Characterization of Cell-Envelope Proteinases from Two Lacticaseibacillus casei Strains Isolated from Parmigiano Reggiano Cheese
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Species Confirmation, Culture Conditions, and Chemicals
2.2. DNA Extraction
2.3. In Silico Analysis, PCR Screening, and Phylogenetic Tree Construction
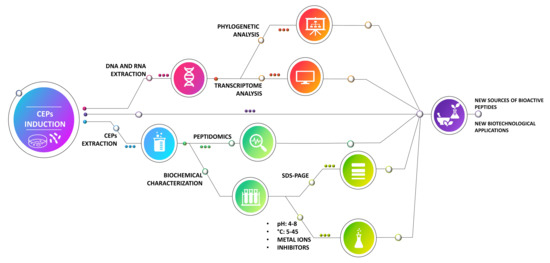
2.4. Induction of Cell-Envelope Proteinases (CEPs)
2.5. RNA Extraction and RT-PCR
2.6. CEP Extraction and Protein Concentration Determination
2.7. CEPs’ Enzymatic Activity Assay
2.8. Effects of Temperature, pH, Metal Ions, and Inhibitors on CEPs’ Activity
2.9. Casein Hydrolysis
2.10. SDS–PAGE
2.11. Identification of Peptides by Ultrahigh-Performance Liquid Chromatography/High-Resolution Mass Spectrometry (UHPLC/HRMS)
2.12. Identification of Bioactive Peptides
2.13. Calculation of the Cleavage Specificity
2.14. Statistical Analysis
3. Results and Discussion
3.1. In Silico Survey of Putative prt Genes in the L. casei Genome
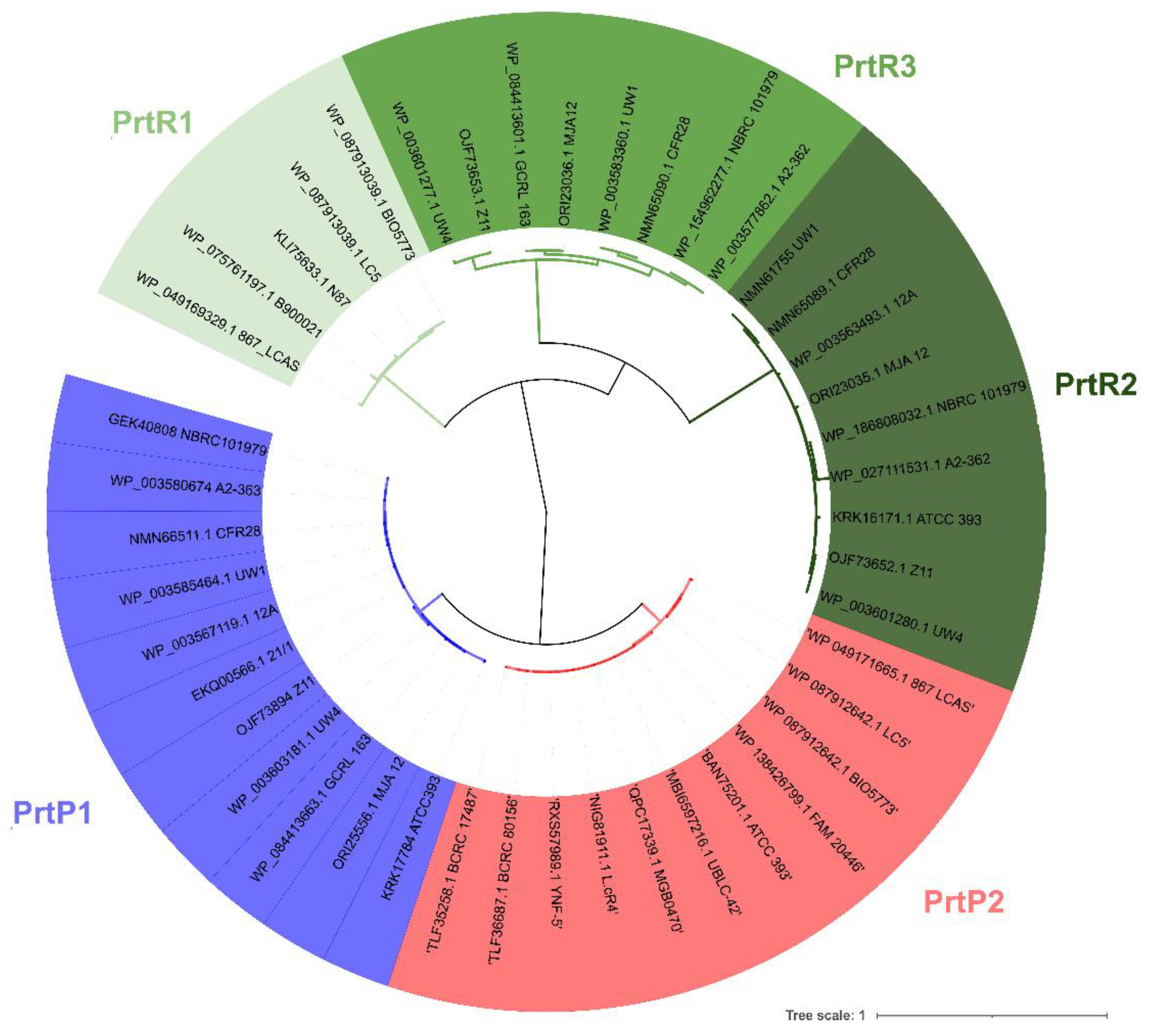
3.2. PCR Screening and Phylogenetic Analysis
3.3. CEP Extraction and prt Gene Expression Profile
3.4. Biochemical Characterization of L. casei PRA205 and 2006 Cell-Envelope Proteinases’ PrtR1
3.5. Caseinolytic Specificity of Extracted PrtR1, and PeptidomicsAnalysis of Casein Hydrolysates
3.6. Analysis of the β-Casein Cleavage Site Specificity
3.7. Analysis of the αS1-Casein Cleavage Site Specificity
3.8. Identification of Bioactive Peptides Using the Milk Bioactive Peptide Database
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christensen, J.E.; Dudley, E.G.; Pederson, J.A.; Steele, J.L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek 1999, 76, 217–246. [Google Scholar] [CrossRef] [PubMed]
- Tagliazucchi, D.; Baldaccini, A.; Martini, S.; Bianchi, A.; Pizzamiglio, V.; Solieri, L. Cultivable non-starter lactobacilli from ripened Parmigiano Reggiano cheeses with different salt content and their potential to release anti-hypertensive peptides. Int. J. Food Microbiol. 2020, 330, 108688. [Google Scholar] [CrossRef]
- Solieri, L.; Bianchi, A.; Giudici, P. Inventory of non starter lactic acid bacteria from ripened Parmigiano Reggiano cheese as assessed by a culture dependent multiphasic approach. Syst. Appl. Microbiol. 2012, 35, 270–277. [Google Scholar] [CrossRef]
