Construction of Light-Responsive Gene Regulatory Network for Growth, Development and Secondary Metabolite Production in Cordyceps militaris
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of the Genome-Scale GRN
2.2. Differentially Expressed Gene (DEG) Analysis and the Light-Responsive GRN
2.3. Identification of Key Regulator Genes
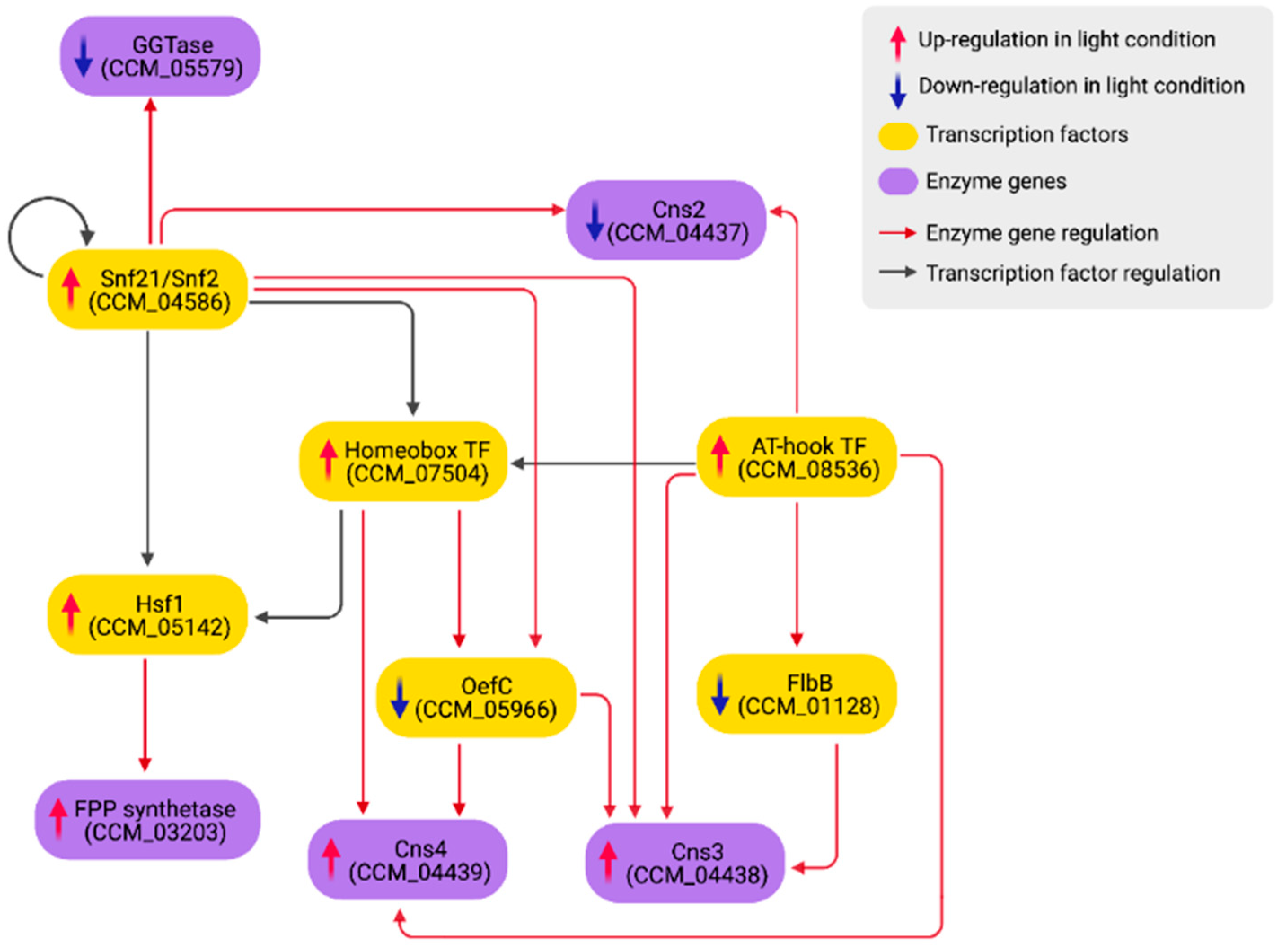
2.4. The Gene Regulatory Sub-Networks of Growth, Development, Carotenoid and Cordycepin Biosynthesis Pathways under Light Response
3. Results and Discussion
3.1. The Genome-Scale GRN
3.2. The Light-Responsive GRN
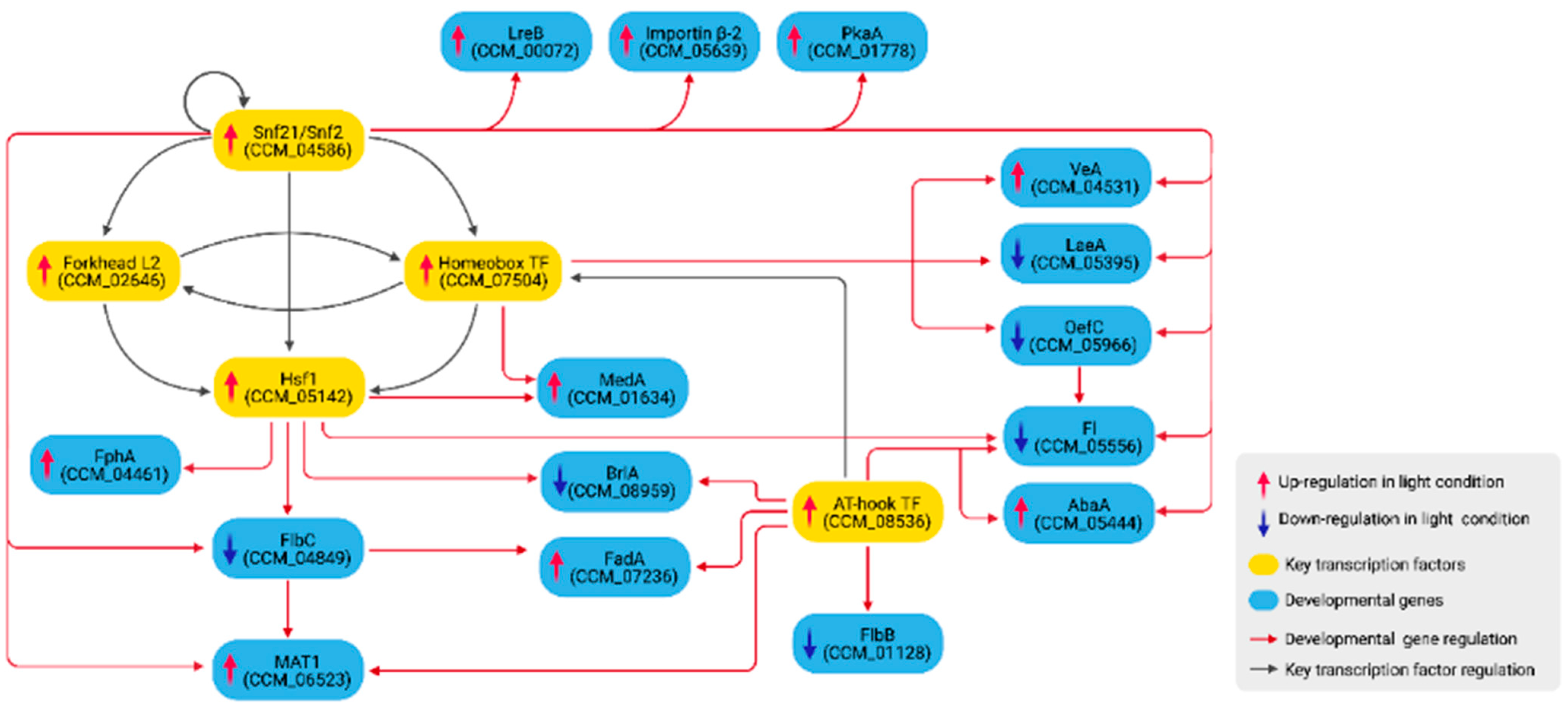
3.3. Growth and Developmental Regulation under Light Exposure
3.4. Light-Mediated Control of Secondary Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [Green Version]
- Das, S.K.; Masuda, M.; Sakurai, A.; Sakakibara, M. Medicinal uses of the mushroom Cordyceps militaris: Current state and prospects. Fitoterapia 2010, 81, 961–968. [Google Scholar] [CrossRef]
- Xiao, J.H.; Zhong, J.J. Secondary metabolites from Cordyceps species and their antitumor activity studies. Recent Pat. Biotechnol. 2008, 1, 123–137. