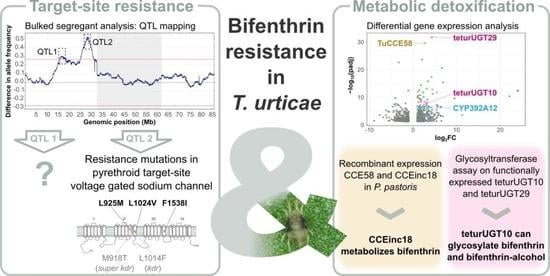
High-Resolution Genetic Mapping Combined with Transcriptome Profiling Reveals That Both Target-Site Resistance and Increased Detoxification Confer Resistance to the Pyrethroid Bifenthrin in the Spider Mite Tetranychus urticae
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials & Methods
2.1. Acaricides
2.2. Mite Strains and Husbandry
2.3. Toxicity Bioassays
2.4. Screening for Resistance Mutations in the VGSC
2.5. Bulked Segregant Analysis
2.5.1. Set-Up Bulk Segregant Analysis
2.5.2. DNA Sequencing and Bioinformatic Analyses
2.5.3. Potential Effect of Variant Alleles in Coding Sequences
2.6. Differential Gene Expression
2.6.1. RNA Extraction and Sequencing
2.6.2. RNA Read Mapping and Principal Component Analysis
2.6.3. Differential Expression (DE) Analysis
2.7. De Novo Transcriptome Assembly of MR-VL and the Mining of Transcripts Encoding CCEs
2.8. Activity of CCE58 and CCEinc18
2.8.1. Recombinant Expression of CCE58 and CCEinc18 in the Pichia pastoris System
2.8.2. Kinetic Analysis of CCE58 and CCEinc18 with Model Substrates
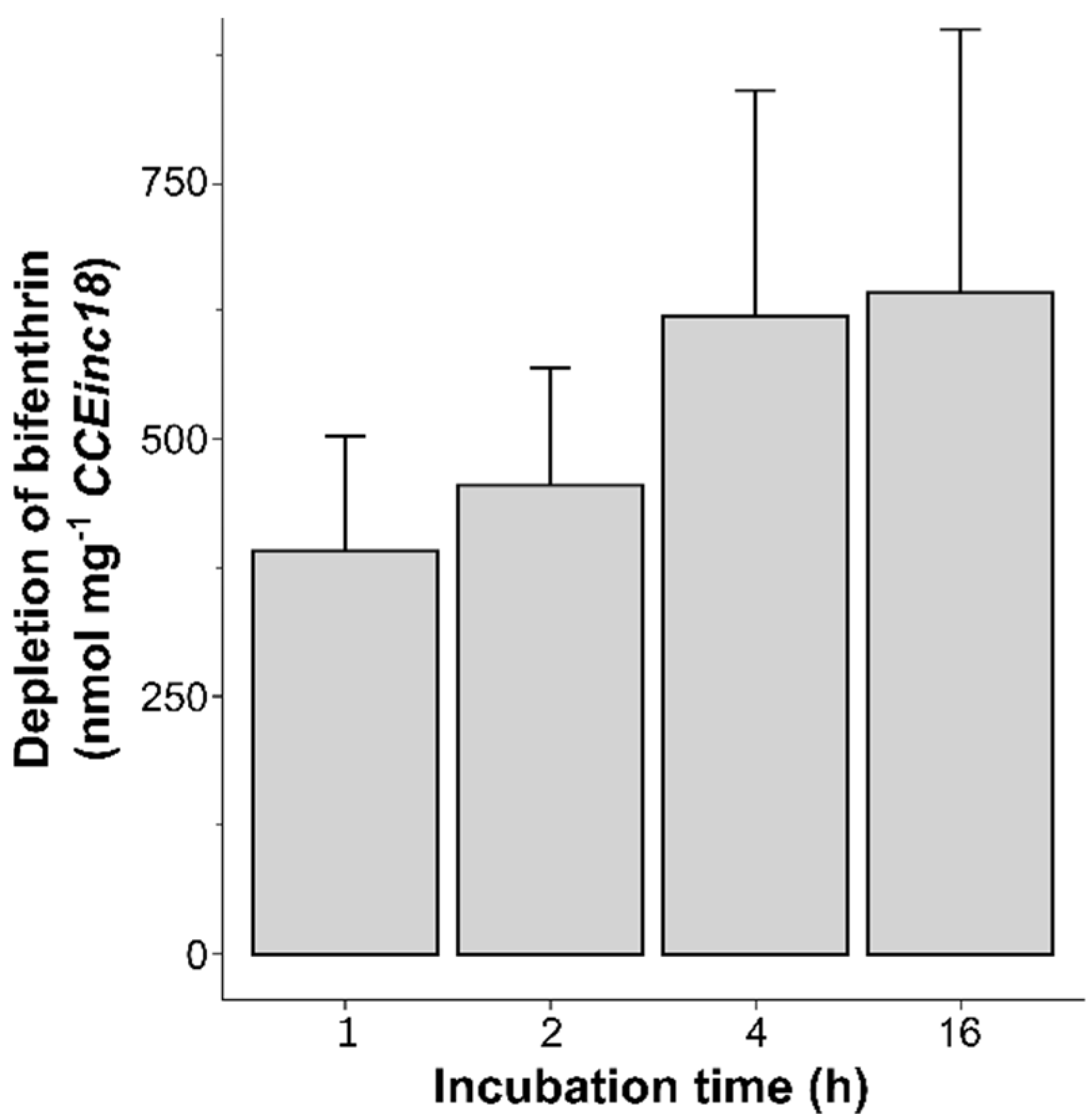
2.8.3. Bifenthrin Metabolism
2.9. UDP-Glo Glycosyltransferase Assay
3. Results
3.1. Toxicity Bioassays
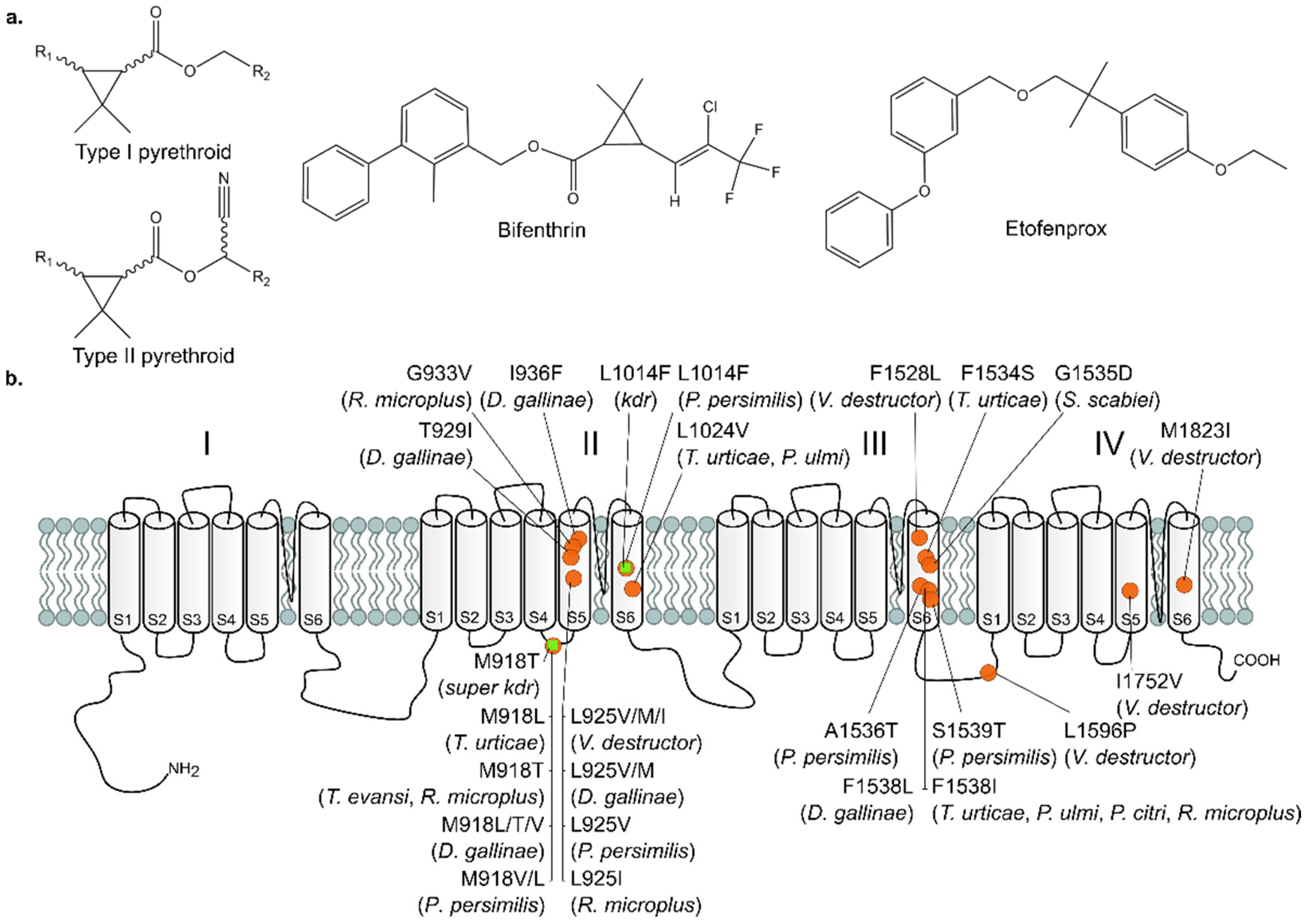
3.2. Presence of Target-Site Mutations in the VGSC
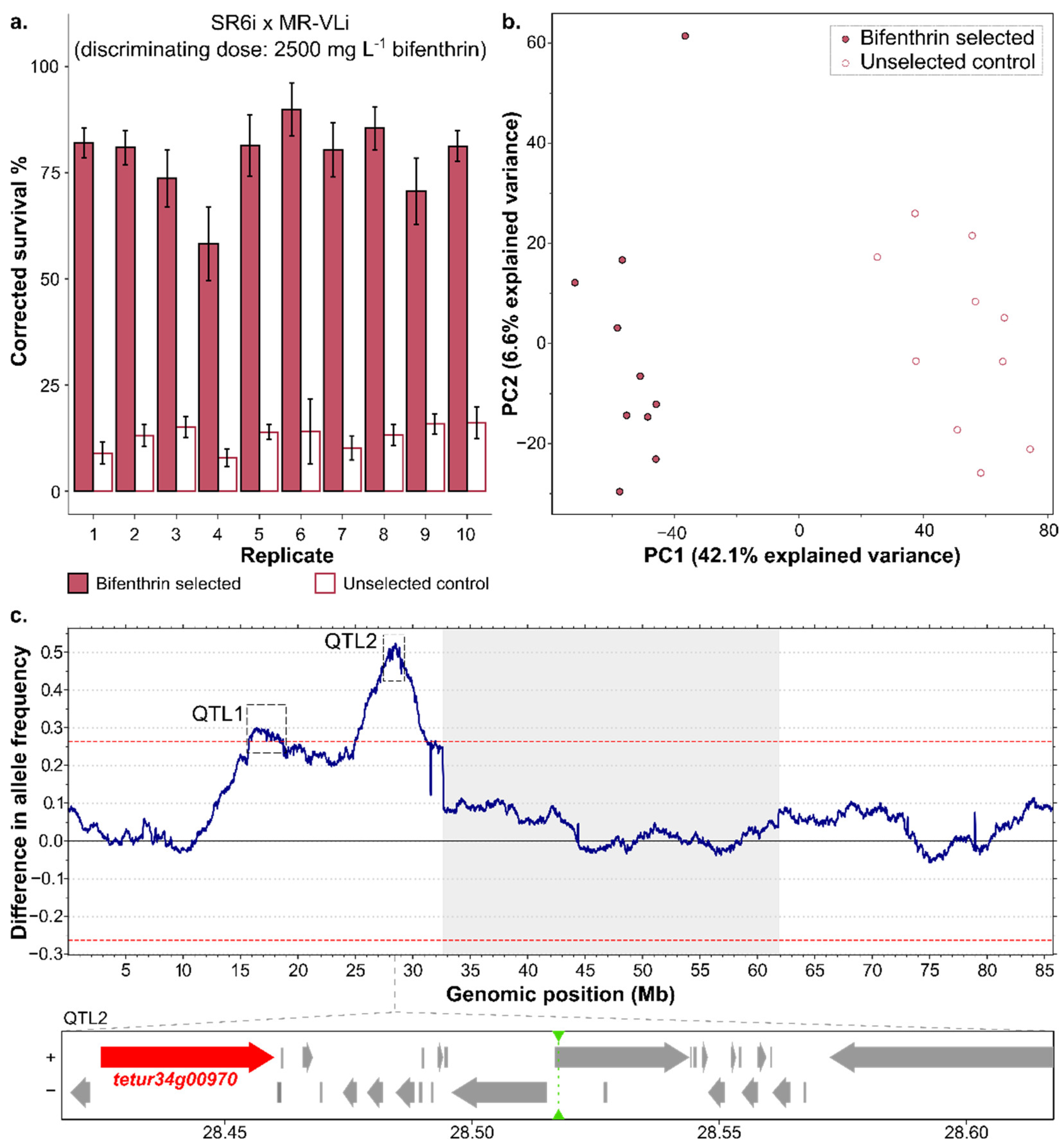
3.3. Bulked Segregant Analysis
3.3.1. Resistance in Parental and Segregating Populations
3.3.2. Genomic Responses to Bifenthrin Selection
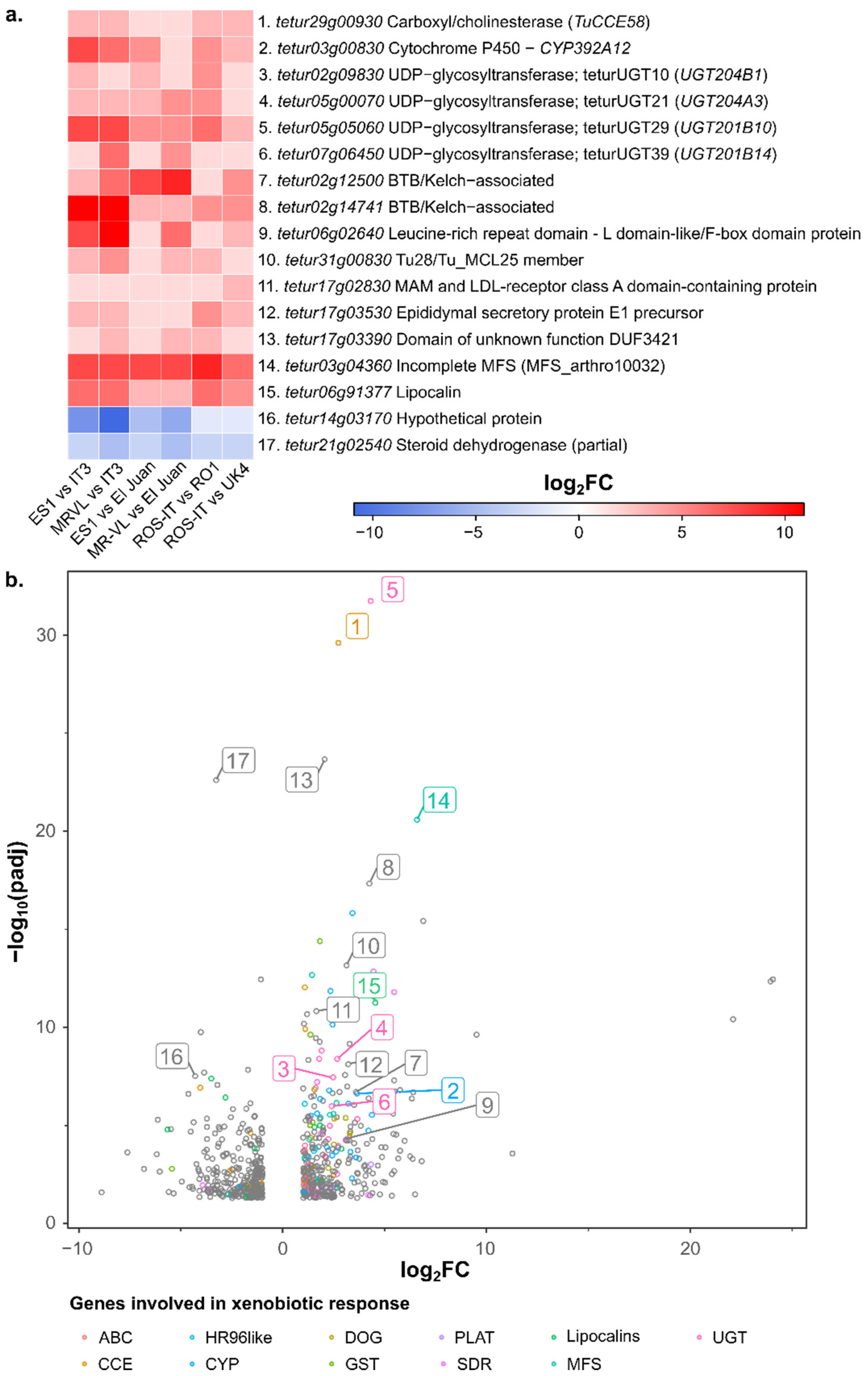
3.4. RNAseq Analysis
3.5. Bifenthrin Metabolizing Activity of Recombinantly Expressed CCE58 and CCEinc18
3.6. Glycosylation Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khambay, B.P.; Jewess, P. Pyrethroids. In Comprehensive Molecular Insect Science; Pergamon: Bergama, Turkey, 2005; Volume 6, pp. 1–29. [Google Scholar] [CrossRef]
- Palmquist, K.; Salatas, J.; Fairbrother, A. Pyrethroid insecticides: Use, environmental fate, and ecotoxicology. In Insecticides—Advances in Integrated Pest Management; Perveen, F., Ed.; InTech: Charlotte, NC, USA, 2012. [Google Scholar]
- Van Leeuwen, T.; Vontas, J.; Tsagkarakou, A.; Tirry, L. Mechanisms of acaricide resistance in the two-spotted spider mite Tetranychus urticae. In Biorational Control of Arthropod Pests; Springer: Berlin/Heidelberg, Germany, 2009; pp. 347–393. [Google Scholar]
- Zhang, W. Global pesticide use: Profile, trend, cost / benefit and more. Proc. Int. Acad. Ecol. Environ. Sci. 2018, 8, 1–27. [Google Scholar]
- Sparks, T.C.; Crossthwaite, A.J.; Nauen, R.; Banba, S.; Cordova, D.; Earley, F.; Ebbinghaus-Kintscher, U.; Fujioka, S.; Hirao, A.; Karmon, D.; et al. Insecticides, biologics and nematicides: Updates to IRAC’s mode of action classification—A tool for resistance management. Pestic. Biochem. Physiol. 2020, 167, 104587. [Google Scholar] [CrossRef] [PubMed]
