COVID-19 Shuts Doors to Flu but Keeps Them Open to Rhinoviruses
Abstract
Simple Summary
Abstract
1. Introduction
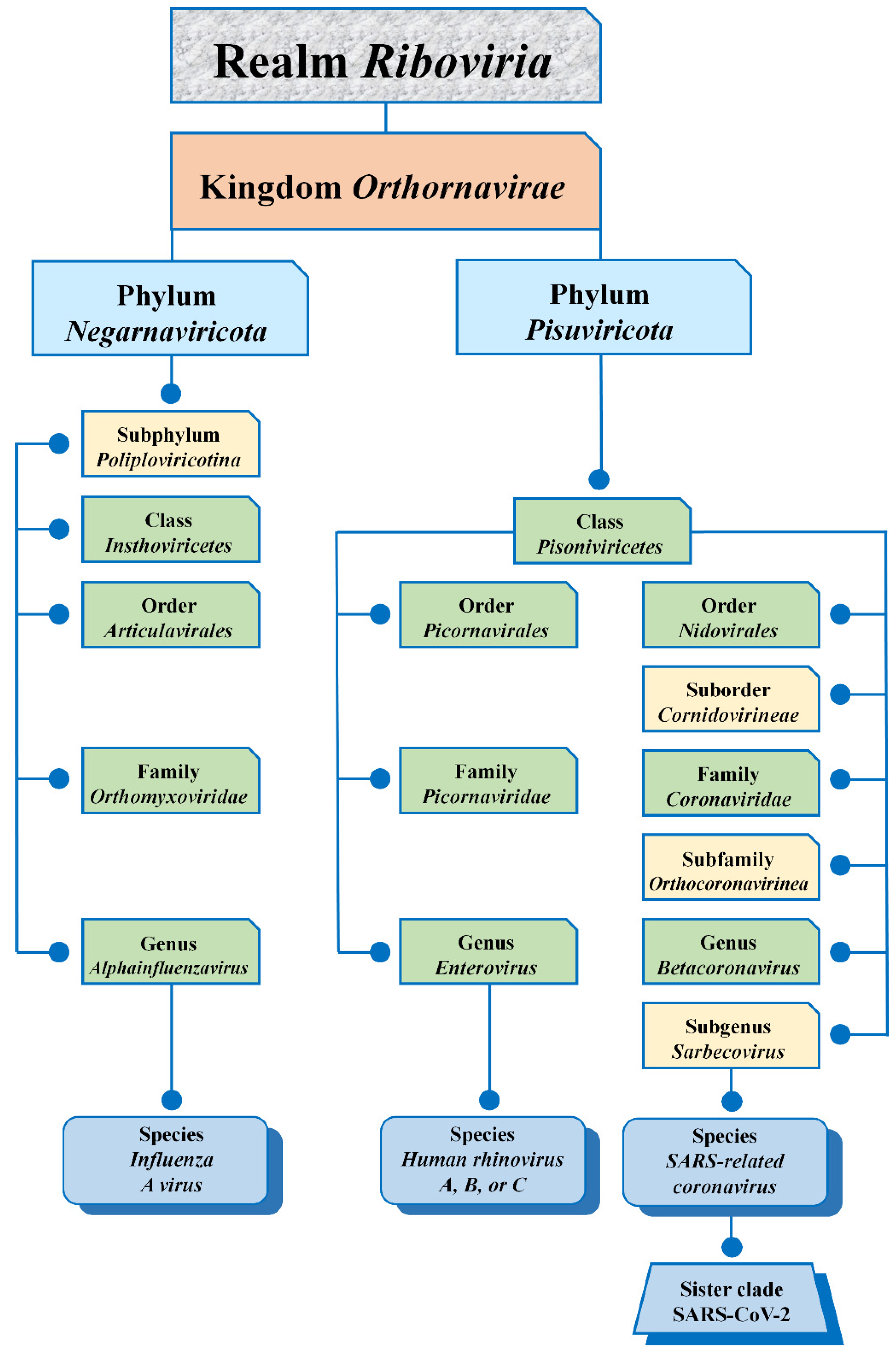
2. Virus Taxonomy
3. Virion Morphology
| Properties [refs] | Causative Agent | ||
|---|---|---|---|
| Rhinovirus | Influenza a Virus | SARS-CoV-2 | |
| VIRION MORPHOLOGY [15,16,17,18,19,20,21] | |||
| Size (nucleocapsid diameter) | Small-sized (~30 nm) | Medium-sized (80–120 nm) | Medium-sized (~120 nm) |
| Nucleocapsid symmetry | Icosahedral symmetry | Icosahedral symmetry | Icosahedral symmetry |
| Shape | Spherical shape | Spherical shape, sometimes filamentous morphology | Spherical shape |
| VIRION STRUCTURE [15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33] | |||
| Capsid (coat protein) | Yes | Yes | Yes |
| Envelop | No | Yes | Yes |
| Glycoprotein spike | No | HA and NA | S glycoprotein |
| Polybasic cleavage site | No | Only in avian IAV | Yes |
| RNA dependent RNA polymerase | Yes | Yes | Yes |
| HA esterase glycoprotein | No | No | Yes |
| PHYSICOCHEMICAL PROPERTIES [2,33,34] | |||
| Thermostability | Environmentally stable to temperature (heat-resistant) | Environmentally labile to temperature (heat-sensitive) | Environmentally labile to temperature (heat-sensitive) |
| Stability to drying | Stable to drying | Disrupted by drying | Disrupted by drying |
| Stability to detergents | Stable to detergents | Disrupted by detergents | Disrupted by detergents |
| Acid stability, pH sensitivity | Unstable below pH 5–6 | Disrupted by acid | Disrupted by acid |
| GENOME ORGANIZATION [22,26,27] | |||
| Nucleic acid structure | RNA | RNA | RNA |
| Size | ~7 kb | ~14 kb | ~30 kb |
| Type of nucleic acid molecule | Linear | Linear | Linear |
| Helix form of nucleic acid | Single-stranded | Single-stranded | Single-stranded |
| Segmented/nonsegmented genome | Nonsegmented | Segmented | Nonsegmented |
| Positive/negative-strand RNA | Positive-sense RNA | Negative-sense RNA | Positive-sense RNA |
| VIRAL REPLICATION [2,22,29,35,36,37,38,39,40,41] | |||
| Recognition and attachment | A canyon in VP1 protein is the site of the attachment of sialic acids to the cell surface | Binding to sialic acids on a host cell by the receptor-binding domain of HA1 | Binding to the cell surface protein ACE2 by S protein |
| Entry into the host-cell | Endocytosis, pinocytosis | Endocytosis | Endocytosis or at plasma membrane fusion |
| Infectivity of naked genomic RNA | Infectious | Not infectious | Infectious |
| Transcription and replication | Cytoplasm | Nucleus | Cytoplasm |
| Synthesis of viral proteins | Cytoplasm | Cytoplasm | Cytoplasm |
| Virion assembly | Cytoplasm | Cytoplasm | Cytoplasm |
| Virion release | Lytic or non-lytic releases | Budding | Exocytosis |
4. Virion Structure
5. Physicochemical Properties
5.1. Environment
5.2. Stability
5.3. Sterilization
6. Genome Organization
7. Viral Replication
8. Pathogenesis
8.1. Temperature Sensitivity of Replication
8.2. Differences and Similarities between Influenza and COVID-19
| Properties [refs] | Causative Agent | ||
|---|---|---|---|
| Rhinovirus | Influenza Virus | SARS-CoV-2 | |
| PATHOGENESIS AND CLINICAL FEATURES [1,2,3,4,21,52,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104] | |||
| “Entrance gate” of infection (the primary route of entry) | Upper respiratory tract (mouth and nose) | Upper respiratory tract (mouth and nose) | Upper respiratory tract (mouth and nose) |
| Upper respiratory tract infection | Common | Common | Common |
| Replication at high temperature | Sometimes | Common | Common |
| Lower respiratory tract infection | Sometimes | Sometimes | Common |
| Incubation period | 1–3 days | 1–4 days | 2–14 days |
| Symptoms | Gradual | Abrupt | Gradual |
| Sudden onset | Smooth onset | Very common early symptom | Sudden or smooth onset |
| Running nose (rhinorrhea) | Common | Common | Common |
| Nasal discharge | Common | Common | Common |
| Nasal congestion | Common | Sometimes | Common |
| Sneezing | Very common | Sometimes | Common |
| Loss of smell and taste | Rare | Sometimes | Very common early symptom |
| Shortness of breath | Mild | Sometimes | Common |
| Sore throat | Very common | Sometimes | Sometimes/common |