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, K.G.; Hutchingson, S.A.; Manson, W.S.F.S. Cordycepin, a metabolic product from cultures of Cordyceps militaris (Linn.) link. Part I. Isolation and characterisation. J. Chem. Soc. 1951, 166, 2299–2300. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00003005 (accessed on 29 December 2021).
- Yang, T.; Sun, J.; Lian, T.; Wang, W.; Dong, C. Process optimization for extraction of carotenoids from medicinal caterpillar fungus, Cordyceps militaris (Ascomycetes). Int. J. Med. Mushrooms 2014, 16, 125–135. [Google Scholar] [CrossRef]
- Avalos, J.; Carmen Limón, M. Biological roles of fungal carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef]
- Stahl, W.; Sies, H. Bioactivity and protective effects of natural carotenoids. Biochim. Biophys. Acta Mol. Basis Dis. 2005, 1740, 101–107. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.Z.; Wang, S.H.; Ai, X.R.; Yao, L.; Sun, Z.W.; Lei, C.; Wang, Y.; Wang, Q. Composition and characterization of cordyxanthins from Cordyceps militaris fruit bodies. J. Funct. Foods 2013, 5, 1450–1455. [Google Scholar] [CrossRef]
- Coherent Market Insights. Available online: https://www.coherentmarketinsights.com/market-insight/cordyceps-sinensis-and-militaris-extract-market-2578 (accessed on 29 December 2021).
