Liver Organoids: Updates on Disease Modeling and Biomedical Applications
Abstract
:Simple Summary
Abstract
1. Introduction
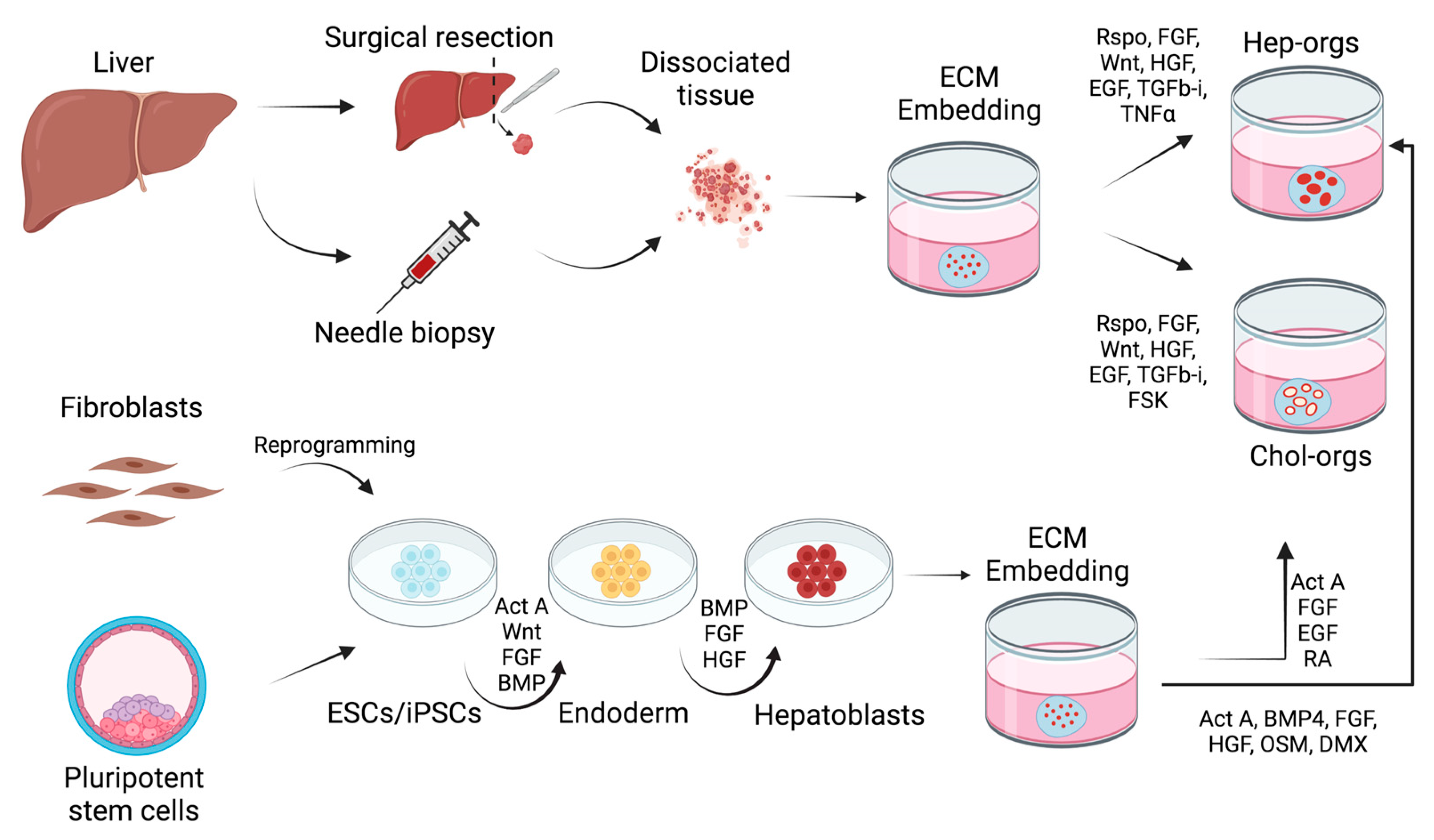
2. Liver Organoids
2.1. Adult Stem Cell-Derived Organoids
2.2. Pluripotent Stem Cell-Derived Liver Organoids
3. Xenofree Liver Organoids
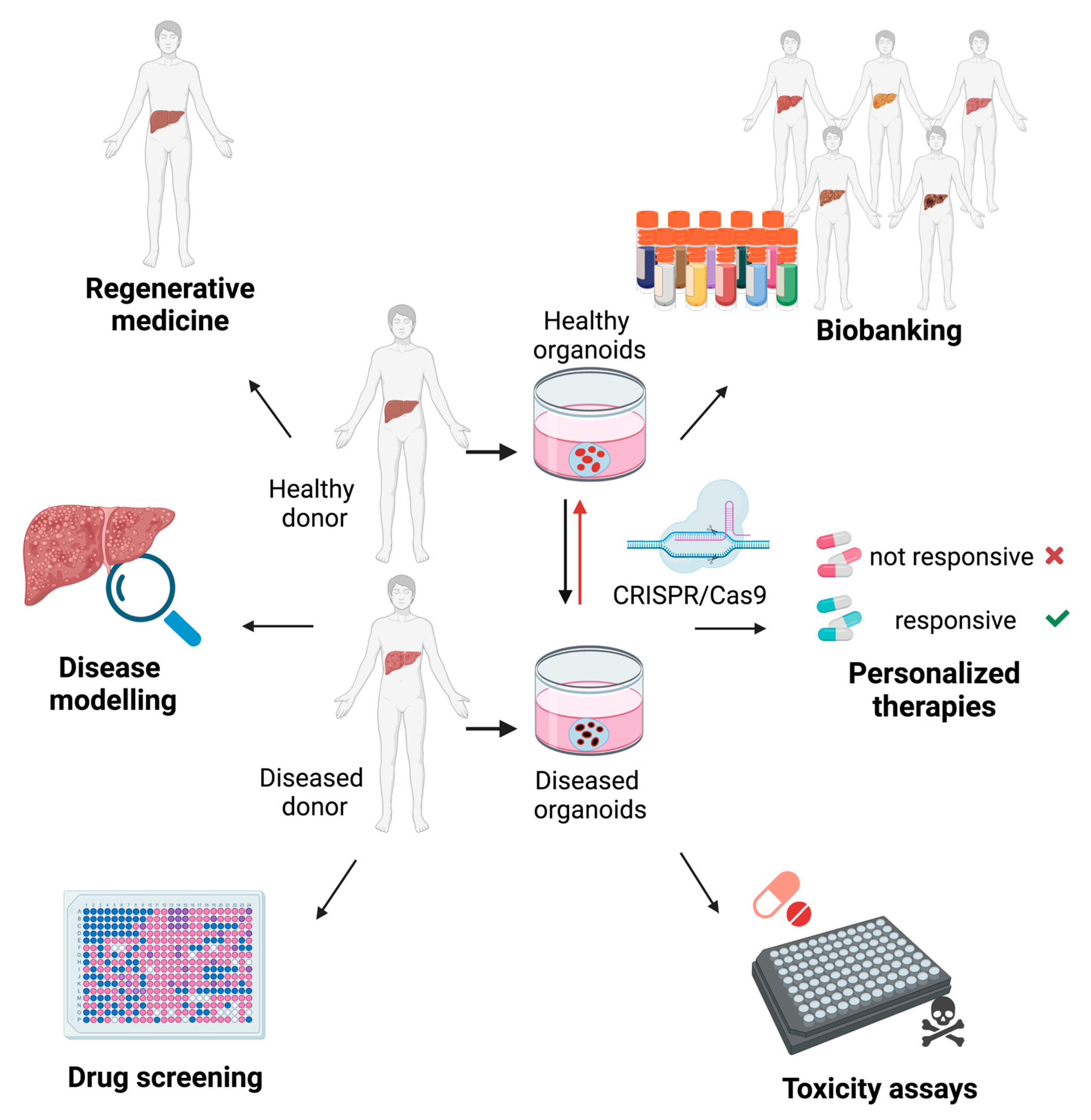
4. Liver Organoids for Disease Modeling
4.1. Monogenic Diseases
4.2. Steatohepatitis
4.3. Liver Infections
4.4. Liver Cancer
5. The Potential Application of Liver Organoids for Regenerative Medicine
6. Liver Organoids as Tools for Drug Discovery
7. Conclusions and Future Perspectives
- (1)
- Differentiation protocols need to be improved in order to reach higher cell maturation and a correct liver cell type representation, therefore increasing the grade of complexity to more accurately resemble the pathophysiological processes;
- (2)
- Development of new bioengineering approaches to guarantee the reproducibility of liver organoid composition and functionality. This goal is fundamental in order to promote liver organoids to drug discovery and pre-clinical testing pipelines;
- (3)
- Development of improved synthetic biomaterials to obtain an animal grade free culturing system for future safe applications in regenerative medicine.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Green, H.; Kehinde, O.; Thomas, J. Growth of Cultured Human Epidermal Cells into Multiple Epithelia Suitable for Grafting. Proc. Natl. Acad. Sci. USA 1979, 76, 5665–5668. [Google Scholar] [CrossRef] [Green Version]
- Weigelt, B.; Ghajar, C.M.; Bissell, M.J. The Need for Complex 3D Culture Models To Unravel Novel Pathways and Identify Accurate Biomarkers in Breast Cancer. Adv. Drug. Deliv. Rev. 2014, 69–70, 42–51. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-Organized Formation of Polarized Cortical Tissues From ESCs and Its Active Manipulation by Extrinsic Signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [Green Version]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of Stem Cells in Small Intestine and Colon by Marker Gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem Cells Build Crypt-Villus Structures In Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Tan, S.H.; Barker, N. Recent Advances in Lgr5(+) Stem Cell Research. Trends Cell Biol. 2018, 28, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The Liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.C.; Dankers, A.C.A.; Lauschke, V.M.; Sison-Young, R.; Jenkins, R.; Rowe, C.; Goldring, C.E.; Park, K.; Regan, S.L.; Walker, T.; et al. Comparison of Hepatic 2D Sandwich Cultures and 3D Spheroids for Long-term Toxicity Applications: A Multicenter Study. Toxicol. Sci. 2018, 162, 655–666. [Google Scholar] [CrossRef] [Green Version]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In Vitro Expansion of Single Lgr5+ Liver Stem Cells Induced by Wnt-Driven Regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef] [Green Version]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Gehart, H.; Artegiani, B.; Carmen, L.-I.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018, 175, 1591–1606.e1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, W.C.; Logan, C.Y.; Fish, M.; Anbarchian, T.; Aguisanda, F.; Alvarez-Varela, A.; Wu, P.; Jin, Y.; Zhu, J.; Li, B.; et al. Inflammatory Cytokine TNFalpha Promotes the Long-Term Expansion of Primary Hepatocytes in 3D Culture. Cell 2018, 175, 1607–1619.e1615. [Google Scholar] [CrossRef] [Green Version]
- Schneeberger, K.; Sanchez-Romero, N.; Ye, S.; van Steenbeek, F.G.; Oosterhoff, L.A.; Pla Palacin, I.; Chen, C.; van Wolferen, M.E.; van Tienderen, G.; Lieshout, R.; et al. Large-Scale Production of LGR5-Positive Bipotential Human Liver Stem Cells. Hepatology 2020, 72, 257–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.R.; Ueno, Y.; Zheng, Y.W.; Koike, N.; et al. Vascularized and Functional Human Liver From an iPSC-Derived Organ Bud Transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef]
- Takebe, T.; Sekine, K.; Kimura, M.; Yoshizawa, E.; Ayano, S.; Koido, M.; Funayama, S.; Nakanishi, N.; Hisai, T.; Kobayashi, T.; et al. Massive and Reproducible Production of Liver Buds Entirely from Human Pluripotent Stem Cells. Cell Rep. 2017, 21, 2661–2670. [Google Scholar] [CrossRef] [Green Version]
- Asai, A.; Aihara, E.; Watson, C.; Mourya, R.; Mizuochi, T.; Shivakumar, P.; Phelan, K.; Mayhew, C.; Helmrath, M.; Takebe, T.; et al. Paracrine Signals Regulate Human Liver Organoid Maturation From Induced Pluripotent Stem Cells. Development 2017, 144, 1056–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human Hepatic Organoids for the Analysis of Human Genetic Diseases. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutolf, M.P.; Hubbell, J.A. Synthetic Biomaterials as Instructive Extracellular Microenvironments for Morphogenesis in Tissue Engineering. Nat. Biotechnol. 2005, 23, 47–55. [Google Scholar] [CrossRef]