- Field, L.M.; Emyr Davies, T.G.; O’Reilly, A.O.; Williamson, M.S.; Wallace, B.A. Voltage-gated sodium channels as targets for pyrethroid insecticides. Eur. Biophys. J. 2017, 46, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H. Pyrethroid chemistry and metabolism. In Hayes’ Handbook of Pesticide Toxicology, 3rd ed.; Krieger, R., Ed.; Academic Press: Cambridge, MA, USA, 2010; pp. 1635–1663. [Google Scholar]
- Soderlund, D.M. Neurotoxicology of pyrethroid insecticides. In Advances in Neurotoxicology; Aschner, M., Costa, L.G., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 4, pp. 113–165. [Google Scholar]
- Dong, K.; Du, Y.Z.; Rinkevich, F.; Nomura, Y.; Xu, P.; Wang, L.X.; Silver, K.; Zhorov, B.S. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rinkevich, F.D.; Du, Y.Z.; Dong, K. Diversity and convergence of sodium channel mutations involved in resistance to pyrethroids. Pestic. Biochem. Physiol. 2013, 106, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Kobayashi, T.; Fujita, T. Symptomatic and neurophysiological activities of new synthetic non-ester pyrethroids, ethofenprox, MTI-800, and related compounds. Pestic. Biochem. Physiol. 1986, 25, 387–395. [Google Scholar] [CrossRef]
- Sun, H.; Nomura, Y.; Du, Y.; Liu, Z.; Zhorov, B.S.; Dong, K. Characterization of two kdr mutations at predicted pyrethroid receptor site 2 in the sodium channels of Aedes aegypti and Nilaparvata lugens. Insect Biochem. Mol. Biol. 2022, 148, 103814. [Google Scholar] [CrossRef] [PubMed]
- Schleier, J.J.; Peterson, R.K. The joint toxicity of type I, II, and nonester pyrethroid insecticides. J. Econ. Entomol. 2012, 105, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, T.; Vontas, J.; Tsagkarakou, A.; Dermauw, W.; Tirry, L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: A review. Insect Biochem. Mol. Biol. 2010, 40, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Davies, T.G.E.; Williamson, M.S. Interactions of pyrethroids with the voltage-gated sodium channel. Bayer Crop Sci. J. 2009, 62, 159–178. [Google Scholar]
- Wu, M.X.; Adesanya, A.W.; Morales, M.A.; Walsh, D.B.; Lavine, L.C.; Lavine, M.D.; Zhu, F. Multiple acaricide resistance and underlying mechanisms in Tetranychus urticae on hops. J. Pest Sci. 2019, 92, 543–555. [Google Scholar] [CrossRef]
- Tsagkarakou, A.; Van Leeuwen, T.; Khajehali, J.; Ilias, A.; Grispou, M.; Williamson, M.S.; Tirry, L.; Vontas, J. Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Insect Mol. Biol. 2009, 18, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Clark, J.M.; Lee, S.H. Cloning of a sodium channel gene and identification of mutations putatively associated with fenpropathrin resistance in Tetranychus urticae. Pestic. Biochem. Physiol. 2010, 97, 93–100. [Google Scholar] [CrossRef]
