MV1035 Overcomes Temozolomide Resistance in Patient-Derived Glioblastoma Stem Cell Lines
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Database Preparation
2.2. RBS Generation
2.3. In Silico Screening and Ranking of the Protein Database
2.4. Synthesis of MV1035
2.5. Cell Free Assay
2.6. Glioma Cell Lines
2.7. Cytotoxicity Assay
2.8. Limiting Dilution Assay (LDA)
2.9. MGMT Expression
2.10. MGMT Methylation Analysis
3. Results
3.1. SPILLO-PBSS Screening
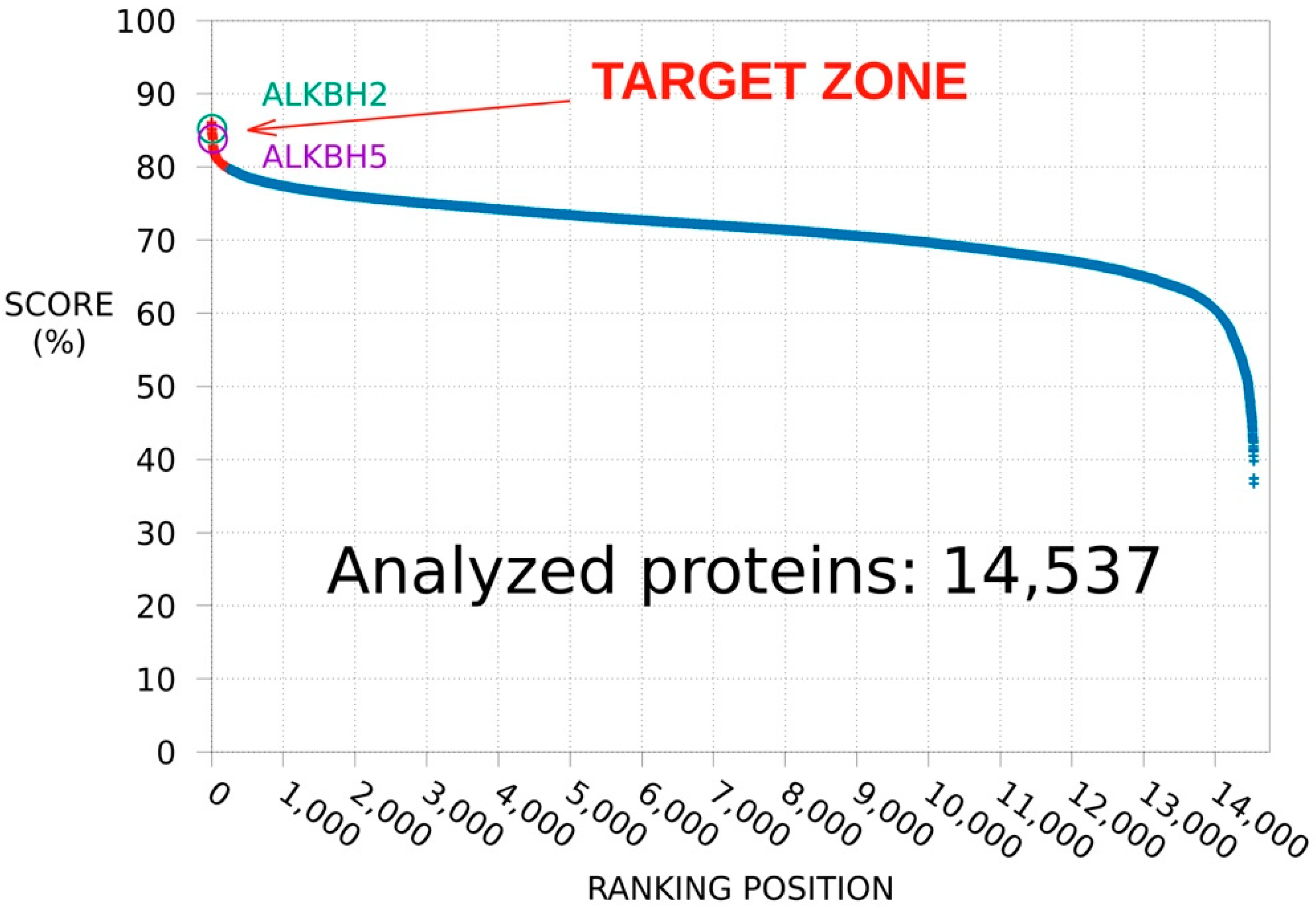
3.1.1. Protein Database Ranking
3.1.2. ALKBH2 as a Further Potential Target of MV1035
3.1.3. Competitive Inhibition Hypothesis
3.2. Target Validation
ALKBH2 Inhibition by MV1035: Cell Free Assay
3.3. Biological Validation
3.3.1. Cytotoxicity Assay
3.3.2. Limiting Dilution Assay
3.3.3. MGMT Expression
3.3.4. MGMT Promoter Methylation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cote, D.J.; Ascha, M.; Kruchko, C.; Barnholtz-Sloan, J.S. Adult Glioma Incidence and Survival by Race or Ethnicity in the United States from 2000 to 2014. JAMA Oncol. 2018, 4, 1254. [Google Scholar] [CrossRef]
- Ricard, D.; Idbaih, A.; Ducray, F.; Lahutte, M.; Hoang-Xuan, K.; Delattre, J.-Y. Primary brain tumours in adults. Lancet 2012, 379, 1984–1996. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Garcia, M.; Álvarez-Linera, J.; Carrato, C.; Ley, L.; Luque, R.; Maldonado, X.; Martínez-Aguillo, M.; Navarro, L.M.; Vaz-Salgado, M.A.; Gil-Gil, M. SEOM clinical guidelines for diagnosis and treatment of glioblastoma (2017). Clin. Transl. Oncol. 2018, 20, 22–28. [Google Scholar] [CrossRef]
- Wu, W.; Klockow, J.L.; Zhang, M.; Lafortune, F.; Chang, E.; Jin, L.; Wu, Y.; Daldrup-Link, H.E. Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharmacol. Res. 2021, 171, 105780. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Christmann, M.; Kaina, B. Epigenetic regulation of DNA repair genes and implications for tumor therapy. Mutat. Res. Rev. Mutat. Res. 2019, 780, 15–28. [Google Scholar] [CrossRef]
- Gutierrez, R.; O’Connor, T.R. DNA direct reversal repair and alkylating agent drug resistance. Cancer Drug Resist. 2021, 4, 414–423. [Google Scholar] [CrossRef]
- Esteller, M.; Hamilton, S.R.; Burger, P.C.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999, 59, 793–797. [Google Scholar]
- Esteller, M.; Corn, P.G.; Baylin, S.B.; Herman, J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001, 61, 3225–3229. [Google Scholar] [PubMed]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 2000, 343, 1350–1354. [Google Scholar] [CrossRef]
- Johannessen, T.-C.A.; Prestegarden, L.; Grudic, A.; Hegi, M.E.; Tysnes, B.B.; Bjerkvig, R. The DNA repair protein ALKBH2 mediates temozolomide resistance in human glioblastoma cells. Neuro-Oncology 2013, 15, 269–278. [Google Scholar] [CrossRef]
- Malacrida, A.; Rivara, M.; Di Domizio, A.; Cislaghi, G.; Miloso, M.; Zuliani, V.; Nicolini, G. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorganic Med. Chem. 2020, 28. [Google Scholar] [CrossRef]