- Ehrbar, M.; Rizzi, S.C.; Schoenmakers, R.G.; Miguel, B.S.; Hubbell, J.A.; Weber, F.E.; Lutolf, M.P. Biomolecular Hydrogels Formed and Degraded via Site-Specific Enzymatic Reactions. Biomacromolecules 2007, 8, 3000–3007. [Google Scholar] [CrossRef]
- Sorrentino, G.; Rezakhani, S.; Yildiz, E.; Nuciforo, S.; Heim, M.H.; Lutolf, M.P.; Schoonjans, K. Mechano-Modulatory Synthetic Niches for Liver Organoid Derivation. Nat. Commun. 2020, 11, 3416. [Google Scholar] [CrossRef]
- Ye, S.C.; Boeter, J.W.B.; Mihajlovic, M.; van Steenbeek, F.G.; van Wolferen, M.E.; Oosterhoff, L.A.; Marsee, A.; Caiazzo, M.; van der Laan, L.J.W.; Penning, L.C.; et al. A Chemically Defined Hydrogel for Human Liver Organoid Culture. Adv. Funct. Mater. 2020, 30. [Google Scholar] [CrossRef]
- Andersson, E.R.; Chivukula, I.V.; Hankeova, S.; Sjoqvist, M.; Tsoi, Y.L.; Ramskold, D.; Masek, J.; Elmansuri, A.; Hoogendoorn, A.; Vazquez, E.; et al. Mouse Model of Alagille Syndrome and Mechanisms of Jagged1 Missense Mutations. Gastroenterology 2018, 154, 1080–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wang, X.; Tan, Z.; Su, Y.; Liu, J.; Chang, M.; Yan, F.; Chen, J.; Chen, T.; Li, C.; et al. Human ESC-Derived Expandable Hepatic Organoids Enable Therapeutic Liver Repopulation and Pathophysiological Modeling of Alcoholic Liver Injury. Cell Res. 2019, 29, 1009–1026. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Ogawa, S.; Bear, C.E.; Ahmadi, S.; Chin, S.; Li, B.; Grompe, M.; Keller, G.; Kamath, B.M.; Ghanekar, A. Directed Differentiation of Cholangiocytes from Human Pluripotent Stem Cells. Nat. Biotechnol. 2015, 33, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.Z.; Zheng, Y.W.; Miyakawa, K.; Murata, S.; Zhang, R.R.; Sekine, K.; Ueno, Y.; Takebe, T.; Wakita, T.; Ryo, A.; et al. Recapitulation of Hepatitis B Virus-Host Interactions in Liver Organoids From Human Induced Pluripotent Stem Cells. EBioMedicine 2018, 35, 114–123. [Google Scholar] [CrossRef] [Green Version]
- Baktash, Y.; Madhav, A.; Coller, K.E.; Randall, G. Single Particle Imaging of Polarized Hepatoma Organoids upon Hepatitis C Virus Infection Reveals an Ordered and Sequential Entry Process. Cell Host Microbe 2018, 23, 382–394. [Google Scholar] [CrossRef] [Green Version]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarro, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human Primary Liver Cancer-Derived Organoid Cultures for Disease Modeling and Drug Screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef]