- Adnan, M.; Ashraf, S.A.; Khan, S.; Alshammari, E.; Awadelkareem, A.M. Effect of pH, temperature and incubation time on cordycepin production from Cordyceps militaris using solid-state fermentation on various substrates. CyTA J. Food 2017, 15, 617–621. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Guo, M.; Yang, H.; Guo, S.; Dong, C. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 2015, 100, 743–755. [Google Scholar] [CrossRef]
- Dong, J.Z.; Lei, C.; Zheng, X.J.; Ai, X.R.; Wang, Y.; Wang, Q. Light wavelengths regulate growth and active components of Cordyceps militaris fruit bodies. J. Food Biochem. 2013, 37, 578–584. [Google Scholar] [CrossRef]
- Kim, S.Y.; Shrestha, B.; Sung, G.H.; Han, S.K.; Sung, J.M. Optimum conditions for artificial fruiting body formation of Cordyceps cardinalis. Mycobiology 2010, 38, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.; Wen, T.C.; Kang, J.C.; Meng, Z.B.; Li, G.R.; Hyde, K.D. Optimization of large-scale culture conditions for the production of cordycepin with Cordyceps militaris by liquid static culture. Sci. World J. 2014, 2014, 20–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vongsangnak, W.; Raethong, N.; Mujchariyakul, W.; Nguyen, N.N.; Leong, H.W.; Laoteng, K. Genome-scale metabolic network of Cordyceps militaris useful for comparative analysis of entomopathogenic fungi. Genes 2017, 626, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Raethong, N.; Wang, H.; Nielsen, J.; Vongsangnak, W. Optimizing cultivation of Cordyceps militaris for fast growth and cordycepin overproduction using rational design of synthetic media. Comput. Struct. Biotechnol. J. 2020, 18, 1–8. [Google Scholar] [CrossRef]
- Sirithep, K.; Xiao, F.; Raethong, N.; Zhang, Y.; Laoteng, K.; Hu, G.; Vongsangnak, W. Probing carbon utilization of Cordyceps militaris by sugar transportome and protein structural analysis. Cells 2020, 9, 401. [Google Scholar] [CrossRef] [Green Version]
- Jiaojiao, Z.; Fen, W.; Kuanbo, L.; Qing, L.; Ying, Y.; Caihong, D. Heat and light stresses affect metabolite production in the fruit body of the medicinal mushroom Cordyceps militaris. Appl. Microbiol. Biotechnol. 2018, 102, 4523–4533. [Google Scholar] [CrossRef]
- Thananusak, R.; Laoteng, K.; Raethong, N.; Zhang, Y.; Vongsangnak, W. Metabolic responses of carotenoid and cordycepin biosynthetic pathways in Cordyceps militaris under light-programming exposure through genome-wide transcriptional analysis. Biology 2020, 9, 242. [Google Scholar] [CrossRef]
- Yu, Z.; Fischer, R. Light sensing and responses in fungi. Nat. Rev. Microbiol. 2019, 17, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Huang, H.; Deng, W.; Li, T. Genome-wide analysis of the Zn(II)2Cys6 Zinc cluster-encoding gene family in Tolypocladium guangdongense and its light-induced expression. Genes 2019, 10, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Han, K.Y.; Kim, K.J.; Han, D.M.; Jahng, K.Y.; Chae, K.S. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 2002, 37, 72–80. [Google Scholar] [CrossRef]