- Sadat-Mekmene, L.; Genay, M.; Atlan, D.; Lortal, S.; Gagnaire, V. Original features of cell-envelope proteinases of Lactobacillus helveticus. A review. Int. J. Food Microbiol. 2011, 146, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Ma, J.; Xu, M.; Agyei, D. Cell-envelope proteinases from lactic acid bacteria: Biochemical features and biotech-nological applications. Compr. Rev. Food Sci. Food Saf. 2021, 20, 369–400. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Huang, J.; Cao, B.; Chen, L.; Song, N.; Lei, N. Study of Extraction and Enzymatic Properties of Cell-Envelope Proteinases from a Novel Wild Lactobacillus plantarum LP69. Catalysts 2018, 8, 325. [Google Scholar] [CrossRef] [Green Version]
- Fira, D.; Kojic, M.; Banina, A.; Spasojevic, I.; Strahinic, I.; Topisirovic, L. Characterization of cell envelope-associated proteinases of thermophilic lactobacilli. J. Appl. Microbiol. 2001, 90, 123–130. [Google Scholar] [CrossRef]
- Liu, M.; Bayjanov, J.R.; Renckens, B.; Nauta, A.; Siezen, R.J. The proteolytic system of lactic acid bacteria revisited: A genomic comparison. BMC Genom. 2010, 11, 36. [Google Scholar] [CrossRef] [Green Version]
- Tagliazucchi, D.; Martini, S.; Solieri, L. Bioprospecting for Bioactive Peptide Production by Lactic Acid Bacteria Isolated from Fermented Dairy Food. Fermentation 2019, 5, 96. [Google Scholar] [CrossRef] [Green Version]
- Von Schillde, M.-A.; Hörmannsperger, G.; Weiher, M.; Alpert, C.-A.; Hahne, H.; Bäuerl, C.; van Huynegem, K.; Steidler, L.; Hrncir, T.; Pérez-Martínez, G.; et al. Lactocepin Secreted by Lactobacillus Exerts Anti-Inflammatory Effects By Selectively Degrading Proinflammatory Chemokines. Cell Host Microbe 2012, 11, 387–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottari, B.; Felis, G.E.; Salvetti, E.; Castioni, A.; Campedelli, I.; Torriani, S.; Bernini, V.; Gatti, M. Effective identification of Lactobacillus casei group species: Genome-based selection of the gene mutL as the target of a novel multiplex PCR assay. Microbiology 2017, 163, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Gala, E.; Landi, S.; Solieri, L.; Nocetti, M.; Pulvirenti, A.; Giudici, P. Diversity of lactic acid bacteria population in ripened Parmigiano Reggiano cheese. Int. J. Food Microbiol. 2008, 125, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kõressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acid. Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Kojic, M.; Fira, D.; Banina, A.; Topisirovic, L. Characterization of the Cell Wall-Bound Proteinase of Lactobacillus casei HN14. Appl. Environ. Microbiol. 1991, 57, 1753–1757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, T.; Ouyang, X.; Xin, Y.; Wang, Y.; Zhang, S.; Kong, J. Characterization of a New Cell Envelope Proteinase PrtP from Lactobacillus rhamnosus CGMCC11055. J. Agric. Food Chem. 2016, 64, 6985–6992. [Google Scholar] [CrossRef] [PubMed]
