A Time-Saving Strategy to Generate Double Maternal Mutants by an Oocyte-Specific Conditional Knockout System in Zebrafish
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Maintenance
2.2. sgRNA Design and Efficiency Test
2.3. Plasmids Construction
2.4. Transgenesis of sgRNA Expression Vector via I-Sce I
2.5. Quality Control before Screening Maternal Mutants
2.6. Ploidy Analysis by Quantitative PCR
2.7. Total RNA Extraction and RT-PCR
3. Results
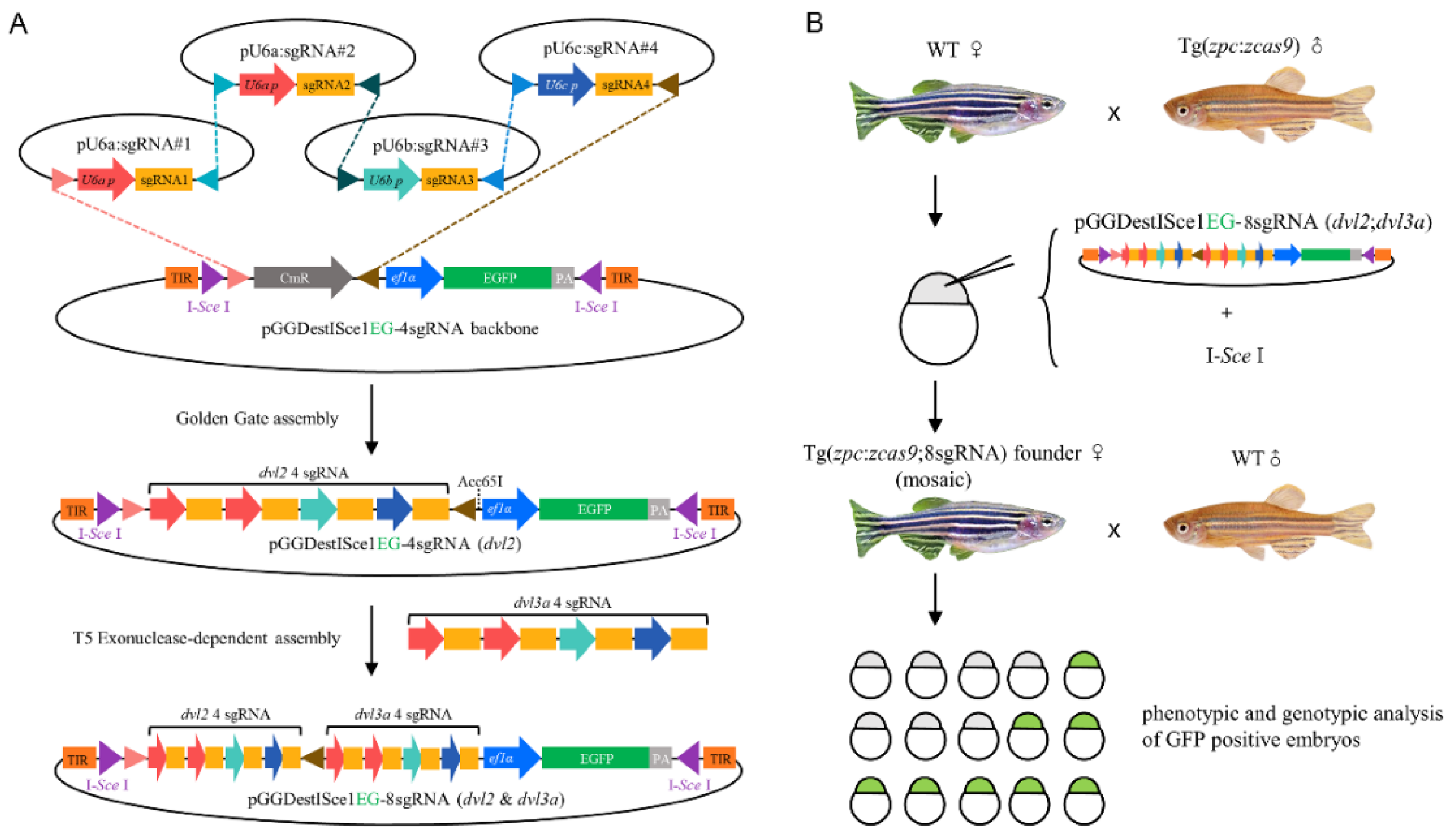
3.1. Experimental Design to Generate Double Maternal Mutants
3.2. Generation of dvl2 and dvl3a Double Mutants by Oocyte-Specific CKO
3.3. Genotyping the Double Maternal Mutants for dvl2 and dvl3a Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marlow, F.L. Maternal Control of Development in Vertebrates; Morgan & Clypool Publishers: San Rafael, CA, USA, 2010; pp. 1–196. [Google Scholar]
- Fuentes, R.; Tajer, B.; Kobayashi, M.; Pelliccia, J.L.; Langdon, Y.; Abrams, E.W.; Mullins, M.C. The maternal coordinate system: Molecular-genetics of embryonic axis formation and patterning in the zebrafish. Curr. Top. Dev. Biol. 2020, 140, 341–389. [Google Scholar] [PubMed]
- Yan, L.; Chen, J.; Zhu, X.; Sun, J.; Wu, X.; Shen, W.; Zhang, W.; Tao, Q.; Meng, A. Maternal Huluwa dictates the embryonic body axis through beta-catenin in vertebrates. Science 2018, 362, eaat1045. [Google Scholar] [CrossRef] [PubMed]
- Abrams, E.W.; Fuentes, R.; Marlow, F.L.; Kobayashi, M.; Zhang, H.; Lu, S.; Kapp, L.; Joseph, S.R.; Kugath, A.; Gupta, T.; et al. Molecular genetics of maternally-controlled cell divisions. PLoS Genet. 2020, 16, e1008652. [Google Scholar] [CrossRef]
- Shao, M.; Wang, M.; Liu, Y.Y.; Ge, Y.W.; Zhang, Y.J.; Shi, D.L. Vegetally localised Vrtn functions as a novel repressor to modulate bmp2b transcription during dorsoventral patterning in zebrafish. Development 2017, 144, 3361–3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escobar-Aguirre, M.; Elkouby, Y.M.; Mullins, M.C. Localization in Oogenesis of Maternal Regulators of Embryonic Development. Adv. Exp. Med. Biol. 2017, 953, 173–207. [Google Scholar] [PubMed]
- Lindeman, R.E.; Pelegri, F. Vertebrate maternal-effect genes: Insights into fertilization, early cleavage divisions, and germ cell determinant localization from studies in the zebrafish. Mol. Reprod. Dev. 2010, 77, 299–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dosch, R.; Wagner, D.S.; Mintzer, K.A.; Runke, G.; Wiemelt, A.P.; Mullins, M.C. Maternal control of vertebrate development before the midblastula transition: Mutants from the zebrafish I. Dev. Cell 2004, 6, 771–780. [Google Scholar] [CrossRef] [Green Version]
- White, R.J.; Collins, J.E.; Sealy, I.M.; Wali, N.; Dooley, C.M.; Digby, Z.; Stemple, D.L.; Murphy, D.N.; Billis, K.; Hourlier, T.; et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 2017, 6, e30860. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.Y.; Cheng, X.N.; Li, Y.L.; Zhang, C.; Saquet, A.; Liu, Y.Y.; Shao, M.; Shi, D.L. Mutational analysis of dishevelled genes in zebrafish reveals distinct functions in embryonic patterning and gastrulation cell movements. PLoS Genet. 2018, 14, e1007551. [Google Scholar] [CrossRef]