- Olmedo, M.; Ruger-Herreros, C.; Corrochano, L.M. Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa. Genetics 2010, 184, 651–658. [Google Scholar] [CrossRef] [Green Version]
- Qiu, L.; Wang, J.J.; Chu, Z.J.; Ying, S.H.; Feng, M.G. Phytochrome controls conidiation in response to red/far-red light and daylight length and regulates multistress tolerance in Beauveria bassiana. Environ. Microbiol. 2014, 16, 2316–2328. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, D.; Suzuki, S.; Kamei, K.; Gonoi, T.; Kawamoto, S. The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2014, 73, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Qin, Y.; Liu, G. Collection and curation of transcriptional regulatory interactions in Aspergillus nidulans and Neurospora crassa reveal structural and evolutionary features of the regulatory networks. Front. Microbiol. 2018, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.H.; Ren, B. Genome-wide analysis of protein-DNA interactions. Annu. Rev. Genom. Hum. Genet. 2006, 7, 81–102. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [Green Version]
- Turatsinze, J.V.; Thomas-Chollier, M.; Defrance, M.; van Helden, J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008, 3, 1578–1588. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Raethong, N.; Laoteng, K.; Vongsangnak, W. Uncovering global metabolic response to cordycepin production in Cordyceps militaris through transcriptome and genome-scale network-driven analysis. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [Green Version]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koschützki, D.; Schreiber, F. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul. Syst. Biol. 2008, 2, GRSB-S702. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Z.; Yao, X.; Huang, Y.; Qu, Q.; Shi, X.; Zhang, H.; Shi, X. Identification of cordycepin biosynthesis-related genes through de novo transcriptome assembly and analysis in Cordyceps cicadae. R. Soc. Open Sci. 2018, 5, 181247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyer, P.S.; O’Gorman, C.M. Sexual development and cryptic sexuality in fungi: Insights from Aspergillus species. FEMS Microbiol. Rev. 2012, 36, 165–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.; Lu, Y.; Wang, C. Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489. [Google Scholar] [CrossRef] [Green Version]
- Bayram, Ö.; Braus, G.H.; Fischer, R.; Rodriguez-Romero, J. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet. Biol. 2010, 47, 900–908. [Google Scholar] [CrossRef]
- Park, H.S.; Yu, J.H. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 2012, 15, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Krijgsheld, P.; Bleichrodt, R.; van Veluw, G.J.; Wang, F.; Müller, W.H.; Dijksterhuis, J.; Wösten, H.A.B. Development in Aspergillus. Stud. Mycol. 2013, 74, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Susumu, G. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Shelest, E. Transcription factors in fungi. FEMS Microbiol. Lett. 2008, 286, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, C.L.; Herskowitz, I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 1992, 68, 573–583. [Google Scholar] [CrossRef]