| Cough | Common (mild to moderate, hacking) | Common (dry cough, can be severe) | Common (dry cough, can be severe) |
| Headache | Rare | Common | Common |
| Muscle pain (body aches) | Sometimes (slight) | Very common (often severe) | Very common (often severe) |
| Chilliness and fever | Rare in adults, possible in children | Very common; may have chills | Very common; may have chills |
| Malaise | Sometimes | Very common | Very common |
| Fatigue, weakness | Sometimes | Very common (can last for weeks) | Very common (can last for weeks) |
| Diarrhea | Rare | Sometimes | Sometimes |
| Loss of appetite | Sometimes | Common | Sometimes |
8.3. Pathogenesis of Rhinovirus Infection
9. Clinical Manifestations
10. Epidemiology
10.1. Origin
10.2. Transmission
10.3. Seasonality
10.4. Morbidity, Hospitalization, and Mortality
| Properties [refs] | Causative Agent | ||
|---|---|---|---|
| Rhinovirus | Influenza Virus | SARS-CoV-2 | |
| EPIDEMIOLOGY [1,10,11,82,84,86,88,89,90,120,123,124,125,126,127,128,129,130,131] | |||
| Origin | Likely not arisen from a recent zoonotic event | Likely not arisen from a recent zoonotic event except pandemic strains | No conclusive evidence yet. Likely zoonotic origin |
| Transmission | Mainly through direct contact with aerosolized particles from an infected individual | Through the spread of droplets or direct contact by sneezing or coughing from an infected individual | Through the spread of droplets or direct contact by sneezing or coughing from an infected individual |
| World distribution | Worldwide | Worldwide | Worldwide |
| Seasonality (circulation pattern) | Cold seasons, but is possible year-round | Cold seasons (fall and winter) | No conclusive evidence yet. Possibly cold seasons |
| Level of transmissibility | Moderate | High | High |
| Annually infected (global), thds | No global data available | ~1,000,000 | ~83,600 |
| Hospitalization rate (global), thds | No global data available | ~9460 | ~15,800 |
| Annual death (global), thds | No global data available | ~410 | ~1800 |
| Prevalence and incidence of the disease | Local outbreaks | Epidemics, pandemics | Pandemic |
| Circulation during an influenza epidemic season | Does not change | Circulation of seasonal influenza viruses | Yes |
| Circulation during an influenza pandemic | Does not change | Decreased circulation of seasonal influenza viruses | Not applicable |
| Circulation during the COVID-19 pandemic | Does not change | Decreased circulation of seasonal influenza viruses | Yes |
10.5. Prevalence and Incidence of the Disease
10.6. Circulation
11. Interactions between Rhinoviruses and Other Respiratory Viruses during Their Co-Circulation
11.1. The Spread of the Influenza A Virus Might Be Interrupted by HRV
11.2. Influenza Viruses Are Delayed in Spread by SARS-CoV-2
11.3. HRV Van Block SARS-CoV-2
11.4. SARS-CoV-2 Can Co-Circulate with Other Respiratory Pathogens
12. A Brief Summary of the Main Similarities and Differences between SARS-CoV-2, IAV, and HRV
13. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eccles, R. Understanding the symptoms of the common cold and influenza. Lancet Infect. Dis. 2005, 5, 718–725. [Google Scholar] [CrossRef]
- Jacobs, S.E.; Lamson, D.M.; St. George, K.; Walsh, T.J. Human rhinoviruses. Clin. Microbiol. Rev. 2013, 26, 135. [Google Scholar] [CrossRef]
- Pappas, D.E.; Hendley, J.O. The common cold and decongestant therapy. Pediatr. Rev. 2011, 32, 47–54. [Google Scholar] [CrossRef]
- Drysdale, S.B.; Mejias, A.; Ramilo, O. Rhinovirus-not just the common cold. J. Infect. 2017, 74 (Suppl. S1), S41–S46. [Google Scholar] [CrossRef]
- Self, W.H.; Williams, D.J.; Zhu, Y.; Ampofo, K.; Pavia, A.T.; Chappell, J.D.; Hymas, W.C.; Stockmann, C.; Bramley, A.M.; Schneider, E.; et al. Respiratory viral detection in children and adults: Comparing asymptomatic controls and patients with community-acquired pneumonia. J. Infect. Dis. 2016, 213, 584–591. [Google Scholar] [CrossRef]
- Charles, C.H.; Yelmene, M.; Luo, G.X. Recent advances in rhinovirus therapeutics. Curr. Drug Targets Infect. Disord. 2004, 4, 331–337. [Google Scholar] [CrossRef]
- ICTV. Virus Taxonomy: 2020 Release. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 30 July 2021).
