptera: Polyphaga) based on larval characters. J. Zool. Syst. Evol. Res. 1995, 33, 145–171. [Google Scholar] [CrossRef]
- Fallon, T.R.; Lower, S.E.; Chang, C.-H.; Bessho-Uehara, M.; Martin, G.J.; Bewick, A.J.; Behringer, M.; Debat, H.J.; Wong, I.; Day, J.C.; et al. Firefly genomes illuminate parallel origins of bioluminescence in beetles. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Costa, C. Note on the bioluminescence of Balgus schnusei (Heller, 1974) (Trixagidae, Coleoptera). Rev. Bras. Entomol. 1984, 28, 397–398. [Google Scholar]
- Oba, Y.; Konishi, K.; Yano, D.; Shibata, H.; Kato, D.; Shirai, T. Resurrecting the ancient glow of the fireflies. Sci. Adv. 2020, 6, eabc5705. [Google Scholar] [CrossRef]
- Traugott, M.; Benefer, C.M.; Blackshaw, R.P.; Van Herk, W.G.; Vernon, R.S. Biology, ecology, and control of Elaterid Beetles in agricultural land. Annu. Rev. Entomol. 2015, 60, 313–334. [Google Scholar] [CrossRef] [Green Version]
- Baalbergen, E.; Schelfhorst, R.; Schilthuizen, M. Drilus larvae in the Netherlands (Coleoptera: Elateridae: Drilini). Entomol. Bericht. 2016, 76, 165–173. [Google Scholar]
- Kondo, A.; Tanaka, F. An experimental study of predation by the larvae of the firefly, Luciola lateralis Motschulsky (Coleoptera: Lampyridae) on the apple snail, Pomacea canaliculata Lamarck (Mesogastropoda: Pilidae). Jpn. J. Appl. Entomol. Zoöl. 1989, 33, 211–216. [Google Scholar] [CrossRef]
- Symondson, W.O.C. Coleoptera (Carabidae, Staphylinidae, Lampyridae, Drilidae and Silphidae) as predators of terrestrial gastro-pods. In Natural Enemies of Terrestrial Molluscs; Barker, G.M., Ed.; Landcare Research: Hamilton, New Zealand, 2004; pp. 37–84. [Google Scholar]
- Traugott, M.; Pázmándi, C.; Kaufmann, R.; Juen, A. Evaluating 15N/14N and 13C/12C isotope ratio analysis to investigate trophic relationships of elaterid larvae (Coleoptera: Elateridae). Soil Biol. Biochem. 2007, 39, 1023–1030. [Google Scholar] [CrossRef]
- Fleutiaux, E. Les élatérides de l’indochine Française. Huitième et dernière partie. Ann. Soc. Entomol. Fr. 1940, 109, 19–40. (In French) [Google Scholar]
- Muona, J.; Chang, H.; Ren, D. The clicking Elateroidea from Chinese Mesozoic deposits (Insecta, Coleoptera). Insects 2020, 11, 875. [Google Scholar] [CrossRef] [PubMed]
- Hyslop, J.A. The phylogeny of the Elateridae based on larval characters. Ann. Entomol. Soc. Am. 1917, 10, 241–263. [Google Scholar] [CrossRef]
- Ôhira, H. Morphological and Taxonomic Study on the Larvae of Elateridae in Japan (Coleoptera); Entomological Laboratory Aichi Gakugei University: Okazaki, Japan, 1962; pp. 1–179. [Google Scholar]
- Stibick, J.N.L. Classification of the Elateridae (Coleoptera). Relationships and classification of the subfamilies and tribes. Pac. Insects 1979, 20, 145–186. [Google Scholar]
- Johnson, P.J. New species of Dioxypterus Fairmaire from Tonga and Fiji, with new distribution records, a tribal reassignment, and key to the species of the region (Coleoptera: Elateridae). Pan Pac. Entomol. 1997, 73, 156–167. [Google Scholar]
- Dolin, V.G. Wing venation of click beetles (Coleoptera, Elateridae) and its importance for taxonomy of the family. Zool. Zhur 1975, 54, 1618–1633. [Google Scholar]
- Calder, A.A. Click Beetles: Genera of the Australian Elateridae (Coleoptera). Monographs on Invertebrate Taxonomy; CSIRO: Canberra, Australia, 1996; Volume 2, pp. 1–401. [Google Scholar]
- Kundrata, R.; Kubaczkova, M.; Prosvirov, A.S.; Douglas, H.B.; Fojtikova, A.; Costa, C.; Bousquet, Y.; Alonso-Zarazaga, M.A.; Bouchard, P. World catalogue of the genus-group names in Elateridae (Insecta, Coleoptera). Part I: Agrypninae, Campyloxeninae, Hemiopinae, Lissominae, Oestodinae, Parablacinae, Physodactylinae, Pityobiinae, Subprotelaterinae, Tetralobinae. ZooKeys 2019, 839, 83–154. [Google Scholar] [CrossRef]
- Kundrata, R.; Bocak, L. Molecular phylogeny reveals the gradual evolutionary transition to soft-bodiedness in click-beetles and identifies sub-Saharan Africa as a cradle of diversity for Drilini (Coleoptera: Elateridae). Zoöl. J. Linn. Soc. 2019, 187, 413–452. [Google Scholar] [CrossRef]