- Laurent, B.C.; Treich, I.; Carlson, M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993, 7, 583–591. [Google Scholar] [CrossRef] [Green Version]
- Hirschhorn, J.N.; Brown, S.A.; Clark, C.D.; Winston, F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992, 6, 2288–2298. [Google Scholar] [CrossRef] [Green Version]
- Yamada, K.; Hirota, K.; Mizuno, K.I.; Shibata, T.; Ohta, K. Essential roles of Snf21, a Swi2/Snf2 family chromatin remodeler, in fission yeast mitosis. Genes Genet. Syst. 2008, 83, 361–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.K.; Sang, Y.; Rodrigues, A.; Wu, M.F.; Rodriguez, P.L.; Wagner, D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 2013, 24, 4892–4906. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Wang, R.; Wang, J.; Hua, K.; Wang, Y.; Liu, X.; Yao, S. ALT1, a Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance through enhanced defense against oxidative stress in rice. PLoS ONE 2014, 9, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.M.; To, T.K.; Nishioka, T.; Seki, M. Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 2010, 33, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Hatori, M.; Hirota, T.; Iitsuka, M.; Kurabayashi, N.; Haraguchi, S.; Kokame, K.; Sato, R.; Nakai, A.; Miyata, T.; Tsutsui, K.; et al. Light-dependent and circadian clock-regulated activation of sterol regulatory element-binding protein, X-box-binding protein 1, and heat shock factor pathways. Proc. Natl. Acad. Sci. USA 2011, 108, 4864–4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorger, P.K.; Pelham, H.R.B. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 1988, 54, 855–864. [Google Scholar] [CrossRef]
- Gallo, G.J.; Prentice, H.; Kingston, R.E. Heat shock factor is required for growth at normal temperatures in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 1993, 13, 749–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purschwitz, J.; Müller, S.; Kastner, C.; Schöser, M.; Haas, H.; Espeso, E.A.; Atoui, A.; Calvo, A.M.; Fischer, R. Functional and physical interaction of blue-and red-light sensors in Aspergillus nidulans. Curr. Biol. 2008, 18, 255–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo, A.M. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 2008, 45, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- López-Berges, M.S.; Schäfer, K.; Hera, C.; Di Pietro, A. Combinatorial function of velvet and AreA in transcriptional regulation of nitrate utilization and secondary metabolism. Fungal Genet. Biol. 2014, 62, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Sari, F.; Braus, G.H.; Irniger, S. The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol. Microbiol. 2009, 71, 1278–1295. [Google Scholar] [CrossRef] [PubMed]
- Sarikaya Bayram, Ö.; Bayram, Ö.; Valerius, O.; Park, H.S.; Irniger, S.; Gerke, J.; Ni, M.; Han, K.-H.; Yu, J.-H.; Braus, G.H. LaeA control of Velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 2010, 6, e1001226. [Google Scholar] [CrossRef] [Green Version]