- Kiseleva, I.; Grigorieva, E.; Larionova, N.; Al Farroukh, M.; Rudenko, L. COVID-19 in light of seasonal respiratory infections. Biology 2020, 9, 240. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, D.W.; et al. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Calderaro, A.; De Conto, F.; Buttrini, M.; Piccolo, G.; Montecchini, S.; Maccari, C.; Martinelli, M.; Di Maio, A.; Ferraglia, F.; Pinardi, F.; et al. Human respiratory viruses, including SARS-CoV-2, circulating in the winter season 2019–2020 in Parma, Northern Italy. Int. J. Infect. Dis. 2020, 102, 79–84. [Google Scholar] [CrossRef]
- Kim, D.; Quinn, J.; Pinsky, B.; Shah, N.H.; Brown, I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA 2020, 323, 2085–2086. [Google Scholar] [CrossRef]
- Laurie, K.L.; Rockman, S. Which influenza viruses will emerge following the SARS-CoV-2 pandemic? Influenza Other Respir. Viruses 2021. [Google Scholar] [CrossRef]
- WHO. Overview of Influenza Activity Globally. Influenza Update N° 397 of 5 July 2021. Available online: https://www.who.int/publications/m/item/influenza-update-n-397 (accessed on 20 July 2021).
- Waman, V.P.; Kolekar, P.S.; Kale, M.M.; Kulkarni-Kale, U. Population structure and evolution of rhinoviruses. PLoS ONE 2014, 9, e88981. [Google Scholar] [CrossRef]
- Griffiths, C.; Drews, S.J.; Marchant, D.J. Respiratory syncytial virus: Infection, detection, and new options for prevention and treatment. Clin. Microbiol. Rev. 2017, 30, 277–319. [Google Scholar] [CrossRef]
- Selvaraj, M.; Yegambaram, K.; Todd, E.; Richard, C.A.; Dods, R.L.; Pangratiou, G.M.; Trinh, C.H.; Moul, S.L.; Murphy, J.C.; Mankouri, J.; et al. The structure of the human respiratory syncytial virus M2-1 protein bound to the interaction domain of the phosphoprotein p defines the orientation of the complex. mBio 2018, 9. [Google Scholar] [CrossRef]
- Barreto-Vieira, D.F.; da Silva, M.A.N.; Garcia, C.C.; Miranda, M.D.; Matos, A.D.R.; Caetano, B.C.; Resende, P.C.; Motta, F.C.; Siqueira, M.M.; Girard-Dias, W.; et al. Morphology and morphogenesis of SARS-CoV-2 in Vero-E6 cells. Mem Inst. Oswaldo Cruz 2021, 116, e200443. [Google Scholar] [CrossRef]
- Hrebík, D.; Füzik, T.; Gondová, M.; Šmerdová, L.; Adamopoulos, A.; Šedo, O.; Zdráhal, Z.; Plevka, P. ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Coscio, F.; Nadra, A.; Ferreiro, D. A Structural Model for the Coronavirus Nucleocapsid. 2020. Available online: https://arxiv.org/pdf/2005.12165.pdf (accessed on 30 July 2021).
- Peng, Y.; Du, N.; Lei, Y.; Dorje, S.; Qi, J.; Luo, T.; Gao, G.F.; Song, H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020, 39, e105938. [Google Scholar] [CrossRef]
- Akram, A.; Mannan, N. Molecular structure, pathogenesis and virology of SARS-CoV-2: A review. Bangladesh J. Infect. Dis. 2020, S36–S40. [Google Scholar] [CrossRef]
- Breen, M.; Nogales, A.; Baker, S.F.; Martínez-Sobrido, L. Replication-competent influenza A viruses expressing reporter genes. Viruses 2016, 8, 179. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Tuthill, T.J.; Groppelli, E.; Hogle, J.M.; Rowlands, D.J. Picornaviruses. Curr. Top. Microbiol. Immunol. 2010, 343, 43–89. [Google Scholar] [CrossRef]
- Cheung, T.K.; Poon, L.L. Biology of influenza a virus. Ann. N. Y. Acad. Sci. 2007, 1102, 1–25. [Google Scholar] [CrossRef]
- De Vlugt, C.; Sikora, D.; Pelchat, M. Insight into influenza: A virus cap-snatching. Viruses 2018, 10, 641. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Stobart, C.C.; Nosek, J.M.; Moore, M.L. Rhinovirus biology, antigenic diversity, and advancements in the design of a human rhinovirus vaccine. Front. Microbiol. 2017, 8, 2412. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Knowles, N.J.; Simmonds, P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J. Gen. Virol. 2013, 94, 1791–1806. [Google Scholar] [CrossRef]
- Sakudo, A.; Onodera, T.; Tanaka, Y. Inactivation of viruses. In Sterilization and Disinfection by Plasma: Sterilization Mechanisms, Biological and Medical Applications (Medical Devices and Equipment), 1st ed.; Sakudo, A., Shintani, H., Hauppauge, N.Y., Eds.; Nova Science Publishers: New York, NY, USA, 2010; pp. 49–60. [Google Scholar]
- Collins, P.L.; Fearns, R.; Graham, B.S. Respiratory syncytial virus: Virology, reverse genetics, and pathogenesis of disease. Curr. Top. Microbiol. Immunol. 2013, 372, 3–38. [Google Scholar] [CrossRef]
- ICTV. Virus Taxonomy. Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses, 9th ed.; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Academic Press: London, UK, 2012. [Google Scholar]
- Tyrrell, D.A.; Bynoe, M.L.; Hitchcock, G.; Pereira, H.G.; Andrewes, C.H. Some virus isolations from common colds. I. Experiments employing human volunteers. Lancet 1960, 1, 235–237. [Google Scholar] [CrossRef]
- Baggen, J.; Thibaut, H.J.; Strating, J.; van Kuppeveld, F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 2018, 16, 368–381. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Trougakos, I.P.; Stamatelopoulos, K.; Terpos, E.; Tsitsilonis, O.E.; Aivalioti, E.; Paraskevis, D.; Kastritis, E.; Pavlakis, G.N.; Dimopoulos, M.A. Insights to SARS-CoV-2 life cycle, pathophysiology, and rationalized treatments that target COVID-19 clinical complications. J. Biomed. Sci. 2021, 28, 9. [Google Scholar] [CrossRef]
- O’Neill, R.E.; Talon, J.; Palese, P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998, 17, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A virus cell entry, replication, virion assembly and movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef]
- Liao, L.E.; Kowal, S.; Cardenas, D.A.; Beauchemin, C.A.A. Exploring virus release as a bottleneck for the spread of influenza A virus infection in vitro and the implications for antiviral therapy with neuraminidase inhibitors. PLoS ONE 2017, 12, e0183621. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Henwood, A.F. Coronavirus disinfection in histopathology. J. Histotechnol. 2020, 43, 102–104. [Google Scholar] [CrossRef]