- Nuciforo, S.; Fofana, I.; Matter, M.S.; Blumer, T.; Calabrese, D.; Boldanova, T.; Piscuoglio, S.; Wieland, S.; Ringnalda, F.; Schwank, G.; et al. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018, 24, 1363–1376. [Google Scholar] [CrossRef] [Green Version]
- Artegiani, B.; van Voorthuijsen, L.; Lindeboom, R.G.H.; Seinstra, D.; Heo, I.; Tapia, P.; Lopez-Iglesias, C.; Postrach, D.; Dayton, T.; Oka, R.; et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell 2019, 24, 927–943.e6. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, Y.; Nilsson-Payant, B.E.; Gupta, V.; Wang, P.; Duan, X.; Tang, X.; Zhu, J.; Zhao, Z.; Jaffre, F.; et al. A Human Pluripotent Stem Cell-Based Platform To Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 2020, 27, 125–136.e7. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, R.; Togo, S.; Kimura, M.; Shinozawa, T.; Koido, M.; Koike, H.; Thompson, W.; Karns, R.A.; Mayhew, C.N.; McGrath, P.S.; et al. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab. 2019, 30, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Kruitwagen, H.S.; Oosterhoff, L.A.; van Wolferen, M.E.; Chen, C.; Assawarachan, N.S.; Schneeberger, K.; Kummeling, A.; van Straten, G.; Akkerdaas, I.C.; Vinke, C.R.; et al. Long-Term Survival of Transplanted Autologous Canine Liver Organoids in a COMMD1-Deficient Dog Model of Metabolic Liver Disease. Cells 2020, 9. [Google Scholar] [CrossRef] [Green Version]
- Kruitwagen, H.S.; Oosterhoff, L.A.; Vernooij, I.; Schrall, I.M.; van Wolferen, M.E.; Bannink, F.; Roesch, C.; van Uden, L.; Molenaar, M.R.; Helms, J.B.; et al. Long-Term Adult Feline Liver Organoid Cultures for Disease Modeling of Hepatic Steatosis. Stem Cell Rep. 2017, 8, 822–830. [Google Scholar] [CrossRef] [Green Version]
- Nantasanti, S.; Spee, B.; Kruitwagen, H.S.; Chen, C.; Geijsen, N.; Oosterhoff, L.A.; van Wolferen, M.E.; Pelaez, N.; Fieten, H.; Wubbolts, R.W.; et al. Disease Modeling and Gene Therapy of Copper Storage Disease in Canine Hepatic Organoids. Stem Cell Rep. 2015, 5, 895–907. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Krantz, I.D.; Deng, Y.; Genin, A.; Banta, A.B.; Collins, C.C.; Qi, M.; Trask, B.J.; Kuo, W.L.; Cochran, J.; et al. Alagille Syndrome Is Caused by Mutations in Human Jagged1, Which Encodes a Ligand for Notch1. Nat. Genet. 1997, 16, 243–251. [Google Scholar] [CrossRef]