- Hebert, E.M.; Mamone, G.; Picariello, G.; Raya, R.R.; Savoy, G.; Ferranti, P.; Addeo, F. Characterization of the pattern of αs1- and β-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581. Appl. Environ. Microbiol. 2008, 74, 3682–3689. [Google Scholar] [CrossRef] [Green Version]
- Martini, S.; Solieri, L.; Cattivelli, A.; Pizzamiglio, V.; Tagliazucchi, D. An Integrated Peptidomics and In Silico Approach to Identify Novel Anti-Diabetic Peptides in Parmigiano-Reggiano Cheese. Biology 2021, 10, 563. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2018, 232, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Solieri, L.; De Vero, L.; Tagliazucchi, D. Peptidomic study of casein proteolysis in bovine milk by Lactobacillus casei PRA205 and Lactobacillus rhamnosus PRA331. Int. Dairy J. 2018, 85, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Kok, J.; Leenhouts, K.J.; Haandrikman, A.J.; Ledeboer, A.M.; Venema, G. Nucleotide-sequence of the cell-wall pro-teinase gene of Streptococcus cremoris WG2. Appl. Environ. Microbiol. 1988, 54, 231–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolić, M.; Tolinački, M.; Fira, D.; Golić, N.; Topisirović, L. Variation in specificity of the PrtP extracellular proteinases in Lactococcus lactis and Lactobacillus paracasei subsp. paracasei. Folia Microbiol. 2009, 54, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Vukotić, G.; Strahinić, I.; Begović, J.; Lukić, J.; Kojić, M.; Fira, D. Survey on proteolytic activity and diversity of pro-teinase genes in mesophilic lactobacilli. Microbiology 2016, 85, 33–41. [Google Scholar] [CrossRef]
- Asahina, Y.; Hagi, T.; Kobayashi, M.; Narita, T.; Sasaki, K.; Tajima, A.; Nomura, M. Expression profiles of milk pro-teolysis-related genes in Lactobacillus paracasei EG9, a non-starter lactic acid bacterial strain, during Gouda-type cheese ripening. Int. Dairy J. 2020, 110, 104812. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E.V. Evolutionary Genomics of Lactic Acid Bacteria. J. Bacteriol. 2007, 189, 1199–1208. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.H.; Chen, C.C.; Liou, J.S.; Lee, A.Y.; Blom, J.; Lin, Y.C.; Huang, L.; Watanabe, K. Genome-based reclassifi-cation of Lactobacillus casei: Emended classification and description of the species Lactobacillus zeae. Int. J. Syst. Evol. Microbiol. 2020, 70, 3755–3762. [Google Scholar] [CrossRef]
- Liu, D.D.; Gu, C.T. Proposal to reclassify Lactobacillus zhaodongensis, Lactobacillus zeae, Lactobacillus argentoratensis and Lactobacillus buchneri subsp. silagei as Lacticaseibacillus zhaodongensis comb. nov., Lacticaseibacillus zeae comb. nov., Lactiplantibacillus argentoratensis comb. nov. and Lentilactobacillus buchneri subsp. silagei comb. nov., respectively and Apilactobacillus kosoi as a later heterotypic synonym of Apilactobacillus micheneri. Int. J. Syst. Evol. Microbiol. 2020, 70, 6414–6417. [Google Scholar] [CrossRef]