- Dehbandi, R.; Zazouli, M.A. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, 1, e145. [Google Scholar] [CrossRef]
- Eslami, H.; Jalili, M. The role of environmental factors to transmission of SARS-CoV-2 (COVID-19). AMB Express 2020, 10, 92. [Google Scholar] [CrossRef]
- Otter, J.A.; Donskey, C.; Yezli, S.; Douthwaite, S.; Goldenberg, S.D.; Weber, D.J. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: The possible role of dry surface contamination. J. Hosp. Infect. 2016, 92, 235–250. [Google Scholar] [CrossRef]
- Hemalatha, M.; Kiran, U.; Kuncha, S.K.; Kopperi, H.; Gokulan, C.G.; Mohan, S.V.; Mishra, R.K. Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: Comprehensive study. Sci. Total Environ. 2021, 768, 144704. [Google Scholar] [CrossRef]
- Mohan, S.V.; Hemalatha, M.; Kopperi, H.; Ranjith, I.; Kumar, A.K. SARS-CoV-2 in environmental perspective: Occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 2021, 405, 126893. [Google Scholar] [CrossRef] [PubMed]
- Sagripanti, J.L.; Lytle, C.D. Inactivation of influenza virus by solar radiation. Photochem. Photobiol. 2007, 83, 1278–1282. [Google Scholar] [CrossRef]
- Hirose, R.; Ikegaya, H.; Naito, Y.; Watanabe, N.; Yoshida, T.; Bandou, R.; Daidoji, T.; Itoh, Y.; Nakaya, T. Survival of SARS-CoV-2 and influenza virus on the human skin: Importance of hand hygiene in COVID-19. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Weber, T.P.; Stilianakis, N.I. Inactivation of influenza A viruses in the environment and modes of transmission: A critical review. J. Infect. 2008, 57, 361–373. [Google Scholar] [CrossRef]
- Hendley, J.O.; Wenzel, R.P.; Gwaltney, J.M., Jr. Transmission of rhinovirus colds by self-inoculation. N. Engl. J. Med. 1973, 288, 1361–1364. [Google Scholar] [CrossRef]
- Papadopoulos, N.G.; Sanderson, G.; Hunter, J.; Johnston, S.L. Rhinoviruses replicate effectively at lower airway temperatures. J. Med. Virol. 1999, 58, 100–104. [Google Scholar] [CrossRef]
- Firquet, S.; Beaujard, S.; Lobert, P.E.; Sané, F.; Caloone, D.; Izard, D.; Hober, D. Survival of enveloped and non-enveloped viruses on inanimate surfaces. Microbes Environ. 2015, 30, 140–144. [Google Scholar] [CrossRef]
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef]
- Mahl, M.C.; Sadler, C. Virus survival on inanimate surfaces. Can. J. Microbiol. 1975, 21, 819–823. [Google Scholar] [CrossRef]
- Oliveira, A.C.; Ishimaru, D.; Gonçalves, R.B.; Smith, T.J.; Mason, P.; Sá-Carvalho, D.; Silva, J.L. Low temperature and pressure stability of picornaviruses: Implications for virus uncoating. Biophys. J. 1999, 76, 1270–1279. [Google Scholar] [CrossRef]
- Pérez, L.; Carrasco, L. Entry of poliovirus into cells does not require a low-pH step. J. Virol. 1993, 67, 4543–4548. [Google Scholar] [CrossRef] [PubMed]
- Geller, C.; Varbanov, M.; Duval, R.E. Human coronaviruses: Insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses 2012, 4, 3044–3068. [Google Scholar] [CrossRef]
- Lamarre, A.; Talbot, P.J. Effect of pH and temperature on the infectivity of human coronavirus 229E. Can. J. Microbiol. 1989, 35, 972–974. [Google Scholar] [CrossRef] [PubMed]
- Kormuth, K.A.; Lin, K.; Qian, Z.; Myerburg, M.M.; Marr, L.C.; Lakdawala, S.S. Environmental persistence of influenza viruses is dependent upon virus type and host origin. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Ju, X.; Zhu, Y.; Wang, Y.; Li, J.; Zhang, J.; Gong, M.; Ren, W.; Li, S.; Zhong, J.; Zhang, L.; et al. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog. 2021, 17, e1009439. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Human coronavirus: Host-pathogen interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef]
- Liu, D.X.; Fung, T.S.; Chong, K.K.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antivirus Res. 2014, 109, 97–109. [Google Scholar] [CrossRef]
- Koonin, E.V.; Gorbalenya, A.E.; Chumakov, K.M. Tentative identification of RNA-dependent RNA polymerases of dsRNA viruses and their relationship to positive strand RNA viral polymerases. FEBS Lett. 1989, 252, 42–46. [Google Scholar] [CrossRef]
- Zanotto, P.M.; Gibbs, M.J.; Gould, E.A.; Holmes, E.C. A reevaluation of the higher taxonomy of viruses based on RNA polymerases. J. Virol. 1996, 70, 6083–6096. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. Overview of viruses and virus infection. In Viruses and Human Disease, 2nd ed.; Strauss, J.H., Strauss, E.G., Eds.; Academic Press: London, UK, 2008; pp. 1–33. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. Minus-strand RNA viruses. In Viruses and Human Disease, 2nd ed.; Strauss, J.H., Strauss, E.G., Eds.; Academic Press: London, UK, 2008; pp. 137–191. [Google Scholar] [CrossRef]
- Strauss, J.H.; Strauss, E.G. Plus-strand RNA viruses. In Viruses and Human Disease, 2nd ed.; Strauss, J.H., Strauss, E.G., Eds.; Academic Press: London, UK, 2008; pp. 63–136. [Google Scholar] [CrossRef]
- Savolainen, C.; Blomqvist, S.; Hovi, T. Human rhinoviruses. Paediatr. Respir. Rev. 2003, 4, 91–98. [Google Scholar] [CrossRef]
- Papadopoulos, N.G.; Bates, P.J.; Bardin, P.G.; Papi, A.; Leir, S.H.; Fraenkel, D.J.; Meyer, J.; Lackie, P.M.; Sanderson, G.; Holgate, S.T.; et al. Rhinoviruses infect the lower airways. J. Infect. Dis. 2000, 181, 1875–1884. [Google Scholar] [CrossRef]
- Kiseleva, I.; Su, Q.; Toner, T.J.; Szymkowiak, C.; Kwan, W.S.; Rudenko, L.; Shaw, A.R.; Youil, R. Cell-based assay for the determination of temperature sensitive and cold adapted phenotypes of influenza viruses. J. Virol. Methods 2004, 116, 71–78. [Google Scholar] [CrossRef]