- Riga, M.; Bajda, S.; Themistokleous, C.; Papadaki, S.; Palzewicz, M.; Dermauw, W.; Vontas, J.; Leeuwen, T.V. The relative contribution of target-site mutations in complex acaricide resistant phenotypes as assessed by marker assisted backcrossing in Tetranychus urticae. Sci. Rep. 2017, 7, 9202. [Google Scholar] [CrossRef] [PubMed]
- Kurlovs, A.H.; De Beer, B.; Ji, M.; Vandenhole, M.; De Meyer, T.; Feyereisen, R.; Clark, R.M.; Van Leeuwen, T. Abundant trans-driven variation in detoxification gene expression in the extreme generalist herbivore Tetranychus urticae. bioRvix 2022. [Google Scholar] [CrossRef]
- Nyoni, B.N.; Gorman, K.; Mzilahowa, T.; Williamson, M.S.; Navajas, M.; Field, L.M.; Bass, C. Pyrethroid resistance in the tomato red spider mite, Tetranychus evansi, is associated with mutation of the para-type sodium channel. Pest Manag. Sci. 2011, 67, 891–897. [Google Scholar] [CrossRef]
- Morgan, J.A.T.; Corley, S.W.; Jackson, L.A.; Lew-Tabor, A.E.; Moolhuijzen, P.M.; Jonsson, N.N. Identification of a mutation in the para-sodium channel gene of the cattle tick Rhipicephalus (Boophilus) microplus associated with resistance to synthetic pyrethroid acaricides. Int. J. Parasitol. 2009, 39, 775–779. [Google Scholar] [CrossRef]
- Jonsson, N.N.; Cutulle, C.; Corley, S.W.; Seddon, J.M. Identification of a mutation in the para-sodium channel gene of the cattle tick Rhipicephalus microplus associated with resistance to flumethrin but not to cypermethrin. Int. J. Parasitol. 2010, 40, 1659–1664. [Google Scholar] [CrossRef]
- Stone, N.E.; Olafson, P.U.; Davey, R.B.; Buckmeier, G.; Bodine, D.; Sidak-Loftis, L.C.; Giles, J.R.; Duhaime, R.; Miller, R.J.; Mosqueda, J.; et al. Multiple mutations in the para-sodium channel gene are associated with pyrethroid resistance in Rhipicephalus microplus from the United States and Mexico. Parasit Vectors 2014, 7, 456. [Google Scholar] [CrossRef]
- Bajda, S.A.; De Clercq, P.; Van Leeuwen, T. Selectivity and molecular stress responses to classical and botanical acaricides in the predatory mite Phytoseiulus persimilis Athias-Henriot (Acari: Phytoseiidae). Pest Manag. Sci. 2021, 78, 881–895. [Google Scholar] [CrossRef]
- Benavent-Albarracín, L.; Alonso, M.; Catalán, J.; Urbaneja, A.; Davies, T.G.E.; Williamson, M.S.; González-Cabrera, J. Mutations in the voltage-gated sodium channel gene associated with deltamethrin resistance in commercially sourced Phytoseiulus persimilis. Insect Mol. Biol. 2020, 29, 373–380. [Google Scholar] [CrossRef]
- González-Cabrera, J.; Rodriguez-Vargas, S.; Davies, T.G.E.; Field, L.M.; Schmehl, D.; Ellis, J.D.; Krieger, K.; Williamson, M.S. Novel mutations in the voltage-gated sodium channel of pyrethroid-resistant Varroa destructor populations from the Southeastern USA. PLoS ONE 2016, 11, e0155332. [Google Scholar] [CrossRef] [PubMed]