- Gritsman, K.; Zhang, J.; Cheng, S.; Heckscher, E.; Talbot, W.S.; Schier, A.F. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 1999, 97, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Hino, H.; Nakanishi, A.; Seki, R.; Aoki, T.; Yamaha, E.; Kawahara, A.; Shimizu, T.; Hibi, M. Roles of maternal wnt8a transcripts in axis formation in zebrafish. Dev. Biol. 2018, 434, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Ciruna, B.; Weidinger, G.; Knaut, H.; Thisse, B.; Thisse, C.; Raz, E.; Schier, A.F. Production of maternal-zygotic mutant zebrafish by germ-line replacement. Proc. Natl. Acad. Sci. USA 2002, 99, 14919–14924. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Li, X.; He, M.; Ye, D.; Xiong, F.; Amin, G.; Zhu, Z.; Sun, Y. Efficient generation of zebrafish maternal-zygotic mutants through transplantation of ectopically induced and Cas9/gRNA targeted primordial germ cells. J. Genet. Genom. 2020, 47, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Cheng, X.N.; Liu, Y.Y.; Li, J.T.; Shi, D.L. Transplantation of Zebrafish Cells by Conventional Pneumatic Microinjector. Zebrafish 2018, 15, 73–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Shen, W.; Zhang, B.; Meng, A. The genetic program of oocytes can be modified in vivo in the zebrafish ovary. J. Mol. Cell Biol. 2018, 10, 479–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhu, Z.; Ho, I.H.T.; Shi, Y.; Xie, Y.; Li, J.; Zhang, Y.; Chan, M.T.V.; Cheng, C.H.K. Germline-specific dgcr8 knockout in zebrafish using a BACK approach. Cell Mol. Life Sci. 2017, 74, 2503–2511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lu, T.; Zhang, Y.Z.; Li, J.G.; Tarique, I.; Wen, F.F.; Chen, A.J.; Wang, J.S.; Zhang, Z.Y.; Zhang, Y.J.; et al. Rapid generation of maternal mutants via oocyte transgenic expression of CRISPR/Cas9 and sgRNAs in zebrafish. Sci. Adv. 2021, 7, eabg4243. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, C.; Zhang, Y.; Lin, S.; Shi, D.L.; Shao, M. Highly efficient genome editing using oocyte-specific zcas9 transgenic zebrafish. J. Genet. Genom. 2018, 45, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Sorrells, T.R.; Johnson, A.D. Making sense of transcription networks. Cell 2015, 161, 714–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, D.L. Decoding Dishevelled-Mediated Wnt Signaling in Vertebrate Early Development. Front. Cell Dev. Biol. 2020, 8, 588370. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Z.; Xiao, A.; Zhang, Y.; Li, W.; Zu, Y.; Yao, S.; Lin, S.; Zhang, B. Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized cas9 and evaluation of off-targeting effect. J. Genet. Genom. 2014, 41, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Maddison, L.A.; Li, M.; Kara, N.; LaFave, M.C.; Varshney, G.K.; Burgess, S.M.; Patton, J.G.; Chen, W. Multiplex Conditional Mutagenesis Using Transgenic Expression of Cas9 and sgRNAs. Genetics 2015, 200, 431–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Li, K.; Li, J.; Wang, T.; Gu, L.; Xun, L. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 2019, 47, e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thermes, V.; Grabher, C.; Ristoratore, F.; Bourrat, F.; Choulika, A.; Wittbrodt, J.; Joly, J.S. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 2002, 118, 91–98. [Google Scholar] [CrossRef]