- Kiseleva, I.; Larionova, N.; Kuznetsov, V.; Rudenko, L. Phenotypic characteristics of novel swine-origin influenza A/California/07/2009 (H1N1) virus. Influenza Other Respir. Viruses 2010, 4, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kiseleva, I.; Rekstin, A.; Al Farroukh, M.; Bazhenova, E.; Katelnikova, A.; Puchkova, L.; Rudenko, L. Non-mouse-adapted H1N1pdm09 virus as a model for influenza research. Viruses 2020, 12, 590. [Google Scholar] [CrossRef] [PubMed]
- Laporte, M.; Raeymaekers, V.; Van Berwaer, R.; Vandeput, J.; Marchand-Casas, I.; Thibaut, H.J.; Van Looveren, D.; Martens, K.; Hoffmann, M.; Maes, P.; et al. The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. PLoS Pathog. 2021, 17, e1009500. [Google Scholar] [CrossRef]
- Chatterjee, S.K.; Saha, S.; Munoz, M.N.M. Molecular pathogenesis, immunopathogenesis and novel therapeutic strategy against COVID-19. Front. Mol. Biosci. 2020, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Coleman, W.; Tsongalis, G. Understanding molecular pathogenesis. In Molecular Pathology, 2nd ed.; Coleman, W., Tsongalis, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 231–242. [Google Scholar] [CrossRef]
- Colman, H.; Aldape, K. Molecular pathogenesis. In Primary Central Nervous System Tumors, Current Clinical Oncology; Norden, A.D., Reardon, D.A.E.A., Eds.; Springer Science: Berlin, Germany, 2011; pp. 27–44. [Google Scholar] [CrossRef]
- Horzinek, M.C. Molecular pathogenesis of virus infections. Experientia 1987, 43, 1193–1196. [Google Scholar] [CrossRef]
- Mattoo, S.; Cherry, J.D. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 2005, 18, 326. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Shinya, K.; Kawaoka, Y. Molecular pathogenesis of H5N1 influenza virus infections. Antivir. Ther. 2007, 12, 617–626. [Google Scholar]
- Polo, J.M.; Davis, N.L.; Rice, C.M.; Huang, H.V.; Johnston, R.E. Molecular analysis of Sindbis virus pathogenesis in neonatal mice by using virus recombinants constructed in vitro. J. Virol. 1988, 62, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020, 109, 102433. [Google Scholar] [CrossRef]
- Patick, A.K. Rhinovirus chemotherapy. Antivir. Res. 2006, 71, 391–396. [Google Scholar] [CrossRef]
- Dawson, P.; Rabold, E.M.; Laws, R.L.; Conners, E.E.; Gharpure, R.; Yin, S.; Buono, S.A.; Dasu, T.; Bhattacharyya, S.; Westergaard, R.P.; et al. Loss of taste and smell as distinguishing symptoms of coronavirus disease 2019. Clin. Infect. Dis. 2021, 72, 682–685. [Google Scholar] [CrossRef]
- Monto, A.S.; Gravenstein, S.; Elliott, M.; Colopy, M.; Schweinle, J. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 2000, 160, 3243–3247. [Google Scholar] [CrossRef]
- Hak, E.; Moons, K.G.; Verheij, T.J.; Hoes, A.W. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 2001, 161, 1351–1352. [Google Scholar] [CrossRef]
- Ortega, H.; Nickle, D.; Carter, L. Rhinovirus and asthma: Challenges and opportunities. Rev. Med. Virol. 2020, e2193. [Google Scholar] [CrossRef]
- Tyrrell, D.A.; Cohen, S.; Schlarb, J.E. Signs and symptoms in common colds. Epidemiol. Infect. 1993, 111, 143–156. [Google Scholar] [CrossRef][Green Version]
- Lechien, J.R.; Chiesa-Estomba, C.M.; Place, S.; Van Laethem, Y.; Cabaraux, P.; Mat, Q.; Huet, K.; Plzak, J.; Horoi, M.; Hans, S.; et al. Clinical and epidemiological characteristics of 1420 European patients with mild-to-moderate coronavirus disease 2019. J. Intern. Med. 2020, 288, 335–344. [Google Scholar] [CrossRef]
- L’Huillier, A.G.; Kaiser, L.; Petty, T.J.; Kilowoko, M.; Kyungu, E.; Hongoa, P.; Vieille, G.; Turin, L.; Genton, B.; D’Acremont, V.; et al. Molecular epidemiology of human rhinoviruses and enteroviruses highlights their diversity in sub-Saharan Africa. Viruses 2015, 7, 6412–6423. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Baid, K.; Mossman, K. Molecular pathogenesis of middle east respiratory syndrome (MERS) coronavirus. Curr. Clin. Microbiol. Rep. 2019, 6, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Berlin, D.A.; Gulick, R.M.; Martinez, F.J. Severe COVID-19. N. Engl. J. Med. 2020, 383, 2451–2460. [Google Scholar] [CrossRef]
- Carod-Artal, F.J. Neurological complications of coronavirus and COVID-19. Rev. Neurol. 2020, 70, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Rothan, H.A.; Natekar, J.P.; Stone, S.; Pathak, H.; Strate, P.G.; Arora, K.; Brinton, M.A.; Kumar, M. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses 2021, 13, 132. [Google Scholar] [CrossRef] [PubMed]
- Beeching, N.J.; Fletcher, T.E.; Beadsworth, M.B.J. Covid-19: Testing times. BMJ 2020, 369, m1403. [Google Scholar] [CrossRef]
- Hui, D.S.; E, I.A.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Winther, B. Rhinoviruses. In International Encyclopedia of Public Health; Heggenhougen, H.K., Ed.; Academic Press: Oxford, UK, 2008; pp. 577–581. [Google Scholar] [CrossRef]
- Winther, B.; Gwaltney, J.M.; Hendley, J.O. Respiratory virus infection of monolayer cultures of human nasal epithelial cells. Am. Rev. Respir. Dis. 1990, 141, 839–845. [Google Scholar] [CrossRef]
- To, K.K.W.; Yip, C.C.Y.; Yuen, K.Y. Rhinovirus-from bench to bedside. J. Med. Assoc. 2017, 116, 496–504. [Google Scholar] [CrossRef]
- Sajjan, U.; Wang, Q.; Zhao, Y.; Gruenert, D.C.; Hershenson, M.B. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am. J. Respir. Crit. Care Med. 2008, 178, 1271–1281. [Google Scholar] [CrossRef]
- Pappas, D.E.; Hendley, J.O.; Hayden, F.G.; Winther, B. Symptom profile of common colds in school-aged children. Pediatric Infect. Dis. J. 2008, 27, 8–11. [Google Scholar] [CrossRef]
- GOV.UK. Guidance COVID-19: Epidemiology, Virology and Clinical Features. Updated 18 February 2021. Available online: https://www.gov.uk/government/publications/wuhan-novel-coronavirus-background-information/wuhan-novel-coronavirus-epidemiology-virology-and-clinical-features (accessed on 17 May 2021).