- Kojic, M.; Strahinic, I.; Topisirovic, L. Proteinase PI and lactococcin A genes are located on the largest plasmid in Lactococcus lactis subsp. lactis bv. diacetylactis S50. Can. J. Microbiol. 2005, 51, 305–314. [Google Scholar] [CrossRef]
- Dandoy, D.; Fremaux, C.; de Frahan, M.H.; Horvath, P.; Boyaval, P.; Hols, P.; Fontaine, L. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb. Cell Factories 2011, 10, S21. [Google Scholar] [CrossRef] [Green Version]
- Selle, K.; Klaenhammer, T.R.; Barrangou, R. CRISPR-based screening of genomic island excision events in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 8076–8081. [Google Scholar] [CrossRef] [Green Version]
- Alcantara, C.; Bäuerl, C.; Revilla-Guarinos, A.; Pérez-Martínez, G.; Monedero, V.; Zúñiga, M. Peptide and amino acid metabolism is controlled by an OmpR-family response regulator in Lactobacillus casei. Mol. Microbiol. 2016, 100, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Hébert, E.M.; Raya, R.R.; de Giori, G.S. Modulation of the Cell-Surface Proteinase Activity of Thermophilic Lactobacilli by the Peptide Supply. Curr. Microbiol. 2002, 45, 385–389. [Google Scholar] [CrossRef]
- Geis, A.; Bockelmann, W.; Teuber, M. Simultaneous extraction and purification of a cell wall-associated peptidase and β-casein specific protease from Streptococcus cremoris AC1. Appl. Microbiol. Biotechnol. 1985, 23, 79–84. [Google Scholar] [CrossRef]
- Blanc, B.; Laloi, P.; Atlan, D.; Gilbert, C.; Portalier, R. Two cell-wall-associated aminopeptidases from Lactobacillus helveticus and the purification and characterization of APII from strain ITGL1. J. Gen. Microbiol. 1993, 139, 1441–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiefer-Partsch, B.; Bockelmann, W.; Geis, A.; Teuber, M. Purification of an X-prolyl-dipeptidyl aminopeptidase from the cell wall proteolytic system of Lactococcus lactis subsp. cremoris. Appl. Microbiol. Biotechnol. 1989, 31, 75–78. [Google Scholar] [CrossRef]
- El Soda, M.; Desmazeaud, M.J.; Le Bars, D.; Zevaco, C. Cell-wall-associated proteinases in Lactobacillus casei and Lac-tobacillus plantarum. J. Food Prot. 1986, 49, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Pastar, I.; Fira, D.; Strahinic, I.; Krstic, K.; Begovic, J.; Topisirovic, L.; Jovanovic, G. Analysis of the presence of prtR proteinase gene in natural isolates of Lactobacillus rhamnsosus. Folia Microbiol. 2006, 51, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Fernández De Palencia, P.; Peláez, C.; Romero, C.; Martín-Hernández, C. Purification and Characterization of the Cell Wall Proteinase of Lactobacillus casei Subsp. casei IFPL 731 Isolated from Raw Goat’s Milk Cheese. J. Agric. Food Chem. 1997, 45, 6985–6992. [Google Scholar] [CrossRef]