- Wu, R.S.; Lam, I.; Clay, H.; Duong, D.N.; Deo, R.C.; Coughlin, S.R. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell 2018, 46, 112–125.e114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [Green Version]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Gunther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.L.; et al. Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef]
- Kushawah, G.; Hernandez-Huertas, L.; Abugattas-Nunez Del Prado, J.; Martinez-Morales, J.R.; DeVore, M.L.; Hassan, H.; Moreno-Sanchez, I.; Tomas-Gallardo, L.; Diaz-Moscoso, A.; Monges, D.E.; et al. CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos. Dev. Cell 2020, 54, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Force, A.; Yan, Y.L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.L.; et al. Zebrafish hox clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshijima, K.; Jurynec, M.J.; Klatt Shaw, D.; Jacobi, A.M.; Behlke, M.A.; Grunwald, D.J. Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev. Cell 2019, 51, 645–657.e644. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, V.; De Santis, F.; Auer, T.O.; Testa, N.; Sanchez-Iranzo, H.; Mercader, N.; Concordet, J.P.; Del Bene, F. 2C-Cas9: A versatile tool for clonal analysis of gene function. Genome Res. 2016, 26, 681–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Fei, F.; Berberoglu, M.A.; Sun, S.; Wang, L.; Dong, Z.; Wang, X. Csy4-based vector system enables conditional chimeric gene editing in zebrafish without interrupting embryogenesis. J. Mol. Cell Biol. 2018, 10, 586–588. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.T.; Ng, A.S.; Ingham, P.W. Ribozyme Mediated gRNA Generation for In Vitro and In Vivo CRISPR/Cas9 Mutagenesis. PLoS ONE 2016, 11, e0166020. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, T.; Kawakami, K. A tRNA-based multiplex sgRNA expression system in zebrafish and its application to generation of transgenic albino fish. Sci. Rep. 2018, 8, 13366. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Li, J.; Tarique, I.; Zhang, Y.; Lu, T.; Wang, J.; Chen, A.; Wen, F.; Zhang, Z.; Zhang, Y.; et al. A Time-Saving Strategy to Generate Double Maternal Mutants by an Oocyte-Specific Conditional Knockout System in Zebrafish. Biology 2021, 10, 777. https://doi.org/10.3390/biology10080777
Zhang C, Li J, Tarique I, Zhang Y, Lu T, Wang J, Chen A, Wen F, Zhang Z, Zhang Y, et al. A Time-Saving Strategy to Generate Double Maternal Mutants by an Oocyte-Specific Conditional Knockout System in Zebrafish. Biology. 2021; 10(8):777. https://doi.org/10.3390/biology10080777
Chicago/Turabian StyleZhang, Chong, Jiaguang Li, Imran Tarique, Yizhuang Zhang, Tong Lu, Jiasheng Wang, Aijun Chen, Fenfen Wen, Zhuoyu Zhang, Yanjun Zhang, and et al. 2021. "A Time-Saving Strategy to Generate Double Maternal Mutants by an Oocyte-Specific Conditional Knockout System in Zebrafish" Biology 10, no. 8: 777. https://doi.org/10.3390/biology10080777
APA StyleZhang, C., Li, J., Tarique, I., Zhang, Y., Lu, T., Wang, J., Chen, A., Wen, F., Zhang, Z., Zhang, Y., & Shao, M. (2021). A Time-Saving Strategy to Generate Double Maternal Mutants by an Oocyte-Specific Conditional Knockout System in Zebrafish. Biology, 10(8), 777. https://doi.org/10.3390/biology10080777