- Jin, Y.; Yang, H.; Ji, W.; Wu, W.; Chen, S.; Zhang, W.; Duan, G. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses 2020, 12, 372. [Google Scholar] [CrossRef]
- Contini, C.; Di Nuzzo, M.; Barp, N.; Bonazza, A.; De Giorgio, R.; Tognon, M.; Rubino, S. The novel zoonotic COVID-19 pandemic: An expected global health concern. J. Infect. Dev. Ctries. 2020, 14, 254–264. [Google Scholar] [CrossRef]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Ghareeb, O.; Ramadhan, S. COVID-19-a novel zoonotic disease: Origin, prevention and control. Pak. J. Med. Health Sci. 2021, 15, 221–223. [Google Scholar]
- Calder, L.J.; Wasilewski, S.; Berriman, J.A.; Rosenthal, P.B. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. USA 2010, 107, 10685–10690. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef]
- Arden, K.E.; Mackay, I.M. Newly identified human rhinoviruses: Molecular methods heat up the cold viruses. Rev. Med. Virol. 2010, 20, 156–176. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Grantham, M.; Pantelic, J.; de Mesquita, J.B.; Albert, B.; Liu, F.; Ehrman, S.; Milton, D.; Consortium, E. Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc. Natl. Acad. Sci. USA 2018, 115. [Google Scholar] [CrossRef] [PubMed]
- Hendley, J.O.; Gwaltney, J.M., Jr. Mechanisms of transmission of rhinovirus infections. Epidemiol. Rev. 1988, 10, 243–258. [Google Scholar] [CrossRef]
- D’Alessio, D.J.; Meschievitz, C.K.; Peterson, J.A.; Dick, C.R.; Dick, E.C. Short-duration exposure and the transmission of rhinoviral colds. J. Infect. Dis. 1984, 150, 189–194. [Google Scholar] [CrossRef]
- Mackay, I.M. Human rhinoviruses: The cold wars resume. J. Clin. Virol. 2008, 42, 297–320. [Google Scholar] [CrossRef]
- Kudo, E.; Song, E.; Yockey, L.J.; Rakib, T.; Wong, P.W.; Homer, R.J.; Iwasaki, A. Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc. Natl. Acad. Sci. USA 2019, 116, 10905–10910. [Google Scholar] [CrossRef] [PubMed]
- Roebuck, M.O. Rhinoviruses in Britain 1963–1973. J. Hyg. 1976, 76, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Winther, B. Rhinovirus infections in the upper airway. Proc. Am. Thorac. Soc. 2011, 8, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Pitkäranta, A.; Hayden, F.G. Rhinoviruses: Important respiratory pathogens. Ann. Med. 1998, 30, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Prim. 2018, 4, 3. [Google Scholar] [CrossRef]
- Troeger, C.E.; Blacker, B.F.; Khalil, I.A.; Zimsen, S.R.M.; Albertson, S.B.; Abate, D.; Abdela, J.; Adhikari, T.B.A.; Aghayan, S.A.; Agrawal, S.; et al. Mortality, morbidity, and hospitalisations due to influenza lower respiratory tract infections, 2017: An analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2019, 7, 69–89. [Google Scholar] [CrossRef]
- Troeger, C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.M.; Albertson, S.; Stanaway, J.D.; Deshpande, A.; Farag, T.; et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [Google Scholar] [CrossRef]
- WHO. Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 20 July 2021).
- Ånestad, G.; Nordbø, S.A. Does rhinovirus inhibit influenza A(H1N1) pandemic? Tidsskr. Nor. Laegeforen. 2010, 130, 1932–1934. [Google Scholar] [CrossRef][Green Version]
- Bulut, C.; Kato, Y. Epidemiology of COVID-19. Turk. J. Med. Sci. 2020, 50, 563–570. [Google Scholar] [CrossRef]
- Casalegno, J.S.; Ottmann, M.; Duchamp, M.B.; Escuret, V.; Billaud, G.; Frobert, E.; Morfin, F.; Lina, B. Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin. Microbiol. Infect. 2010, 16, 326–329. [Google Scholar] [CrossRef]
- Dee, K.; Goldfarb, D.M.; Haney, J.; Amat, J.A.R.; Herder, V.; Stewart, M.; Szemiel, A.M.; Baguelin, M.; Murcia, P.R. Human rhinovirus infection blocks SARS-CoV-2 replication within the respiratory epithelium: Implications for COVID-19 epidemiology. J. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Nickbakhsh, S.; Ho, A.; Marques, D.F.P.; McMenamin, J.; Gunson, R.N.; Murcia, P.R. Epidemiology of seasonal coronaviruses: Establishing the context for COVID-19 emergence. J. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Wu, A.; Mihaylova, V.T.; Landry, M.L.; Foxman, E.F. Interference between rhinovirus and influenza A virus: A clinical data analysis and experimental infection study. Lancet Microbe 2020, 1, e254–e262. [Google Scholar] [CrossRef]
- Zlateva, K.T.; van Rijn, A.L.; Simmonds, P.; Coenjaerts, F.E.J.; van Loon, A.M.; Verheij, T.J.M.; de Vries, J.J.C.; Little, P.; Butler, C.C.; van Zwet, E.W.; et al. Molecular epidemiology and clinical impact of rhinovirus infections in adults during three epidemic seasons in 11 European countries (2007–2010). Thorax 2020, 75, 882–890. [Google Scholar] [CrossRef]
- Nowak, M.D.; Sordillo, E.M.; Gitman, M.R.; Paniz Mondolfi, A.E. Co-infection in SARS-CoV-2 infected patients: Where are influenza virus and rhinovirus/enterovirus? J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- PAHO. Flu Net Home Page. 2010–2021. Available online: http://ais.paho.org/phip/viz/ed_flu.asp (accessed on 4 June 2021).