- Solieri, L.; Rutella, G.S.; Tagliazucchi, D. Impact of non-starter lactobacilli on release of peptides with angioten-sin-converting enzyme inhibitory and antioxidant activities during bovine milk fermentation. Food Microbiol. 2015, 51, 108–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozo, J.; Strahinic, I.; Dalgalarrondo, M.; Chobert, J.-M.; Haertlé, T.; Topisirovic, C. Comparative analysis of β-casein proteolysis by PrtP proteinase from Lactobacillus paracasei subsp. paracasei BGHN14, PrtR proteinase from Lactobacillus rhamnosus BGT10 and PrtH proteinase from Lactobacillus helveticus BGRA43. Int. Dairy J. 2011, 21, 863–868. [Google Scholar] [CrossRef]
- Monnet, V.; Ley, J.P.; Gonzàlez, S. Substrate specificity of the cell envelope-located proteinase of Lactococcus lactis subsp. lactis NCDO 763. Int. J. Biochem. 1992, 24, 707–718. [Google Scholar] [CrossRef]
- Sowmya, K.; Mala, D.; Bhat, M.I.; Kumar, N.; Bajaj, R.K.; Kapila, S.; Kapila, R. Bio-accessible milk casein derived tripeptide (LLY) mediates overlapping anti- inflammatory and anti-oxidative effects under cellular (Caco-2) and in vivo milieu. J. Nutr. Biochem. 2018, 62, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Coste, M.; Rochet, V.; Léonil, J.; Mollé, D.; Bouhallab, S.; Tomé, D. Identification of C-terminal peptides of bovine β-casein that enhance proliferation of rat lymphocytes. Immunol. Lett. 1992, 33, 41–46. [Google Scholar] [CrossRef]
- Tagliazucchi, D.; Helal, A.; Verzelloni, E.; Bellesia, A.; Conte, A. Composition and properties of peptides that survive standardised in vitro gastro-pancreatic digestion of bovine milk. Int. Dairy J. 2016, 61, 196–204. [Google Scholar] [CrossRef]
- Boutrou, R.; Gaudichon, C.; Dupont, D.; Jardin, J.; Airinei, G.; Marsset-Baglieri, A.; Benamouzig, R.; Tome’, D.; Leonil, J. Sequential release of milk protein–derived bioactive peptides in the jejunum in healthy humans. Am. J. Clin. Nutr. 2013, 97, 1314–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| L. casei PRA205 PrtR1 | L. casei 2006 PrtR1 | |||||||
|---|---|---|---|---|---|---|---|---|
| Amino Acid 1 | P1 Subsite | P1′ Subsite | P1 Subsite | P1′ Subsite | ||||
| Number of Cleaved Bonds 2 (%P1 3) | Kn | Number of Cleaved Bonds 2 (%P1′ 3) | Kn | Number of Cleaved Bonds 2 (%P1 3) | Kn | Number of Cleaved Bonds 2 (%P1′ 3) | Kn | |
| Aliphatic | ||||||||
| A (5) | 2 (40.0) | 0.08 | 4 (80.0) | 0.86 | 3 (60.0) | 0.69 | 5 (100.0) | 1.42 |
| G (5) | 1 (20.0) | −0.46 | 3 (60.0) | 0.40 | 2 (40.0) | 0.13 | 3 (60.0) | 0.45 |