- He, H.Q.; Chen, A.C.; Davey, R.B.; Ivie, G.W.; George, J.E. Identification of a point mutation in the para-type sodium channel gene from a pyrethroid-resistant cattle tick. Biochem. Biophys. Res. Commun. 1999, 261, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Katsavou, E.; Vlogiannitis, S.; Karp-Tatham, E.; Blake, D.P.; Ilias, A.; Strube, C.; Kioulos, I.; Dermauw, W.; Van Leeuwen, T.; Vontas, J. Identification and geographical distribution of pyrethroid resistance mutations in the poultry red mite Dermanyssus gallinae. Pest Manag. Sci. 2020, 76, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Rameshgar, F.; Khajehali, J.; Nauen, R.; Bajda, S.; Jonckheere, W.; Dermauw, W.; Van Leeuwen, T. Point mutations in the voltage-gated sodium channel gene associated with pyrethroid resistance in Iranian populations of the European red mite Panonychus ulmi. Pestic. Biochem. Physiol. 2019, 157, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, T.; Dermauw, W. The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu. Rev. Entomol. 2016, 61, 475–498. [Google Scholar] [CrossRef] [PubMed]
- Pasay, C.; Arlian, L.; Morgan, M.; Vyszenski-Moher, D.; Rose, A.; Holt, D.; Walton, S.; McCarthy, J. High-resolution melt analysis for the detection of a mutation associated with permethrin resistance in a population of scabies mites. Med. Vet. Entomol. 2008, 22, 82–88. [Google Scholar] [CrossRef]
- Bhardwaj, K.; Sharma, R.; Abraham, J.; Sharma, P. Pyrethroids: A natural product for crop protection. In Natural Bioactive Products in Sustainable Agriculture; Singh, J., Yadav, A.N., Eds.; Springer: Singapore, 2020; pp. 113–130. [Google Scholar]
- Strode, C.; Donegan, S.; Garner, P.; Enayati, A.A.; Hemingway, J. The impact of pyrethroid resistance on the efficacy of insecticide-treated bed nets against African anopheline mosquitoes: Systematic review and meta-analysis. PLoS Med. 2014, 11, e1001619. [Google Scholar] [CrossRef]
- Vontas, J.; Kioulos, E.; Pavlidi, N.; Morou, E.; della Torre, A.; Ranson, H. Insecticide resistance in the major dengue vectors Aedes albopictus and Aedes aegypti. Pestic. Biochem. Physiol. 2012, 104, 126–131. [Google Scholar] [CrossRef]
- Anadon, A.; Martinez-Larranaga, M.R.; Martinez, M.A. Use and abuse of pyrethrins and synthetic pyrethroids in veterinary medicine. Vet. J. 2009, 182, 7–20. [Google Scholar] [CrossRef]
- Agramonte, N.M.; Bloomquist, J.R.; Bernier, U.R. Pyrethroid resistance alters the blood-feeding behavior in Puerto Rican Aedes aegypti mosquitoes exposed to treated fabric. PLoS Negl. Trop. Dis. 2017, 11, e0005954. [Google Scholar] [CrossRef] [PubMed]
- Mota-Sanchez, D.; Wise, J.C. The Arthropod Pesticide Resistance Database. Available online: http://www.pesticideresistance.org (accessed on 2 September 2022).