- Audi, A.; AlIbrahim, M.; Kaddoura, M.; Hijazi, G.; Yassine, H.M.; Zaraket, H. Seasonality of respiratory viral infections: Will COVID-19 follow suit? Front. Public Health 2020, 8, 567184. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.J.; Fitchett, J.M.; Engelbrecht, F.A.; Scholes, R.J.; Dzhivhuho, G.; Sweijd, N.A. Winter is coming: A Southern Hemisphere perspective of the environmental drivers of SARS-CoV-2 and the potential seasonality of COVID-19. Int. J. Environ. Res. Public Health 2020, 17, 5634. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Nair, H. Global seasonality of human seasonal coronaviruses: A clue for postpandemic circulating season of severe acute respiratory syndrome coronavirus 2? J. Infect. Dis. 2020, 222, 1090–1097. [Google Scholar] [CrossRef]
- COVID-10 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Available online: https://coronavirus.jhu.edu/map.html (accessed on 20 July 2021).
- Monto, A.S. The seasonality of rhinovirus infections and its implications for clinical recognition. Clin. Ther. 2002, 24, 1987–1997. [Google Scholar] [CrossRef]
- Kamau, E.; Onyango, C.O.; Otieno, G.P.; Kiyuka, P.K.; Agoti, C.N.; Medley, G.F.; Cane, P.A.; Nokes, D.J.; Munywoki, P.K. An intensive, active surveillance reveals continuous invasion and high diversity of rhinovirus in households. J. Infect. Dis. 2019, 219, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Marí, J.M.; Pérez-Ruiz, M.; Galán Montemayor, J.C.; Marcos Maeso, M.; Reina, J.; de Oña Navarro, M.; Cilla Eguiluz, C.G. Circulation of other respiratory viruses and viral co-infection during the 2009 pandemic influenza. Enferm. Infecc. Y Microbiol. Clin. 2012, 30 (Suppl. S4), 25–31. [Google Scholar] [CrossRef]
- Costa, L.F.; Queiróz, D.A.; da Silveira, H.L.; Neto, M.B.; de Paula, N.T.; Oliveira, T.F.; Tolardo, A.L.; Yokosawa, J. Human rhinovirus and disease severity in children. Pediatrics 2014, 133, e312–e321. [Google Scholar] [CrossRef] [PubMed]
- Leotte, J.; Trombetta, H.; Faggion, H.Z.; Almeida, B.M.; Nogueira, M.B.; Vidal, L.R.; Raboni, S.M. Impact and seasonality of human rhinovirus infection in hospitalized patients for two consecutive years. J. Pediatr. 2017, 93, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Gardinassi, L.G.; Simas, P.V.M.; Salomão, J.B.; Durigon, E.L.; Trevisan, D.M.Z.; Cordeiro, J.A.; Lacerda, M.N.; Rahal, P.; de Souz, F.P. Seasonality of viral respiratory infections in southeast of Brazil: The influence of temperature and air humidity. Braz. J. Microbiol. 2012, 43, 98–108. [Google Scholar] [CrossRef]
- Marcone, D.N.; Culasso, A.; Carballal, G.; Campos, R.; Echavarría, M. Genetic diversity and clinical impact of human rhinoviruses in hospitalized and outpatient children with acute respiratory infection, Argentina. J. Clin. Virol. 2014, 61, 558–564. [Google Scholar] [CrossRef]
- Pierangeli, A.; Scagnolari, C.; Selvaggi, C.; Verzaro, S.; Spina, M.T.; Bresciani, E.; Antonelli, G.; Bertazzoni, G. Rhinovirus frequently detected in elderly adults attending an emergency department. J. Med. Virol. 2011, 83, 2043–2047. [Google Scholar] [CrossRef]
- Pisareva, M.M.; Eder, V.A.; Buzitskaya, Z.V.; Musaeva, T.D.; Afanaseva, V.S.; Go, A.A.; Obraztsova, E.A.; Sukhovetskaya, V.F.; Komissarov, A.B. Etiological structure of influenza and other ARVI in St. Petersburg during epidemic seasons 2012–2016. Vopr. Virusol. 2018, 63, 233–239. [Google Scholar] [CrossRef]
- Turunen, R.; Jartti, T.; Bochkov, Y.A.; Gern, J.E.; Vuorinen, T. Rhinovirus species and clinical characteristics in the first wheezing episode in children. J. Med. Virol. 2016, 88, 2059–2068. [Google Scholar] [CrossRef] [PubMed]
- Jartti, T.; Lehtinen, P.; Vuorinen, T.; Osterback, R.; van den Hoogen, B.; Osterhaus, A.D.; Ruuskanen, O. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg. Infect. Dis. 2004, 10, 1095–1101. [Google Scholar] [CrossRef]
- Longtin, J.; Marchand-Austin, A.; Winter, A.L.; Patel, S.; Eshaghi, A.; Jamieson, F.; Low, D.E.; Gubbay, J.B. Rhinovirus outbreaks in long-term care facilities, Ontario, Canada. Emerg. Infect. Dis. 2010, 16, 1463–1465. [Google Scholar] [CrossRef] [PubMed]
- Pappas, D.E.; Hendley, J.O.; Schwartz, R.H. Respiratory viral RNA on toys in pediatric office waiting rooms. Pediatric Infect. Dis. J. 2010, 29, 102–104. [Google Scholar] [CrossRef]
- Arruda, E.; Pitkäranta, A.; Witek, T.J., Jr.; Doyle, C.A.; Hayden, F.G. Frequency and natural history of rhinovirus infections in adults during autumn. J. Clin. Microbiol. 1997, 35, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Kraft, C.S.; Jacob, J.T.; Sears, M.H.; Burd, E.M.; Caliendo, A.M.; Lyon, G.M. Severity of human rhinovirus infection in immunocompromised adults is similar to that of 2009 H1N1 influenza. J. Clin. Microbiol. 2012, 50, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Lambert, S.B.; McErlean, P.K.; Faux, C.E.; Arden, K.E.; Nissen, M.D.; Sloots, T.P. Prior evidence of putative novel rhinovirus species, Australia. Emerg. Infect. Dis. 2008, 14, 1823–1824. [Google Scholar] [CrossRef]
- Hai le, T.; Bich, V.T.; Le, K.N.; Diep, N.T.; Phuc, P.H.; Hung, V.P.; Taylor, W.R.; Horby, P.; Liem, N.T.; Wertheim, H.F. Fatal respiratory infections associated with rhinovirus outbreak, Vietnam. Emerg. Infect. Dis. 2012, 18, 1886–1888. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Meng, L.; Zhu, W.; Liu, X.; Yang, M.; Yu, D.; Niu, L.; Shen, X. Viral etiologies and epidemiology of patients with acute respiratory infections based on sentinel hospitals in Gansu Province, Northwest China, 2011–2015. J. Med. Virol. 2018, 90, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shen, J.; Wu, B.; Liu, G.; Lu, R.; Tan, W. Genotypic diversity and epidemiology of human rhinovirus among children with severe acute respiratory tract infection in Shanghai, 2013–2015. Front. Microbiol. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Mak, G.C.; Wong, A.H.; Ho, W.Y.; Lim, W. The impact of pandemic influenza A (H1N1) 2009 on the circulation of respiratory viruses 2009–2011. Influenza Other Respir. Viruses 2012, 6, e6–e10. [Google Scholar] [CrossRef] [PubMed]