- Di Domizio, A.; Vitriolo, A.; Vistoli, G.; Pedretti, A. SPILLO-PBSS: Detecting hidden binding sites within protein 3D-structures through a flexible structure-based approach. J. Comput. Chem. 2014, 35, 2005–2017. [Google Scholar] [CrossRef]
- Raghavendra, N.M.; Pingili, D.; Kadasi, S.; Mettu, A.; Prasad, S.V.U.M. Dual or multi-targeting inhibitors: The next generation anticancer agents. Eur. J. Med. Chem. 2018, 143, 1277–1300. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, R.B.; Homayouni, T.S.; Baluch, N.; Morgatskaya, E.; Kumar, S.; Das, B.; Yeger, H.; Mokhtari, R.B.; Homayouni, T.S.; Baluch, N.; et al. Combination therapy in combating cancer. Oncotarget 2017, 8, 38022–38043. [Google Scholar] [CrossRef]
- Fantini, M.; Zuliani, V.; Spotti, M.A.; Rivara, M. Microwave assisted efficient synthesis of imidazole-based privileged structures. J. Comb. Chem. 2010, 12, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Rivara, M.; Fantini, M.; Acquotti, D.; Zuliani, V. NMR analysis of a series of imidazobenzoxazines. Magn. Reson. Chem. 2010, 48, 500–503. [Google Scholar] [CrossRef]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef]
- Baronchelli, S.; Bentivegna, A.; Redaelli, S.; Riva, G.; Butta, V.; Paoletta, L.; Isimbaldi, G.; Miozzo, M.; Tabano, S.; Daga, A.; et al. Delineating the cytogenomic and epigenomic landscapes of glioma stem cell lines. PLoS ONE 2013, 8, e57462. [Google Scholar] [CrossRef]
- Giambra, M.; Messuti, E.; Di Cristofori, A.; Cavandoli, C.; Bruno, R.; Buonanno, R.; Marzorati, M.; Zambuto, M.; Rodriguez-Menendez, V.; Redaelli, S.; et al. Characterizing the Genomic Profile in High-Grade Gliomas: From Tumor Core to Peritumoral Brain Zone, Passing through Glioma-Derived Tumorspheres. Biology 2021, 10, 1157. [Google Scholar] [CrossRef]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef]
- Ianevski, A.; Giri, A.K.; Aittokallio, T. SynergyFinder 2.0: Visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020, 48, W488–W493. [Google Scholar] [CrossRef]
- Hu, Y.; Smyth, G.K. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 2009, 347, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Giatti, S.; Di Domizio, A.; Diviccaro, S.; Falvo, E.; Caruso, D.; Contini, A.; Melcangi, R.C. Three-Dimensional Proteome-Wide Scale Screening for the 5-Alpha Reductase Inhibitor Finasteride: Identification of a Novel Off-Target. J. Med. Chem. 2021, 64, 4553–4566. [Google Scholar] [CrossRef]
- Malacrida, A.; Semperboni, S.; Di Domizio, A.; Palmioli, A.; Broggi, L.; Airoldi, C.; Meregalli, C.; Cavaletti, G.; Nicolini, G. Tubulin binding potentially clears up Bortezomib and Carfilzomib differential neurotoxic effect. Sci. Rep. 2021, 11, 10523. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Lan, F.; Yang, Y.; Han, J.; Wu, Q.; Yu, H.; Yue, X. Sulforaphane reverses chemo-resistance to temozolomide in glioblastoma cells by NF-κB-dependent pathway downregulating MGMT expression. Int. J. Oncol. 2016, 48, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Aguado, T.; Romero-Revilla, J.A.; Granados, R.; Campuzano, S.; Torrente-Rodríguez, R.M.; Cuesta, Á.M.; Albiñana, V.; Botella, L.M.; Santamaría, S.; Garcia-Sanz, J.A.; et al. 11PS04 is a new chemical entity identified by microRNA-based biosensing with promising therapeutic potential against cancer stem cells. Sci. Rep. 2019, 9, 11916. [Google Scholar] [CrossRef]