- Dajoz, R. Anatomie et importance taxinomique des voies génitales femelles d’origine ectodermique chez les Elateridae (Insectes, Coléoptères). Cah. Nat. Bull. Nat. Paris 1964, 20, 55–72. (In French) [Google Scholar]
- Rosa, S.P.; Németh, T.; Kundrata, R. Comparative morphology of immature stages of Ludioctenus cyprius (Baudi di Selve, 1871) (Coleoptera: Elateridae: Agrypninae), with discussion on the monophyly of Hemirhipini. Zool. Anz. 2019, 283, 33–39. [Google Scholar] [CrossRef]
- Cate, P.C. Elateridae Leach, 1815 (- Cebrioninae, Lissominae, Subprotelaterinae). In Catalogue of Palaearctic Coleoptera; Löbl, I., Smetana, A., Eds.; Apollo Books: Stenstrup, Denmark, 2007; Volume 4, pp. 89–209. [Google Scholar]
- Von Hayek, C.M.F. A reclassification of the subfamily Agrypninae (Coleoptera: Elateridae). Bull. Brit. Mus. Nat. Hist. 1973, 20, 1–309. [Google Scholar]
- Johnson, P.J. Elateridae Leach 1815. In American Beetles. Polyphaga: Scarabaeoidea through Curculionoidea; Arnett, R.H., Thomas, M.C., Skelley, P.E., Frank, J.H., Eds.; CRC Press: Boca Raton, FL, USA, 2002; Volume 2, pp. 160–173. [Google Scholar]
- Schimmel, R.; Tarnawski, D.; Han, T.; Platia, G. Monograph of the new tribe Selatosomini from China (Elateridae: Denticollinae). Part I: Genera Pristilophus Latreille, 1834 stat. nov., Selatosomus Stephens, 1830, Warchalowskia (Tarnawski, 1995) stat. nov., and Sinophotistus gen. nov. Pol. Entomol. Monogr. 2015, 11, 1–328. [Google Scholar]
- Li, Y.-D.; Kundrata, R.; Tihelka, E.; Liu, Z.; Huang, D.; Cai, C. Cretophengodidae, a new Cretaceous beetle family, sheds light on the evolution of bioluminescence. Proc. R. Soc. B Biol. Sci. 2021, 288, 20202730. [Google Scholar] [CrossRef] [PubMed]




| Clades/Analyses | CU aa | CP aa | CU n | CP n | CPF n | CPF no3 | CU no3 | CP no3 | AS aa | AS n |
|---|---|---|---|---|---|---|---|---|---|---|
| Lissominae + (Elateridae + lampyroids) | 79/71 | 89/75 | 89/91 | 85/82 | 91/89 | 83/79 | 67/39 | 77/67 | 0.95 | NR |
| Elateridae | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Lissominae | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | NR |
| Elateridae + lampyroids (minus Lissominae) | 100/93 | 100/96 | 100/100 | 100/100 | 100/100 | 100/100 | 100/97 | 100/99 | 0.87 | NR |
| Oestodes + lampyroids | 98/98 | 99/97 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | NR | NR |
| Oestodes + lampyroids + Elaterinae | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | NR | NR |
| Oestodes + lampyroids + Hemiops + Elaterinae | 100/100 | 100/100 | 99/99 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 0.68 | 0.81 |
| Elaterinae (incl. Cebrionini, Aplastini, Eudicronychini) | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | 1 |
| Elaterinae: “Ampedus clade” | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/99 | 0.71 | 1 |
| Elaterinae: “Elater clade” | 100/100 | 100/99 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | NR | 1 |
| Each of Agriotini; Ampedini; Dicrepidiini; Elaterini; Megapenthini; and Physorhinini | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Each of Eudicronychini and Synaptini + Agriotini + Pomachiliini | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | 1 |
| Pityobius + Tibionema (=Pityobiinae) | 99/99 | 100/99 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 0.67 | 1 |
| Pityobius + Tibionema + Hapatesus | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 0.84 | 1 |
| Pityobius + Tibionema + Hapatesus + Omalisus + Agrypninae + Tetralobinae + Cardiophorinae + Negastriinae + Hypnoidini + Dendrometrinae* | 100/100 | 100/99 | NR | 100/99 | 100/100 | 100/100 | 100/99 | 100/100 | 0.99 | NR |
| Omalisus + Agrypninae + Tetralobinae + Cardiophorinae + Negastriinae + Hypnoidini + Dendrometrinae* | 100/96 | 100/93 | NR | 100/99 | 100/100 | 100/100 | 100/99 | 100/100 | 0.49 | NR |
| Omalisus + Agrypninae + Tetralobinae | 97/95 | 96/90 | NR | NR | NR | 100/100 | 100/99 | 100/100 | 0.54 | NR |