- Migeon, A.; Dorkeld, F. Spider Mites Web: A Comprehensive Database for the Tetranychidae. Available online: https://www1.montpellier.inra.fr/CBGP/spmweb/ (accessed on 18 May 2022).
- Helle, W.; Sabelis, M.W. Spider Mites: Their Biology, Natural Enemies and Control; Elsevier: Amsterdam, The Netherlands, 1986; Volume 1A. [Google Scholar]
- Busvine, J.R. Mechanism of resistance to insecticide in houseflies. Nature 1951, 168, 193–195. [Google Scholar] [CrossRef] [PubMed]
- Farnham, A.W.; Murray, A.W.A.; Sawicki, R.M.; Denholm, I.; White, J.C. Characterization of the structure activity relationship of kdr and 2 variants of super-kdr to pyrethroids in the housefly (Musca domestica L). Pestic Sci. 1987, 19, 209–220. [Google Scholar] [CrossRef]
- Ay, R.; Gurkan, M.O. Resistance to bifenthrin and resistance mechanisms of different strains of the two-spotted spider mite (Tetranychus urticae) from Turkey. Phytoparasitica 2005, 33, 237–244. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Van Pottelberge, S.; Tirry, L. Comparative acaricide susceptibility and detoxifying enzyme activities in field-collected resistant and susceptible strains of Tetranychus urticae. Pest Manag. Sci. 2005, 61, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, T.; Tirry, L. Esterase-mediated bifenthrin resistance in a multiresistant strain of the two-spotted spider mite, Tetranychus urticae. Pest Manag. Sci. 2007, 63, 150–156. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Stillatus, V.; Tirry, L. Genetic analysis and cross-resistance spectrum of a laboratory-selected chlorfenapyr resistant strain of two-spotted spider mite (Acari: Tetranychidae). Exp. Appl. Acarol. 2004, 32, 249–261. [Google Scholar] [CrossRef]
- Susurluk, H.; Gurkan, M.O. Mode of inheritance and biochemical mechanisms underlying lambda-cyhalothrin and bifenthrin resistance in the laboratory-selected two-spotted spider mite, Tetranychus urticae. Crop Prot. 2020, 137, 105280. [Google Scholar] [CrossRef]
- Yang, X.; Zhu, K.Y.; Buschman, L.L.; Margolies, D.C. Comparative susceptibility and possible detoxification mechanisms for selected miticides in banks grass mite and two-spotted spider mite (Acari: Tetranychidae). Exp. Appl. Acarol. 2001, 25, 293–299. [Google Scholar] [CrossRef]
- Yang, X.M.; Buschman, L.L.; Zhu, K.Y.; Margolies, D.C. Susceptibility and detoxifying enzyme activity in two spider mite species (Acari: Tetranychidae) after selection with three insecticides. J. Econ. Entomol. 2002, 95, 399–406. [Google Scholar] [CrossRef]
- Wei, P.; Li, J.H.; Liu, X.Y.; Nan, C.; Shi, L.; Zhang, Y.C.; Li, C.Z.; He, L. Functional analysis of four upregulated carboxylesterase genes associated with fenpropathrin resistance in Tetranychus cinnabarinus (Boisduval). Pest Manag. Sci. 2019, 75, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Auger, P.; Migeon, A.; Ueckermann, E.A.; Tiedt, L.; Navajas, M. Evidence for synonymy between Tetranychus urticae and Tetranychus cinnabarinus (Acari, Prostigmata, Tetranychidae): Review and new data. Acarologia 2013, 53, 383–415. [Google Scholar] [CrossRef]
- De Beer, B.; Villacis-Perez, E.; Khalighi, M.; Saalwaechter, C.; Vandenhole, M.; Jonckheere, W.; Ismaeil, I.; Geibel, S.; Van Leeuwen, T.; Dermauw, W. QTL mapping suggests that both cytochrome P450-mediated detoxification and target-site resistance are involved in fenbutatin oxide resistance in Tetranychus urticae. Insect Biochem. Mol Biol. 2022, 145, 103757. [Google Scholar] [CrossRef]
- Xue, W.; Mermans, C.; Papapostolou, K.M.; Lamprousi, M.; Christou, I.K.; Inak, E.; Douris, V.; Vontas, J.; Dermauw, W.; Van Leeuwen, T. Untangling a Gordian knot: The role of a GluCl3 I321T mutation in abamectin resistance in Tetranychus urticae. Pest Manag. Sci. 2020, 77, 1581–1593. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Snoeck, S.; Njiru, C.; Inak, E.; Dermauw, W.; Van Leeuwen, T. Geographical distribution and molecular insights into abamectin and milbemectin cross-resistance in European field populations of Tetranychus urticae. Pest Manag. Sci. 2020, 76, 2569–2581. [Google Scholar] [CrossRef]
- Bryon, A.; Kurlovs, A.H.; Dermauw, W.; Greenhalgh, R.; Riga, M.; Grbic, M.; Tirry, L.; Osakabe, M.; Vontas, J.; Clark, R.M.; et al. Disruption of a horizontally transferred phytoene desaturase abolishes carotenoid accumulation and diapause in Tetranychus urticae. Proc. Natl. Acad. Sci. USA 2017, 114, E5871–E5880. [Google Scholar] [CrossRef] [PubMed]
- Van Pottelberge, S.; Van Leeuwen, T.; Nauen, R.; Tirry, L. Resistance mechanisms to mitochondrial electron transport inhibitors in a field-collected strain of Tetranychus urticae Koch (Acari: Tetranychidae). Bull. Entomol. Res. 2009, 99, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdottir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Van Leeuwen, T.; Demaeght, P.; Osborne, E.J.; Dermauw, W.; Gohlke, S.; Nauen, R.; Grbic, M.; Tirry, L.; Merzendorfer, H.; Clark, R.M. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc. Natl. Acad. Sci. USA 2012, 109, 4407–4412. [Google Scholar] [CrossRef]
- Grbic, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouze, P.; Grbic, V.; Osborne, E.J.; Dermauw, W.; Ngoc, P.C.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef]