| V (19) | 3 (15.8) | −0.58 | 9 (47.4) | 0.10 | 3 (15.8) | −0.56 | 8 (42.1) | 0.02 |
| L (22) | 10 (45.5) | 0.22 | 10 (45.5) | 0.06 | 9 (40.9) | 0.15 | 8 (36.4) | −0.12 |
| I (10) | 1 (10.0) | −0.73 | 4 (40.0) | −0.07 | 1 (10.0) | −0.72 | 3 (30.0) | −0.27 |
| M (6) | 5 (83.3) | 1.24 | 2 (33.3) | −0.22 | 4 (66.7) | 0.88 | 2 (33.3) | −0.19 |
| Polar un-charged | ||||||||
| T (9) | 2 (22.2) | −0.40 | 2 (22.2) | −0.48 | 1 (11.1) | −0.69 | 1 (11.1) | −0.73 |
| S (16) | 8 (50.0) | 0.35 | 7 (43.8) | 0.02 | 7 (43.8) | 0.23 | 7 (43.8) | 0.06 |
| Q (20) | 10 (50.0) | 0.35 | 2 (10.0) | −0.77 | 10 (50.0) | 0.41 | 3 (15.0) | −0.64 |
| N (5) | 4 (80.0) | 1.15 | 2 (40.0) | −0.07 | 3 (60.0) | 0.69 | 1 (20.0) | −0.52 |
| P (35) | 8 (22.9) | −0.38 | 4 (11.4) | −0.73 | 5 (14.3) | −0.60 | 4 (11.4) | −0.72 |
| Positively charged | ||||||||
| R (4) | 2 (50.0) | 0.35 | 1 (25.0) | −0.42 | 2 (50.0) | 0.41 | 1 (25.0) | −0.40 |
| K (11) | 5 (45.5) | 0.22 | 4 (36.4) | −0.15 | 6 (54.6) | 0.54 | 5 (45.5) | 0.10 |
| H (5) | 1 (20.0) | −0.46 | 3 (60.0) | 0.40 | 1 (20.0) | −0.44 | 3 (60.0) | 0.45 |
| Negatively charged | ||||||||
| E (19) | 3 (15.8) | −0.58 | 2 (10.5) | −0.76 | 5 (26.3) | −0.26 | 2 (10.5) | −0.75 |
| D (4) | 1 (25.0) | −0.33 | 4 (100.0) | 1.33 | 0 (0) | −1.00 | 4 (100.0) | 1.42 |
| Aromatic | ||||||||
| F (9) | 2 (22.2) | −0.40 | 4 (44.4) | 0.03 | 2 (22.2) | −0.37 | 3 (33.3) | −0.19 |
| Y (4) | 1 (25.0) | −0.33 | 2 (50.0) | 0.16 | 1 (25.0) | −0.30 | 2 (50.0) | 0.21 |
| Sequence | Fragment | Bioactivity | PrtR1 |
|---|---|---|---|
| VVPP | β-casein f(83–86) | ACE inhibition | L. casei PRA205 |
| VKEAMAPK | β-casein f(98–105) | Antioxidant Antimicrobial | L. casei 2006 |
| EAMAPK | β-casein f(100–105) | Antimicrobial | L. casei 2006 |
| SQSKVLPVPQ | β-casein f(166–175) | ACE inhibition | L. casei PRA205 L. casei 2006 |
| KVLPVPQ | β-casein f(169–175) | ACE inhibition | L. casei 2006 |
| VLPVPQKAVPYPQR | β-casein f(170–183) | Antimicrobial | L. casei PRA205 L. casei 2006 |
| VLPVPQK | β-casein f(170–176) | ACE inhibition Antioxidant Antimicrobial | L. casei 2006 |
| LPVPQ | β-casein f(171–175) | DPP-IV inhibition | L. casei PRA205 L. casei 2006 |
| PYPQ | β-casein f(179–182) | Antioxidant | L. casei PRA205 L. casei 2006 |
| RDMPIQAF | β-casein f(183–190) | ACE inhibition | L. casei PRA205 L. casei 2006 |
| LLY | β-casein f(191–193) | Immunomodulation | L. casei PRA205 L. casei 2006 |
| LLYQEPVLGPVRGPFPIIV | β-casein f(191–209) | ACE inhibition | L. casei PRA205 L. casei 2006 |
| LYQEPVLGPVRGPFPIIV | β-casein f(192–209) | Immunomodulation | L. casei PRA205 L. casei 2006 |
| YQEPVLGPVR | β-casein f(193-202) | ACE inhibition Immunomodulation | L. casei PRA205 L. casei 2006 |