- Crosnier, C.; Attie-Bitach, T.; Encha-Razavi, F.; Audollent, S.; Soudy, F.; Hadchouel, M.; Meunier-Rotival, M.; Vekemans, M. JAGGED1 Gene Expression During Human Embryogenesis Elucidates the Wide Phenotypic Spectrum of Alagille Syndrome. Hepatology 2000, 32, 574–581. [Google Scholar] [CrossRef]
- Greene, C.M.; Marciniak, S.J.; Teckman, J.; Ferrarotti, I.; Brantly, M.L.; Lomas, D.A.; Stoller, J.K.; McElvaney, N.G. Alpha1-Antitrypsin Deficiency. Nat. Rev. Dis. Primers 2016, 2, 16051. [Google Scholar] [CrossRef] [PubMed]
- Fiorotto, R.; Strazzabosco, M. Pathophysiology of Cystic Fibrosis Liver Disease: A Channelopathy Leading to Alterations in Innate Immunity and in Microbiota. Cell Mol. Gastroenterol. Hepatol. 2019, 8, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Schwank, G.; Koo, B.K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef] [Green Version]
- Ala, A.; Walker, A.P.; Ashkan, K.; Dooley, J.S.; Schilsky, M.L. Wilson’s Disease. Lancet 2007, 369, 397–408. [Google Scholar] [CrossRef]
- Parisi, S.; Polishchuk, E.V.; Allocca, S.; Ciano, M.; Musto, A.; Gallo, M.; Perone, L.; Ranucci, G.; Iorio, R.; Polishchuk, R.S.; et al. Characterization of the Most Frequent ATP7B Mutation Causing Wilson Disease in Hepatocytes From Patient Induced Pluripotent Stem Cells. Sci. Rep. 2018, 8, 6247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Concilli, M.; Petruzzelli, R.; Parisi, S.; Catalano, F.; Sirci, F.; Napolitano, F.; Renda, M.; Galietta, L.J.V.; Di Bernardo, D.; Polishchuk, R.S. Pharmacoproteomics Pinpoints HSP70 Interaction for Correction of the Most Frequent Wilson Disease-Causing Mutant of ATP7B. Proc. Natl. Acad. Sci. USA 2020, 117, 32453–32463. [Google Scholar] [CrossRef]
- Overeem, A.W.; Klappe, K.; Parisi, S.; Kloters-Planchy, P.; Matakovic, L.; du Teil Espina, M.; Drouin, C.A.; Weiss, K.H.; van IJzendoorn, S.C.D. Pluripotent Stem Cell-Derived Bile Canaliculi-Forming Hepatocytes to Study Genetic Liver Diseases Involving Hepatocyte Polarity. J. Hepatol. 2019, 71, 344–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beinhardt, S.; Leiss, W.; Stattermayer, A.F.; Graziadei, I.; Zoller, H.; Stauber, R.; Maieron, A.; Datz, C.; Steindl-Munda, P.; Hofer, H.; et al. Long-Term Outcomes of Patients With Wilson Disease in a Large Austrian Cohort. Clin. Gastroenterol. Hepatol. 2014, 12, 683–689. [Google Scholar] [CrossRef]