- Wybouw, N.; Kosterlitz, O.; Kurlovs, A.H.; Bajda, S.; Greenhalgh, R.; Snoeck, S.; Bui, H.Y.; Bryon, A.; Dermauw, W.; Van Leeuwen, T.; et al. Long-term population studies uncover the genome structure and genetic basis of xenobiotic and host plant adaptation in the herbivore Tetranychus urticae. Genetics 2019, 211, 1409–1427. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media: Sebastopol, CA, USA, 2020. [Google Scholar]
- Kurlovs, A.H.; Snoeck, S.; Kosterlitz, O.; Van Leeuwen, T.; Clark, R.M. Trait mapping in diverse arthropods by bulked segregant analysis. Curr. Opin. Insect Sci. 2019, 36, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Hahne, F.; Ivanek, R. Visualizing genomic data using Gviz and Bioconductor. In Statistical Genomics. Methods in Molecular Biology; Mathé, E., Davis, S., Eds.; Humana Press: New York, NY, USA, 2016; Volume 1418. [Google Scholar]
- Snoeck, S.; Kurlovs, A.H.; Bajda, S.; Feyereisen, R.; Greenhalgh, R.; Villacis-Perez, E.; Kosterlitz, O.; Dermauw, W.; Clark, R.M.; Van Leeuwen, T. High-resolution QTL mapping in Tetranychus urticae reveals acaricide-specific responses and common target-site resistance after selection by different METI-I acaricides. Insect Biochem. Mol. Biol. 2019, 110, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2 Elegant Graphics for Data Analysis, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Sterck, L.; Billiau, K.; Abeel, T.; Rouze, P.; Van de Peer, Y. ORCAE: Online resource for community annotation of eukaryotes. Nat. Methods 2012, 9, 1041. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Ruden, D.M.; Lu, X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012, 3, 35. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 March 2022).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Proc, G.P.D. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Wei, P.; Demaeght, P.; De Schutter, K.; Grigoraki, L.; Labropoulou, V.; Riga, M.; Vontas, J.; Nauen, R.; Dermauw, W.; Van Leeuwen, T. Overexpression of an alternative allele of carboxyl/choline esterase 4 (CCE04) of Tetranychus urticae is associated with high levels of resistance to the keto-enol acaricide spirodiclofen. Pest Manag. Sci. 2020, 76, 1142–1153. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Snoeck, S.; Pavlidi, N.; Pipini, D.; Vontas, J.; Dermauw, W.; Van Leeuwen, T. Substrate specificity and promiscuity of horizontally transferred UDP-glycosyltransferases in the generalist herbivore Tetranychus urticae. Insect Biochem. Mol. Biol. 2019, 109, 116–127. [Google Scholar] [CrossRef]
- Hubert, J.; Nesvorna, M.; Kamler, M.; Kopecky, J.; Tyl, J.; Titera, D.; Stara, J. Point mutations in the sodium channel gene conferring tau-fluvalinate resistance in Varroa destructor. Pest Manag. Sci. 2014, 70, 889–894. [Google Scholar] [CrossRef]
- Kennedy, C.J.; Tierney, K.B. Xenobiotic protection/resistance mechanisms in organisms. In Environmental Toxicology; Laws, E.A., Ed.; Springer: New York, NY, USA, 2013; pp. 689–721. [Google Scholar]
- Amezian, D.; Nauen, R.; Le Goff, G. Transcriptional regulation of xenobiotic detoxification genes in insects—An overview. Pestic. Biochem. Physiol. 2021, 174, 104822. [Google Scholar] [CrossRef] [PubMed]
- Duce, I.R.; Khan, T.R.; Green, A.C.; Thompson, A.J.; Warburton, S.P.M.; Wong, J. Calcium channels in insects. In Progress in Neuropharmacology and Neurotoxicology of Pesticides and Drugs; Beadle, D.J., Ed.; Royal Society of Chemistry: Cambridge, UK, 1999; pp. 56–66. [Google Scholar]
- Clark, J.M.; Symington, S.B. Neurotoxic implications of the agonistic action of CS-syndrome pyrethroids on the N-type Cav2.2. calcium channel. Pest Manag. Sci. 2008, 64, 628–638. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, W.B.; Si, F.L.; Yan, Z.T.; Zhang, Y.J.; He, Q.Y.; Chen, B. UDP-glycosyltransferase genes and their association and mutations associated with pyrethroid resistance in Anopheles sinensis (Diptera: Culicidae). Malar. J. 2019, 18, 62. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.G.; Small, G.J.; Nikou, D.C.; Ranson, H.; Hemingway, J. Purification, molecular cloning and heterologous expression of a glutathione S-transferase involved in insecticide resistance from the rice brown planthopper, Nilaparvata lugens. Biochem. J. 2002, 362, 329–337. [Google Scholar] [CrossRef]
- Liao, C.Y.; Feng, Y.C.; Li, G.; Shen, X.M.; Liu, S.H.; Dou, W.; Wang, J.J. Antioxidant role of PcGSTd1 in fenpropathrin resistant population of the citrus red mite, Panonychus citri (McGregor). Front. Physiol. 2018, 9, 314. [Google Scholar] [CrossRef]
- David, J.P.; Ismail, H.M.; Chandor-Proust, A.; Paine, M.J.I. Role of cytochrome P450s in insecticide resistance: Impact on the control of mosquito-borne diseases and use of insecticides on Earth. Philos. Trans. R Soc. Lond. B Biol. Sci. 2013, 368, 20120429. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Li, M.; Liu, N. Carboxylesterase genes in pyrethroid resistant house flies, Musca domestica. Insect Biochem. Mol Biol. 2018, 92, 30–39. [Google Scholar] [CrossRef]
- Inak, E.; Alpkent, Y.N.; Cobanoglu, S.; Dermauw, W.; Van Leeuwen, T. Resistance incidence and presence of resistance mutations in populations of Tetranychus urticae from vegetable crops in Turkey. Exp. Appl. Acarol. 2019, 78, 343–360. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, D.D.; Zhang, Y.J.; Wu, Q.J.; Xie, W.; Guo, Z.J.; Wang, S.L. Frequencies and mechanisms of pesticide resistance in Tetranychus urticae field populations in China. Insect Sci. 2021, 29, 827–839. [Google Scholar] [CrossRef]