- Nakauchi, M.; Nagata, N.; Takayama, I.; Saito, S.; Kubo, H.; Kaida, A.; Oba, K.; Odagiri, T.; Kageyama, T. Propagation of rhinovirus C in differentiated immortalized human airway HBEC3-KT epithelial cells. Viruses 2019, 11, 216. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Daleno, C.; Baggi, E.; Ciarmoli, E.; Lavizzari, A.; Pierro, M.; Semino, M.; Groppo, M.; Scala, A.; Terranova, L.; et al. Circulation of different rhinovirus groups among children with lower respiratory tract infection in Kiremba, Burundi. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3251–3256. [Google Scholar] [CrossRef] [PubMed]
- Famoroti, T.; Sibanda, W.; Ndung’u, T. Prevalence and seasonality of common viral respiratory pathogens, including Cytomegalovirus in children, between 0–5 years of age in KwaZulu-Natal, an HIV endemic province in South Africa. BMC Pediatr. 2018, 18, 240. [Google Scholar] [CrossRef]
- Warshauer, D.M.; Dick, E.C.; Mandel, A.D.; Flynn, T.C.; Jerde, R.S. Rhinovirus infections in an isolated antarctic station. Transmission of the viruses and susceptibility of the population. Am. J. Epidemiol. 1989, 129, 319–340. [Google Scholar] [CrossRef]
- WHO. Influenza Updates. Available online: https://www.who.int/influenza/surveillance_monitoring/updates/en/ (accessed on 11 February 2021).
- Poole, S.; Brendish, N.J.; Clark, T.W. SARS-CoV-2 has displaced other seasonal respiratory viruses: Results from a prospective cohort study. J. Infect. 2020. [Google Scholar] [CrossRef]
- Ånestad, G.; Nordbø, S.A. Virus interference. Did rhinoviruses activity hamper the progress of the 2009 influenza A (H1N1) pandemic in Norway? Med. Hypotheses 2011, 77, 1132–1134. [Google Scholar] [CrossRef]
- Casalegno, J.S.; Ottmann, M.; Bouscambert-Duchamp, M.; Valette, M.; Morfin, F.; Lina, B. Impact of the 2009 influenza A(H1N1) pandemic wave on the pattern of hibernal respiratory virus epidemics, France, 2009. Euro Surveill. 2010, 15, 19485. [Google Scholar] [CrossRef]
- Anderson, R.M.; Fraser, C.; Ghani, A.C.; Donnelly, C.A.; Riley, S.; Ferguson, N.M.; Leung, G.M.; Lam, T.H.; Hedley, A.J. Epidemiology, transmission dynamics and control of SARS: The 2002-2003 epidemic. Philos. Trans. R. Soc. 2004, 359, 1091–1105. [Google Scholar] [CrossRef]
- Nickbakhsh, S.; Mair, C.; Matthews, L.; Reeve, R.; Johnson, P.C.D.; Thorburn, F.; von Wissmann, B.; Reynolds, A.; McMenamin, J.; Gunson, R.N.; et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. USA 2019, 116, 27142–27150. [Google Scholar] [CrossRef]
- Jartti, T.; Jartti, L.; Peltola, V.; Waris, M.; Ruuskanen, O. Identification of respiratory viruses in asymptomatic subjects: Asymptomatic respiratory viral infections. Pediatric Infect. Dis. J. 2008, 27, 1103–1107. [Google Scholar] [CrossRef]
- Wolsk, H.M.; Følsgaard, N.V.; Birch, S.; Brix, S.; Hansel, T.T.; Johnston, S.L.; Kebadze, T.; Chawes, B.L.; Bønnelykke, K.; Bisgaard, H. Picornavirus-induced airway mucosa immune profile in asymptomatic neonates. J. Infect. Dis. 2016, 213, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Makris, S.; Johnston, S. Recent advances in understanding rhinovirus immunity. F1000Research 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the numbers. eLife 2020, 9, e57309. [Google Scholar] [CrossRef]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [PubMed]
- WHO. Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 18 May 2021).
- Mandal, A. COVID-19 Pandemic Is “One Big Wave” Says WHO. Available online: https://www.news-medical.net/news/20200730/COVID-19-pandemic-is-one-big-wave-says-WHO.aspx (accessed on 22 July 2021).
- Aboubakr, H.A.; Sharafeldin, T.A.; Goyal, S.M. Stability of SARS-CoV-2 and other coronaviruses in the environment and on common touch surfaces and the influence of climatic conditions: A review. Transbound. Emerg. Dis. 2021, 68, 296–312. [Google Scholar] [CrossRef] [PubMed]
- Cimolai, N. Complicating infections associated with common endemic human respiratory coronaviruses. Health Secur. 2021, 19, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Dreschers, S.; Dumitru, C.A.; Adams, C.; Gulbins, E. The cold case: Are rhinoviruses perfectly adapted pathogens? Cell. Mol. Life Sci. 2007, 64, 181–191. [Google Scholar] [CrossRef]
- Fendrick, A.M.; Monto, A.S.; Nightengale, B.; Sarnes, M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch. Intern. Med. 2003, 163, 487–494. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiseleva, I.; Ksenafontov, A. COVID-19 Shuts Doors to Flu but Keeps Them Open to Rhinoviruses. Biology 2021, 10, 733. https://doi.org/10.3390/biology10080733
Kiseleva I, Ksenafontov A. COVID-19 Shuts Doors to Flu but Keeps Them Open to Rhinoviruses. Biology. 2021; 10(8):733. https://doi.org/10.3390/biology10080733
Chicago/Turabian StyleKiseleva, Irina, and Andrey Ksenafontov. 2021. "COVID-19 Shuts Doors to Flu but Keeps Them Open to Rhinoviruses" Biology 10, no. 8: 733. https://doi.org/10.3390/biology10080733
APA StyleKiseleva, I., & Ksenafontov, A. (2021). COVID-19 Shuts Doors to Flu but Keeps Them Open to Rhinoviruses. Biology, 10(8), 733. https://doi.org/10.3390/biology10080733