- Chien, C.-H.; Hsueh, W.-T.; Chuang, J.-Y.; Chang, K.-Y. Dissecting the mechanism of temozolomide resistance and its association with the regulatory roles of intracellular reactive oxygen species in glioblastoma. J. Biomed. Sci. 2021, 28, 18. [Google Scholar] [CrossRef]
- Costa, B.M.; Caeiro, C.; Guimarães, I.; Martinho, O.; Jaraquemada, T.; Augusto, I.; Castro, L.; Osório, L.; Linhares, P.; Honavar, M.; et al. Prognostic value of MGMT promoter methylation in glioblastoma patients treated with temozolomide-based chemoradiation: A Portuguese multicentre study. Oncol. Rep. 2010, 23, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, L.; Wei, Q.; Shao, A. O6-Methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front. Oncol. 2020, 9, 1547. [Google Scholar] [CrossRef]
- Erasimus, H.; Gobin, M.; Niclou, S.; Van Dyck, E. DNA repair mechanisms and their clinical impact in glioblastoma. Mutat. Res. Mutat. Res. 2016, 769, 19–35. [Google Scholar] [CrossRef]
- Allen, M.; Bjerke, M.; Edlund, H.; Nelander, S.; Westermark, B. Origin of the U87MG glioma cell line: Good news and bad news. Sci. Transl. Med. 2016, 8, 354re3. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef]
- Liu, G.; Yuan, X.; Zeng, Z.; Tunici, P.; Ng, H.; Abdulkadir, I.R.; Lu, L.; Irvin, D.; Black, K.L.; Yu, J.S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Pistollato, F.; Abbadi, S.; Rampazzo, E.; Persano, L.; Della Puppa, A.; Frasson, C.; Sarto, E.; Scienza, R.; D’avella, D.; Basso, G. Intratumoral hypoxic gradient drives stem cells distribution and MGMT expression in glioblastoma. Stem Cells 2010, 28, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Chumakova, A.; Lathia, J.D. Outlining involvement of stem cell program in regulation of O6-methylguanine DNA methyltransferase and development of temozolomide resistance in glioblastoma: An Editorial Highlight for “Transcriptional control of O6-methylguanine DNA methyltransferase expression and temozolomide resistance in glioblastoma” on page 780. J. Neurochem. 2018, 144, 688–690. [Google Scholar] [CrossRef]
- Happold, C.; Stojcheva, N.; Silginer, M.; Weiss, T.; Roth, P.; Reifenberger, G.; Weller, M. Transcriptional control of O6-methylguanine DNA methyltransferase expression and temozolomide resistance in glioblastoma. J. Neurochem. 2018, 144, 780–790. [Google Scholar] [CrossRef]
- Cabrini, G.; Fabbri, E.; Lo Nigro, C.; Dechecchi, M.C.; Gambari, R. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review). Int. J. Oncol. 2015, 47, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Tang, Q.; Ren, L.; Liu, J.; Li, W.; Fu, W.; Wang, J.; Du, G. A narrative review of research progress on drug therapies for glioblastoma multiforme. Ann. Transl. Med. 2021, 9, 943. [Google Scholar] [CrossRef]