- Tian, T.; Wu, M.M.; Zhang, Y.; Xu, D.D.; Wu, M.Y.; Xie, W.; Su, Q.; Wang, S.L. Pesticide resistance and related mutation frequencies of Tetranychus urticae in Hainan, China. Horticulturae 2022, 8, 590. [Google Scholar] [CrossRef]
- Mitina, G.V.; Tulaeva, I.A.; Malysh, S.M.; Tokarev, Y.S. Molecular genetic analysis of resistance-associated mutations in the experimental lines of spider mite Tetranychus urticae Koch, selected for resistance to bifenthrin and abamectin. Int. J. Acarol. 2021, 47, 721–725. [Google Scholar] [CrossRef]
- Tan, J.G.; Liu, Z.Q.; Wang, R.W.; Huang, Z.Y.; Chen, A.C.; Gurevitz, M.; Dong, K. Identification of amino acid residues in the insect sodium channel critical for pyrethroid binding. Mol. Pharmacol. 2005, 67, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, N.J.; Bass, C.; Dixon, A.; Neve, P. The evolutionary origins of pesticide resistance. Biol. Rev. 2019, 94, 135–155. [Google Scholar] [CrossRef]
- Gross, A.D.; Bloomquist, J.R. Characterizing permethrin and etofenprox resistance in two common laboratory strains of Anopheles gambiae (Diptera: Culicidae). Insects 2018, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Mutunga, J.M.; Anderson, T.D.; Craft, D.T.; Gross, A.D.; Swale, D.R.; Tong, F.; Wong, D.M.; Carlier, P.R.; Bloomquist, J.R. Carbamate and pyrethroid resistance in the akron strain of Anopheles gambiae. Pestic. Biochem. Physiol. 2015, 121, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.N.; Du, Y.Z.; Nomura, Y.; Dong, K. A sodium channel mutation identified in Aedes aegypti selectively reduces cockroach sodium channel sensitivity to type I, but not type II pyrethroids. Insect Biochem. Mol. Biol. 2011, 41, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Nomura, Y.; Luo, N.; Liu, Z.; Lee, J.E.; Khambay, B.; Dong, K. Molecular determinants on the insect sodium channel for the specific action of type II pyrethroid insecticides. Toxicol. Appl. Pharmacol. 2009, 234, 266–272. [Google Scholar] [CrossRef]
- Dermauw, W.; Van Leeuwen, T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 2014, 45, 89–110. [Google Scholar] [CrossRef]
- He, Q.Y.; Yan, Z.T.; Si, F.L.; Zhou, Y.; Fu, W.B.; Chen, B. ATP-binding cassette (ABC) transporter genes involved in pyrethroid resistance in the malaria vector Anopheles sinensis: Genome-wide identification, characteristics, phylogenetics, and expression profile. Int. J. Mol Sci. 2019, 20, 1409. [Google Scholar] [CrossRef]
- Epis, S.; Porretta, D.; Mastrantonio, V.; Comandatore, F.; Sassera, D.; Rossi, P.; Cafarchia, C.; Otranto, D.; Favia, G.; Genchi, C.; et al. ABC transporters are involved in defense against permethrin insecticide in the malaria vector Anopheles stephensi. Parasit Vectors 2014, 7, 1–7. [Google Scholar] [CrossRef]
- Shi, L.; Wei, P.; Wang, X.Z.; Shen, G.M.; Zhang, J.; Xiao, W.; Xu, Z.F.; Xu, Q.; He, L. Functional analysis of esterase TCE2 gene from Tetranychus cinnabarinus (Boisduval) involved in acaricide resistance. Sci. Rep. 2016, 6, srep18646. [Google Scholar] [CrossRef]
- Ding, Q.; Xu, X.; Sang, Z.; Wang, R.; Ullah, F.; Gao, X.; Song, D. Characterization of the insecticide detoxification carboxylesterase Boest1 from Bradysia odoriphaga Yang et Zhang (Diptera: Sciaridae). Pest. Manag. Sci. 2022, 78, 591–602. [Google Scholar] [CrossRef]
- Li, Y.; Sun, H.; Tian, Z.; Li, Y.; Ye, X.; Li, R.; Li, X.; Zheng, S.; Liu, J.; Zhang, Y. Identification of key residues of carboxylesterase PxEst-6 involved in pyrethroid metabolism in Plutella xylostella (L.). J. Hazard Mater. 2021, 407, 124612. [Google Scholar] [CrossRef]
- Li, R.; Zhu, B.; Hu, X.P.; Shi, X.Y.; Qi, L.L.; Liang, P.; Gao, X.W. Overexpression of PxαE14 contributing to detoxification of multiple insecticides in Plutella xylostella (L.). J. Agric. Food Chem. 2022, 70, 5794–5804. [Google Scholar] [CrossRef] [PubMed]
- Devorshak, C.; Roe, R.M. The role of esterases in insecticide resistance. Rev. Toxicol. 1998, 2, 501–537. [Google Scholar]
- Chen, G. Xenobiotic metabolism and disposition. In An Introduction to Interdisciplinary Toxicology; Pope, C.N., Liu, J., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 31–42. [Google Scholar]
- Fecko, A. Environmental fate of bifenthrin. In Environmental Monitoring and Pest Management Branch; Department of Pesticide Regulation: Sacramento, CA, USA, 1999. [Google Scholar]
- Kamble, S.T.; Saran, R.K. Effect of concentration on the adsorption of three termiticides in soil. Bull. Environ. Contam. Toxicol. 2005, 75, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Conclusion on the peer review of the pesticide risk assessment of the active substance bifenthrin. EFSA J. 2011, 9, 2159. [Google Scholar] [CrossRef]
- Ji, C.; Tanabe, P.; Shi, Q.; Qian, L.; McGruer, V.; Magnuson, J.T.; Wang, X.; Gan, J.; Gadepalli, R.S.; Rimoldi, J.; et al. Stage dependent enantioselective metabolism of bifenthrin in embryos of zebrafish (Danio rerio) and Japanese medaka (Oryzias latipes). Environ. Sci. Technol. 2021, 55, 9087–9096. [Google Scholar] [CrossRef]
- Nallani, G.C.; Chandrasekaran, A.; Kassahun, K.; Shen, L.; ElNaggar, S.F.; Liu, Z.W. Age dependent in vitro metabolism of bifenthrin in rat and human hepatic microsomes. Toxicol. Appl. Pharmacol. 2018, 338, 65–72. [Google Scholar] [CrossRef]