- Suparmin, A.; Kato, T.; Takemoto, H.; Park, E.Y. Metabolic comparison of aerial and submerged mycelia formed in the liquid surface culture of Cordyceps militaris. Microbiologyopen 2019, 8, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Görlich, D. Nuclear protein import. Curr. Opin. Cell Biol. 1997, 9, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.Y.; Han, S.Y.; Han, G.C.; Jee, H.K.; Han, K.H.; Han, D.M. Screening of growth- or development-related genes by using genomic library with inducible promoter in Aspergillus nidulans. J. Microbiol. 2005, 43, 523–528. [Google Scholar]
- Kwon, N.J.; Garzia, A.; Espeso, E.A.; Ugalde, U.; Yu, J.H. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 2010, 77, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Xia, Y.; Luo, F.; Dong, C.; Wang, C. Functional convergence and divergence of mating-type genes fulfilling in Cordyceps militaris. Fungal Genet. Biol. 2016, 88, 35–43. [Google Scholar] [CrossRef]
- Shimizu, K.; Keller, N.P. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 2001, 157, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.J.; Shin, K.S.; Yu, J.H. Characterization of the developmental regulator FlbE in Aspergillus fumigatus and Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 981–993. [Google Scholar] [CrossRef]
- Busby, T.M.; Miller, K.Y.; Miller, B.L. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics 1996, 143, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, P.; Fujita, M.; Jensen, S.T.; Conlon, E.M.; Rudner, D.Z.; Wang, S.T.; Ferguson, C.; Haga, K.; Sato, T.; Liu, J.S.; et al. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004, 2, e328. [Google Scholar] [CrossRef] [PubMed]
- Kalir, S.; Alon, U. Using a quantitative blueprint to reprogram the dynamics of the flagella gene network. Cell 2004, 117, 713–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalir, S.; Mangan, S.; Alon, U. A coherent feed-forward loop with a SUM input function prolongs flagella expression in Escherichia coli. Mol. Syst. Biol. 2005, 1, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noyes, M.B.; Christensen, R.G.; Wakabayashi, A.; Stormo, G.D.; Brodsky, M.H.; Wolfe, S.A. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 2008, 133, 1277–1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, S.; Gisselbrecht, S.S.; Rogers, J.M.; Hartl, D.L.; Bulyk, M.L. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12349–12354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonner, J.J.; Ballou, C.; Fackenthal, D.L. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol. Cell. Biol. 1994, 14, 501–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, J.; Ananthan, J.; Voellmy, R. Key features of heat shock regulatory elements. Mol. Cell. Biol. 1988, 8, 3761–3769. [Google Scholar] [CrossRef]
- Avalos, J.; Pardo-Medina, J.; Parra-Rivero, O.; Ruger-Herreros, M.; Rodríguez-Ortiz, R.; Hornero-Méndez, D.; Limón, M.C. Carotenoid biosynthesis in Fusarium. J. Fungi 2017, 3, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyartchuk, V.L.; Ashby, M.N.; Rine, J. Modulation of ras and a-factor function by carboxyl-terminal proteolysis. Science 1997, 275, 1796–1800. [Google Scholar] [CrossRef]