- Reiner, Z.; Guardamagna, O.; Nair, D.; Soran, H.; Hovingh, K.; Bertolini, S.; Jones, S.; Coric, M.; Calandra, S.; Hamilton, J.; et al. Lysosomal Acid Lipase Deficiency-An Under-Recognized Cause of Dyslipidaemia and Liver Dysfunction. Atherosclerosis 2014, 235, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Dash, A.; Figler, R.A.; Blackman, B.R.; Marukian, S.; Collado, M.S.; Lawson, M.J.; Hoang, S.A.; Mackey, A.J.; Manka, D.; Cole, B.K.; et al. Pharmacotoxicology of Clinically-Relevant Concentrations of Obeticholic Acid in an Organotypic Human Hepatocyte System. Toxicol. Vitr. 2017, 39, 93–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haaker, M.W.; Kruitwagen, H.S.; Vaandrager, A.B.; Houweling, M.; Penning, L.C.; Molenaar, M.R.; van Wolferen, M.E.; Oosterhoff, L.A.; Spee, B.; Helms, J.B. Identification of Potential Drugs for Treatment of Hepatic Lipidosis in Cats Using an In Vitro Feline Liver Organoid System. J. Vet. Intern. Med. 2020, 34, 132–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic Liver Disease. Nat. Rev. Dis. Primers 2018, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, J.U.; Andersen, J.B. Liver Cancer Oncogenomics: Opportunities and Dilemmas for Clinical Applications. Hepat. Oncol. 2015, 2, 79–93. [Google Scholar] [CrossRef]
- Arellanes-Robledo, J.; Hernández, C.; Camacho, J.; Pérez-Carreón, J.I. Chapter 42-In Vitro Models of HCC. In Liver Pathophysiology; Muriel, P., Ed.; Academic Press: Boston, MA, USA, 2017; pp. 563–579. [Google Scholar]
- He, S.; Hu, B.; Li, C.; Lin, P.; Tang, W.G.; Sun, Y.F.; Feng, F.Y.; Guo, W.; Li, J.; Xu, Y.; et al. PDXliver: A Database of Liver Cancer Patient Derived Xenograft Mouse Models. BMC Cancer 2018, 18, 550. [Google Scholar] [CrossRef] [PubMed]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of Liver Diseases in the World. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Kang, L.I.; Mars, W.M.; Michalopoulos, G.K. Signals and Cells Involved in Regulating Liver Regeneration. Cells 2012, 1, 1261–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raven, A.; Lu, W.Y.; Man, T.Y.; Ferreira-Gonzalez, S.; O’Duibhir, E.; Dwyer, B.J.; Thomson, J.P.; Meehan, R.R.; Bogorad, R.; Koteliansky, V.; et al. Cholangiocytes Act As Facultative Liver Stem Cells During Impaired Hepatocyte Regeneration. Nature 2017, 547, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Markova, S.M.; De Marco, T.; Bendjilali, N.; Kobashigawa, E.A.; Mefford, J.; Sodhi, J.; Le, H.; Zhang, C.; Halladay, J.; Rettie, A.E.; et al. Association of CYP2C9*2 With Bosentan-Induced Liver Injury. Clin. Pharmacol Ther. 2013, 94, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Shinozawa, T.; Kimura, M.; Cai, Y.; Saiki, N.; Yoneyama, Y.; Ouchi, R.; Koike, H.; Maezawa, M.; Zhang, R.R.; Dunn, A.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell-Derived Organoids. Gastroenterology 2021, 160, 831–846. [Google Scholar] [CrossRef]