- Quinn, J.A.; Jiang, S.X.; Reardon, D.A.; Desjardins, A.; Vredenburgh, J.J.; Gururangan, S.; Sampson, J.H.; McLendon, R.E.; Herndon, J.E.; Friedman, H.S. Phase 1 trial of temozolomide plus irinotecan plus O6-benzylguanine in adults with recurrent malignant glioma. Cancer 2009, 115, 2964–2970. [Google Scholar] [CrossRef][Green Version]
- Sabharwal, A.; Middleton, M.R. Exploiting the role of O6-methylguanine-DNA-methyltransferase (MGMT) in cancer therapy. Curr. Opin. Pharmacol. 2006, 6, 355–363. [Google Scholar] [CrossRef]
- Yoshino, A.; Ogino, A.; Yachi, K.; Ohta, T.; Fukushima, T.; Watanabe, T.; Katayama, Y.; Okamoto, Y.; Naruse, N.; Sano, E. Effect of IFN-beta on human glioma cell lines with temozolomide resistance. Int. J. Oncol. 2009, 35, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Bobustuc, G.C.; Baker, C.H.; Limaye, A.; Jenkins, W.D.; Pearl, G.; Avgeropoulos, N.G.; Konduri, S.D. Levetiracetam enhances p53-mediated MGMT inhibition and sensitizes glioblastoma cells to temozolomide. Neuro. Oncol. 2010, 12, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, S.; Wang, L.; Yachi, K.; Mahabir, R.; Narita, T.; Itoh, T.; Tanino, M.; Kimura, T.; Nishihara, H.; Tanaka, S. STAT3 inhibition overcomes temozolomide resistance in glioblastoma by downregulating MGMT expression. Mol. Cancer Ther. 2012, 11, 1289–1299. [Google Scholar] [CrossRef] [PubMed]






| TMZ IC50 µM | MV1035 | |||
|---|---|---|---|---|
| 0 µM | 10 µM | 25 µM | 50 µM | |
| U87-MG | 1578 ± 44 | 1295 ± 31 | 941 ± 79 | 465 ± 26 |
| G179 | 1984 ± 198 | 1770 ± 30 | 1289 ± 112 | 869 ± 36 |
| GSC7 | 1951 ± 142 | 988 ± 82 | 631 ± 36 | 357 ± 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malacrida, A.; Di Domizio, A.; Bentivegna, A.; Cislaghi, G.; Messuti, E.; Tabano, S.M.; Giussani, C.; Zuliani, V.; Rivara, M.; Nicolini, G. MV1035 Overcomes Temozolomide Resistance in Patient-Derived Glioblastoma Stem Cell Lines. Biology 2022, 11, 70. https://doi.org/10.3390/biology11010070
Malacrida A, Di Domizio A, Bentivegna A, Cislaghi G, Messuti E, Tabano SM, Giussani C, Zuliani V, Rivara M, Nicolini G. MV1035 Overcomes Temozolomide Resistance in Patient-Derived Glioblastoma Stem Cell Lines. Biology. 2022; 11(1):70. https://doi.org/10.3390/biology11010070
Chicago/Turabian StyleMalacrida, Alessio, Alessandro Di Domizio, Angela Bentivegna, Giacomo Cislaghi, Eleonora Messuti, Silvia Maria Tabano, Carlo Giussani, Valentina Zuliani, Mirko Rivara, and Gabriella Nicolini. 2022. "MV1035 Overcomes Temozolomide Resistance in Patient-Derived Glioblastoma Stem Cell Lines" Biology 11, no. 1: 70. https://doi.org/10.3390/biology11010070
APA StyleMalacrida, A., Di Domizio, A., Bentivegna, A., Cislaghi, G., Messuti, E., Tabano, S. M., Giussani, C., Zuliani, V., Rivara, M., & Nicolini, G. (2022). MV1035 Overcomes Temozolomide Resistance in Patient-Derived Glioblastoma Stem Cell Lines. Biology, 11(1), 70. https://doi.org/10.3390/biology11010070