- Schmidt, W.K.; Tam, A.; Fujimura-Kamada, K.; Michaelis, S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc. Natl. Acad. Sci. USA 1998, 95, 11175–11180. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Li, S.-L.; Chen, H.-Y.; Zou, Y.; Zheng, Q.-W.; Guo, L.-Q.; Wu, G.-H.; Lu, J.; Lin, J.-F.; Ye, Z.-W. Enhancement of carotenoid production and its regulation in edible mushroom Cordyceps militaris by abiotic stresses. Enzym. Microb. Technol. 2021, 148, 109808. [Google Scholar] [CrossRef]

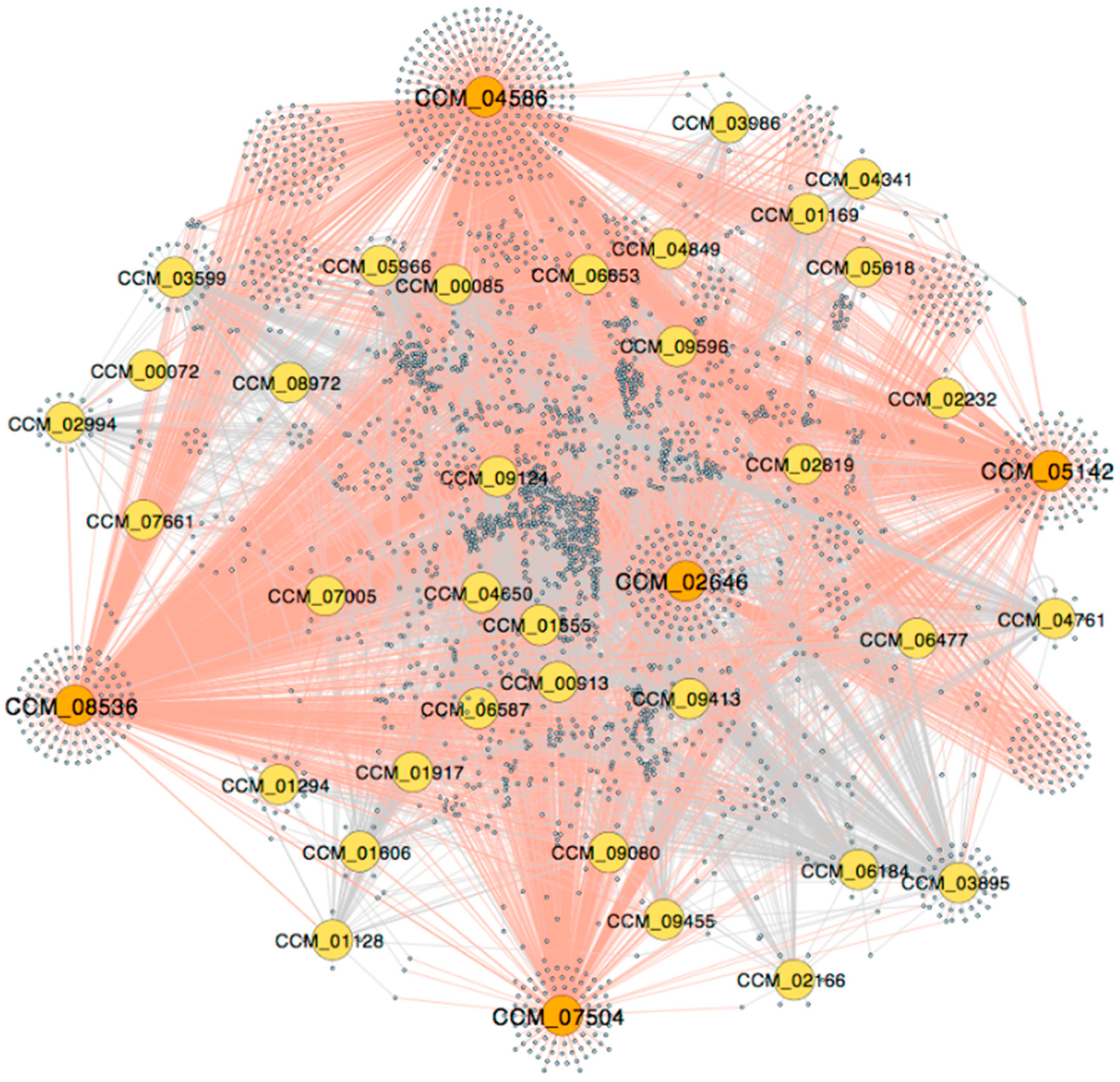
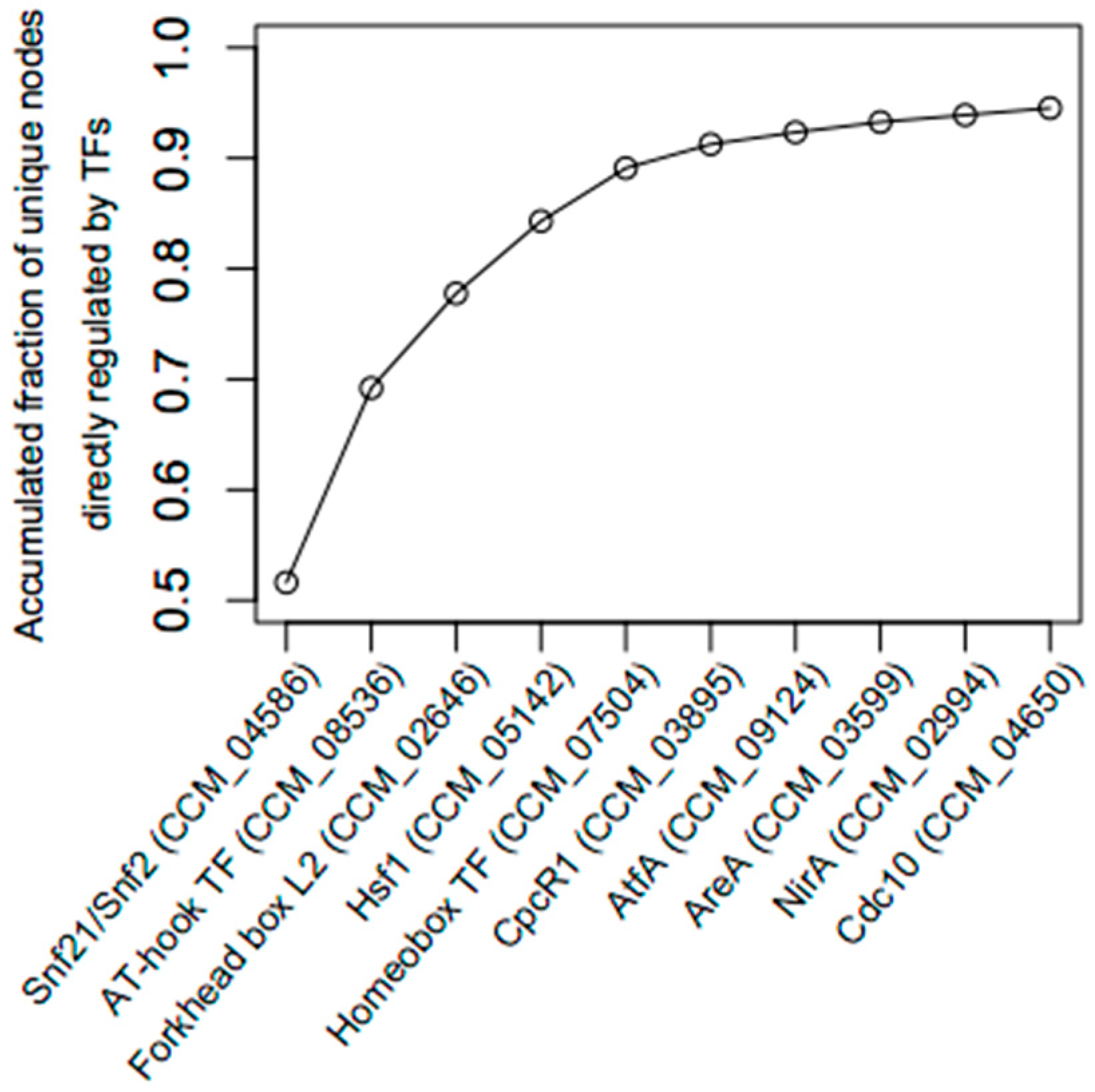
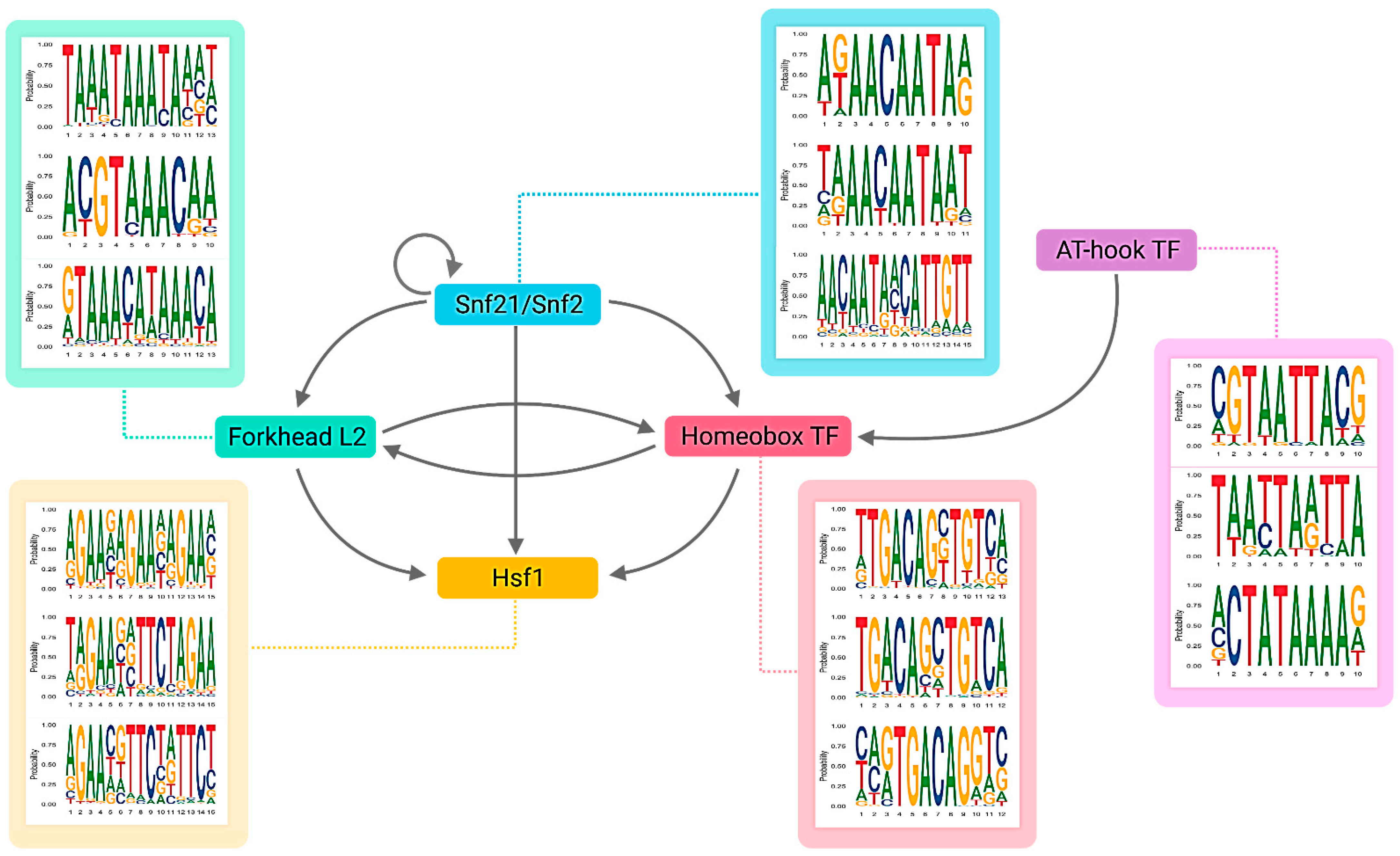

| Gene ID | TFs | Outdegree | Master | Intermediate | Target | Betweenness | DEG |
|---|---|---|---|---|---|---|---|
| CCM_04586 | Snf21/Snf2 family helicase (putative) | 1388 | 1407 | 0 | 0 | 0.00 | up |
| CCM_08536 | AT-hook DNA-binding motif TF | 1044 | 460 | 9 | 0 | 0.06 | up |
| CCM_02646 | Forkhead box protein L2 | 787 | 497 | 561 | 1 | 0.25 | up |
| CCM_05142 | Heat shock factor Hsf1 (putative) | 566 | 110 | 521 | 5 | 1.00 | up |
| CCM_07504 | Homeobox TF (putative) | 507 | 285 | 568 | 1 | 0.49 | up |
| CCM_03895 | Cephalosporin C regulator 1 (CpcR1) | 259 | 26 | 177 | 1 | 0.33 | up |
| CCM_09124 | bZIP TF (AtfA) (putative) | 159 | 2 | 6 | 0 | 0.15 | down |
| CCM_03599 | Nitrogen regulatory protein AreA | 152 | 0 | 11 | 0 | 0.27 | up |
| CCM_02994 | Nitrogen assimilation TF NirA | 70 | 20 | 52 | 1 | 0.49 | up |
| CCM_04650 | Start control protein Cdc10 | 72 | 0 | 55 | 3 | 0.10 | up |
| Gene ID | Description | Gene Symbol | DEG | Ortholog |
|---|---|---|---|---|
| CCM_00072 | Cutinase palindrome-binding protein | lreB | Up | lreB, white collar-2 (Neurospora crassa OR74A), Cutinase palindrome-binding protein (Cordyceps fumosorosea ARSEF 2679) |
| CCM_00560 | Sexual development transcription factor NsdD | nsdD | - | nsdD (Cordyceps javanica, Beauveria brongniartii RCEF 3172) |
| CCM_01106 | C2H2 transcription factor (Egr2) putative | egr2 | - | Conidial separation-1 (Beauveria bassiana ARSEF_2860), Transcriptional repressor (C. javanica) |
| CCM_01128 | Hypothetical protein | flbB | Down | BZIP-type transcription factor (C. fumosorosea ARSEF 2679, B. bassiana ARSEF 2860) |
| CCM_01444 | Transcription factor SteA | steA | - | brlA, steA (Akanthomyces lecanii RCEF 1005, C. fumosorosea ARSEF 2679) |
| CCM_01634 | Transcriptional regulator Medusa | medA | Up | Transcriptional regulator Medusa, medA (A. lecanii RCEF 1005, C. javanica) |
| CCM_01778 | cAMP-dependent protein kinase type 3 | pkaA | Up | pkaA cAMP dependent protein kinase A catalytic subunit (A. lecanii RCEF 1005, C. javanica) |
| CCM_04022 | DNA-binding protein creA | creA | - | creA (C. javanica), Carbon catabolite repressor (A. lecanii RCEF 1005) |
| CCM_04461 | Sensor histidine kinase/response regulator putative | fphA | Up | Sensor histidine kinase/response regulator (C. fumosorosea ARSEF 2679, C. javanica) |