- Morris, E.J.; Jha, S.; Restaino, C.R.; Dayananth, P.; Zhu, H.; Cooper, A.; Carr, D.; Deng, Y.; Jin, W.; Black, S.; et al. Discovery of a Novel ERK Inhibitor With Activity in Models of Acquired Resistance to BRAF and MEK Inhibitors. Cancer Discov. 2013, 3, 742–750. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Knutsdottir, H.; Hui, K.; Weiss, M.J.; He, J.; Philosophe, B.; Cameron, A.M.; Wolfgang, C.L.; Pawlik, T.M.; Ghiaur, G.; et al. Human Primary Liver Cancer Organoids Reveal Intratumor and Interpatient Drug Response Heterogeneity. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
| Disease | Species | Source | References |
|---|---|---|---|
| Alagille syndrome | Human | Biopsy | [12] |
| iPSCs | [19] | ||
| Mouse | Adult tissue | [24] | |
| Alpha-1 antitrypsin deficiency | Human | Biopsy | [12] |
| Alcohol-related steatohepatitis | Human | ESCs | [25] |
| Cystic fibrosis | Human | iPSCs | [26] |
| HBV infection | Human | iPSCs | [27] |
| HCV infection | Human | iPSCs | [28] |
| Liver cancer | Human | Biopsy | [29,30] |
| Human | Adult tissue (GE) | [31] | |
| Sars-CoV-2 infection | Human | PSCs | [32] |
| Steatosis and steatohepatitis | Human | iPSCs | [33] |
| Cat | Adult tissue | [34,35] | |
| Wilson’s disease | Dog | Adult tissue | [34,36] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caiazza, C.; Parisi, S.; Caiazzo, M. Liver Organoids: Updates on Disease Modeling and Biomedical Applications. Biology 2021, 10, 835. https://doi.org/10.3390/biology10090835
Caiazza C, Parisi S, Caiazzo M. Liver Organoids: Updates on Disease Modeling and Biomedical Applications. Biology. 2021; 10(9):835. https://doi.org/10.3390/biology10090835
Chicago/Turabian StyleCaiazza, Carmen, Silvia Parisi, and Massimiliano Caiazzo. 2021. "Liver Organoids: Updates on Disease Modeling and Biomedical Applications" Biology 10, no. 9: 835. https://doi.org/10.3390/biology10090835
APA StyleCaiazza, C., Parisi, S., & Caiazzo, M. (2021). Liver Organoids: Updates on Disease Modeling and Biomedical Applications. Biology, 10(9), 835. https://doi.org/10.3390/biology10090835