| YQEPVLGPVRGPFPI | β-casein f(193–207) | Antimicrobial | L. casei PRA205 L. casei 2006 |
| YQEPVLGPVRGPFPIIV | β-casein f(193–209) | ACE inhibition Immunomodulation Antimicrobial | L. casei PRA205 L. casei 2006 |
| QEPVLGPVRGPFPIIV | β-casein f(194–209) | ACE inhibition | L. casei PRA205 L. casei 2006 |
| EPVLGPVRGPFP | β-casein f(195–206) | ACE inhibition | L. casei PRA205 L. casei 2006 |
| RPKHPIK | αS1-casein f(1–7) | Antimicrobial | L. casei 2006 |
| RPKHPIKHQ | αS1-casein f(1–9) | ACE inhibition | L. casei 2006 |
| RPKHPIKHQGLPQEVLNENLLRF | αS1-casein f(1–23) | Immunomodulation Antimicrobial | L. casei PRA205 L. casei 2006 |
| RPKHPIKHQGLPQEVLNENLLRFF | αS1-casein f(1–24) | Antimicrobial | L. casei PRA205 L. casei 2006 |
| RPKHPIKHQGLPQEVLNENLLRFFVAPFPEVFGKEK | αS1-casein f(1–36) | Antimicrobial | L. casei 2006 |
| VLNENLLR | αS1-casein f(15–22) | Antimicrobial | L. casei PRA205 L. casei 2006 |
| YLGYLEQLLR | αS1-casein f(91–100) | Anxiolytic | L. casei PRA205 |
| LGY | αS1-casein f(92–94) | ACE inhibition Antioxidant | L. casei PRA205 L. casei 2006 |
| LGYLEQLLRL | αS1-casein f(92–101) | Antimicrobial | L. casei PRA205 L. casei 2006 |
| YLEQLLR | αS1-casein f(94–100) | Antimicrobial | L. casei PRA205 L. casei 2006 |
| PEL | αS1-casein f(147–149) | Antioxidant | L. casei PRA205 L. casei 2006 |
| GTQYTDAPSFSDIPNPIGSENSEKTTMPLW | αS1-casein f(170–199) | ACE inhibition Antioxidant | L. casei PRA205 L. casei 2006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solieri, L.; Sola, L.; Vaccalluzzo, A.; Randazzo, C.L.; Martini, S.; Tagliazucchi, D. Characterization of Cell-Envelope Proteinases from Two Lacticaseibacillus casei Strains Isolated from Parmigiano Reggiano Cheese. Biology 2022, 11, 139. https://doi.org/10.3390/biology11010139
Solieri L, Sola L, Vaccalluzzo A, Randazzo CL, Martini S, Tagliazucchi D. Characterization of Cell-Envelope Proteinases from Two Lacticaseibacillus casei Strains Isolated from Parmigiano Reggiano Cheese. Biology. 2022; 11(1):139. https://doi.org/10.3390/biology11010139
Chicago/Turabian StyleSolieri, Lisa, Laura Sola, Amanda Vaccalluzzo, Cinzia Lucia Randazzo, Serena Martini, and Davide Tagliazucchi. 2022. "Characterization of Cell-Envelope Proteinases from Two Lacticaseibacillus casei Strains Isolated from Parmigiano Reggiano Cheese" Biology 11, no. 1: 139. https://doi.org/10.3390/biology11010139
APA StyleSolieri, L., Sola, L., Vaccalluzzo, A., Randazzo, C. L., Martini, S., & Tagliazucchi, D. (2022). Characterization of Cell-Envelope Proteinases from Two Lacticaseibacillus casei Strains Isolated from Parmigiano Reggiano Cheese. Biology, 11(1), 139. https://doi.org/10.3390/biology11010139