| CCM_04514 | GATA transcription factor LreA | lreA | Down | Vivid PAS protein VVD (C. javanica), lreA (C. fumosorosea ARSEF 2679) |
| CCM_04531 | Sexual development activator VeA | veA | Up | veA (B. brongniartii RCEF 3172, C. javanica) |
| CCM_04849 | Uncharacterized protein | flbC | Down | C2H2 zinc finger protein flbC (C. javanica, A. lecanii RCEF 1005) |
| CCM_05395 | Methyltransferase LaeA | laeA | Down | Methyltransferase laeA (C. fumosorosea ARSEF 267, C. javanica) |
| CCM_05444 | Transcription factor AbaA (Conidiophore development regulator) | abaA | Up | abaA (C. fumosorosea ARSEF 2679, B. brongniartii RCEF 3172) |
| CCM_05556 | Fungal transcriptional regulatory protein | fl | Down | Fluffy (B. brongniartii RCEF 3172) |
| CCM_05639 | Importin β-2 subunit | kapB | Up | Importin beta-2 subunit (B. bassiana ARSEF 2860, C. javanica) |
| CCM_05697 | Glutamine synthetase | fluG | - | Glutamine synthetase (C. fumosorosea ARSEF 2679, C. javanica) |
| CCM_05966 | OefC protein | oefC | Down | oefC (B. bassiana ARSEF 2860, C. javanica) |
| CCM_06523 | Mating-type protein MAT 1-1-1 | mat1 | Up | Mating-type protein MAT1-1-1 (B. bassiana ARSEF 2860, A. lecanii RCEF 1005) |
| CCM_07203 | Developmental regulator FlbA | flbA | - | flbA (C. fumosorosea ARSEF 2679, C. javanica) |
| CCM_07236 | Guanine nucleotide-binding protein alpha subunit | fadA | Up | fadA (B. bassiana ARSEF 2860, Aspergillus fumigatus Af293) |
| CCM_08391 | Hypothetical protein | wetA | - | Developmental regulatory protein WetA (Fusarium phyllophilum) |
| CCM_08959 | Hypothetical protein | brlA | Down | brlA (B. bassiana ARSEF_2860, C. fumosorosea ARSEF 2679) |
| CCM_09566 | DNA-binding protein eta putative | flbD | - | flbD, Myb-like DNA-binding domain (Madurella mycetomatis) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
In-on, A.; Thananusak, R.; Ruengjitchatchawalya, M.; Vongsangnak, W.; Laomettachit, T. Construction of Light-Responsive Gene Regulatory Network for Growth, Development and Secondary Metabolite Production in Cordyceps militaris. Biology 2022, 11, 71. https://doi.org/10.3390/biology11010071
In-on A, Thananusak R, Ruengjitchatchawalya M, Vongsangnak W, Laomettachit T. Construction of Light-Responsive Gene Regulatory Network for Growth, Development and Secondary Metabolite Production in Cordyceps militaris. Biology. 2022; 11(1):71. https://doi.org/10.3390/biology11010071
Chicago/Turabian StyleIn-on, Ammarin, Roypim Thananusak, Marasri Ruengjitchatchawalya, Wanwipa Vongsangnak, and Teeraphan Laomettachit. 2022. "Construction of Light-Responsive Gene Regulatory Network for Growth, Development and Secondary Metabolite Production in Cordyceps militaris" Biology 11, no. 1: 71. https://doi.org/10.3390/biology11010071
APA StyleIn-on, A., Thananusak, R., Ruengjitchatchawalya, M., Vongsangnak, W., & Laomettachit, T. (2022). Construction of Light-Responsive Gene Regulatory Network for Growth, Development and Secondary Metabolite Production in Cordyceps militaris. Biology, 11(1), 71. https://doi.org/10.3390/biology11010071