| Agrypninae + Tetralobinae | 100/100 | 100/100 | NR | NR | NR | 96/85 | 87/82 | 99/96 | 0.91 | NR |
| Omalisus + Agrypninae | NR | NR | 100/100 | 100/100 | 100/100 | NR | NR | NR | NR | NR |
| Each of Agrypninae and Tetrolobinae | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | 1 |
| Each of Agrypnini (incl. Lacon Laporte, 1838 and Elasmosomus Schwarz, 1902); and Hemirhipini (incl. Ludioctenus Fairmaire, 1893); and Oophorini | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Each of Oophorini (excl. Pachyderes Guérin-Méneville, 1829) and Drilini | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | 1 |
| Cardiophorinae + Negastriinae + Hypnoidini + Dendrometrinae * | 100/100 | 100/100 | NR | NR | NR | 100/100 | 100/100 | 100/100 | 0.49 | NR |
| Cardiophorinae + Negastriinae + Hypnoidini | 100/100 | 100/100 | NR | NR | NR | 100/100 | 100/100 | 100/100 | NR | NR |
| Cardiophorinae + Negastriinae + Tetralobinae | NR | NR | 100/100 | 100/100 | 100/100 | NR | NR | NR | NR | NR |
| Cardiophorinae + Negastriinae | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | NR |
| Each of Cardiophorinae and Hypnoidini | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | 1 |
| Negastriinae | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 0.99 | 1 |
| Hypnoidini + Dendrometrinae* | NR | NR | 100/ 100 | 100/ 100 | 100/ 100 | NR | NR | NR | 1 | 1 |
| Dendrometrinae* (without Hypnoidini) | 98/96 | 99/97 | NR | NR | NR | NR | NR | NR | 0.67 | 0.92 |
| Dendrometrinae (without Plastocerus) | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Oxynopterini + Plastocerus + Dimini | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | NR | NR |
| Each of Oxynopterini + Plastocerus and Dimini | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 1 | 1 |
| Dendrometrini + Semiotini + Prosternini + Selatosomini + Hypnoidini | NR | NR | 100/100 | 100/100 | 100/100 | NR | NR | NR | 1 | NR |
| Prosternini + Selatosomini + Ctenicerini | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Dendrometrini excl. Semiotini | NR | NR | NR | NR | NR | NR | NR | NR | 0.39 | NR |
| Dendrometrini incl. Semiotini | 99/99 | 100/99 | 94/90 | 94/89 | 89/87 | 96/87 | 96/93 | 99/96 | NR | NR |
| Each of Denticollina + Athous Eschscholtz, 1829 + Hemicrepidiina and Dendrometrina (minus Athous) | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 0.79 | 1 |
| Denticollina + Athous + Hemicrepidiina + Semiotini | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | 100/100 | NR | NR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Douglas, H.B.; Kundrata, R.; Brunke, A.J.; Escalona, H.E.; Chapados, J.T.; Eyres, J.; Richter, R.; Savard, K.; Ślipiński, A.; McKenna, D.; et al. Anchored Phylogenomics, Evolution and Systematics of Elateridae: Are All Bioluminescent Elateroidea Derived Click Beetles? Biology 2021, 10, 451. https://doi.org/10.3390/biology10060451
Douglas HB, Kundrata R, Brunke AJ, Escalona HE, Chapados JT, Eyres J, Richter R, Savard K, Ślipiński A, McKenna D, et al. Anchored Phylogenomics, Evolution and Systematics of Elateridae: Are All Bioluminescent Elateroidea Derived Click Beetles? Biology. 2021; 10(6):451. https://doi.org/10.3390/biology10060451
Chicago/Turabian StyleDouglas, Hume B., Robin Kundrata, Adam J. Brunke, Hermes E. Escalona, Julie T. Chapados, Jackson Eyres, Robin Richter, Karine Savard, Adam Ślipiński, Duane McKenna, and et al. 2021. "Anchored Phylogenomics, Evolution and Systematics of Elateridae: Are All Bioluminescent Elateroidea Derived Click Beetles?" Biology 10, no. 6: 451. https://doi.org/10.3390/biology10060451
APA StyleDouglas, H. B., Kundrata, R., Brunke, A. J., Escalona, H. E., Chapados, J. T., Eyres, J., Richter, R., Savard, K., Ślipiński, A., McKenna, D., & Dettman, J. R. (2021). Anchored Phylogenomics, Evolution and Systematics of Elateridae: Are All Bioluminescent Elateroidea Derived Click Beetles? Biology, 10(6), 451. https://doi.org/10.3390/biology10060451