- Adesanya, A.W.; Cardenas, A.; Lavine, M.D.; Walsh, D.B.; Lavine, L.C.; Zhu, F. RNA interference of NADPH-cytochrome P450 reductase increases susceptibilities to multiple acaricides in Tetranychus urticae. Pestic. Biochem. Physiol. 2020, 165. [Google Scholar] [CrossRef]
- Zimmer, C.T.; Nauen, R. Cytochrome P450 mediated pyrethroid resistance in European populations of Meligethes aeneus (Coleoptera: Nitidulidae). Pestic. Biochem. Physiol. 2011, 100, 264–272. [Google Scholar] [CrossRef]
- Riga, M.; Myridakis, A.; Tsakireli, D.; Morou, E.; Stephanou, E.G.; Nauen, R.; Van Leeuwen, T.; Douris, V.; Vontas, J. Functional characterization of the Tetranychus urticae CYP392A11, a cytochrome P450 that hydroxylates the METI acaricides cyenopyrafen and fenpyroximate. Insect Biochem. Mol. Biol. 2015, 65, 91–99. [Google Scholar] [CrossRef]
- Reinking, J.; Lam, M.M.S.; Pardee, K.; Sampson, H.M.; Liu, S.Y.; Yang, P.; Williams, S.; White, W.; Lajoie, G.; Edwards, A.; et al. The Drosophila nuclear receptor E75 contains heme and is gas responsive. Cell 2005, 122, 195–207. [Google Scholar] [CrossRef]
- King-Jones, K.; Thummel, C.S. Nuclear receptors—A perspective from Drosophila. Nat. Rev. Genet. 2005, 6, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, P.C.; Greenhalgh, R.; Dermauw, W.; Rombauts, S.; Bajda, S.; Zhurov, V.; Grbic, M.; Van de Peer, Y.; Van Leeuwen, T.; Rouze, P.; et al. Complex evolutionary dynamics of massively expanded chemosensory receptor families in an extreme generalist chelicerate herbivore. Genome Biol. Evol. 2016, 8, 3323–3339. [Google Scholar] [CrossRef] [PubMed]
- Wicher, D.; Marion-Poll, F. Editorial: Function and regulation of chemoreceptors. Front. Cell. Neurosci. 2018, 12, 496. [Google Scholar] [CrossRef] [PubMed]
- Ingham, V.A.; Anthousi, A.; Douris, V.; Harding, N.J.; Lycett, G.; Morris, M.; Vontas, J.; Ranson, H. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 2020, 577, 376–380. [Google Scholar] [CrossRef]
| Bifenthrin | Etofenprox | VGSC Mutations | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | n | χ² (df) | Slope (±SE) | LC50 (95% CI) | RR (95%CI) | n | χ² (df) | Slope (±SE) | LC50 (95% CI) | RR (95% CI) | M918L | L925M | L1024V | F1534S | F1538I |
| El Juan | 490 | 43.27 (18) | 1.82 (±0.19) | 2.9 (2.0–3.9) | 2.2 a (1.4–3.4) | 585 | 54.58 (18) | 3.13 (±0.46) | 24 (17–30) | 1.02 a (0.84–1.2) | - | - | - | - | - |
| SR6i | 1120 | 81.58 (38) | 1.76 (±0.12) | 5.4 (4.4–6.4) | 4.0 a (2.6–6.2) | 520 | 8.74 (18) | 2.61 (±0.24) | 6.9 (5.9–7.9) | 0.30 a (0.24–0.37) | - | - | - | - | - |
| MR-VL | 506 | 18.65 (18) | 2.08 (±0.31) | 3400 (2700–4600) | 2500 a (1500–4100) | 479 | 55.26 (18) | 3.36 (±0.34) | 220 (170–270) | 9.4 a (7.8–11) | - | - | 100% | - | - |
| MR-VLi | 727 | 53.83 (26) | 1.59 (±0.17) | 2400 (1800–3600) | 1800 a (1100–2900) | 445 | 26.83 (18) | 2.19 (±0.21) | 60 (47 -74) | 2.6 a (2.1–3.2) | - | - | 100% | - | - |
| ES1 | 1082 | 59.18 (38) | 2.06 (±0.48) | 6300 (4200–22,000) | 9500 a (2500–35,000) | 512 | 26.79 (18) | 4.00 (±0.71) | 190 (120–240) | 8.3 a (6.4–11) | - | - | 33% | - | 25% |
| IT3 | 446 | 24.77 (18) | 0.94 (±0.16) | 1.35 (0.62–2.1) | / | 479 | 14.02 (18) | 2.60 (±0.23) | 23 (20–26) | / | - | - | - | - | - |
| ROS-IT | 950 | 28.09 (38) | 0.678 (±0.081) | 3500 (2100–7200) | 1470 b (770–2800) | 542 | 8.31 (18) | 2.59 (±0.71) | 93 (20–150) | 9.0 b (4.2–19) | - | 25% | - | - | - |
| ROS-ITi | 482 | 26.74 (21) | 2.02 (±0.22) | 3.2 (2.4–4.1) | 1.4 b (1.0–1.8) | 536 | 47.89 (21) | 3.03 (±0.36) | 12.5 (9.3–15) | 1.2 b (1.0–1.5) | - | - | - | - | - |
| UK4 | 1056 | 70.27 (38) | 1.397 (±0.099) | 5.5 (4.0–7.0) | 2.3 b (1.7–3.1) | 458 | 19.98 (18) | 2.99 (±0.27) | 21 (18–24) | 2.0 b (1.7–2.4) | - | - | - | - | 20% |
| RO1 | 911 | 52.23 (38) | 1.65 (±0.13) | 2.4 (1.7–3.0) | / | 426 | 43.21 (18) | 4.23 (±0.45) | 10.3 (8.3–12) | / | - | - | - | - | - |
| Reference Substrate | CCE58 | CCEinc18 | ||
|---|---|---|---|---|
| Vmax (±SE) | Km (±SE) | Vmax (±SE) | Km (±SE) | |
| 4-Nitrophenyl acetate a | 76.2 (±2.2) | 0.379 (±0.028) | 92.0 (±4.6) | 0.633 (±0.071) |
| 1-Naphthyl acetate b | 48.6 (±2.2) | 0.446 (±0.051) | 62.7 (±2.6) | 0.325 (±0.038) |
| Protein | Activity | Bifenthrin Depletion Rate a (Mean ±SE) | Corrected Bifenthrin Depletion Rate b (Mean ±SE) |
|---|---|---|---|
| CCE58 | Active | 0.78 ± 0.78 | 0.20 ± 0.59 |
| Inactivated | 0.58 ± 0.43 | ||
| CCEinc18 | Active | 3.79 ± 0.95 * | 3.5 ± 1.1 * |
| Inactivated | 0.29 ± 0.24 |
| Substrate | Free-UDP ± SE (µM) | |
|---|---|---|
| teturUGT29 (tetur05g05060) | teturUGT10 (tetur02g09830) | |
| Bifenthrin | −0.76 ± 0.21 | 5.1 ± 3.4 |
| TFP-acid | 0.55 ± 0.23 | −2.5 ± 3.7 |
| Bifenthrin-alcohol | 0.924 ± 0.020 | 21.1 ± 2.1 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Beer, B.; Vandenhole, M.; Njiru, C.; Spanoghe, P.; Dermauw, W.; Van Leeuwen, T. High-Resolution Genetic Mapping Combined with Transcriptome Profiling Reveals That Both Target-Site Resistance and Increased Detoxification Confer Resistance to the Pyrethroid Bifenthrin in the Spider Mite Tetranychus urticae. Biology 2022, 11, 1630. https://doi.org/10.3390/biology11111630
De Beer B, Vandenhole M, Njiru C, Spanoghe P, Dermauw W, Van Leeuwen T. High-Resolution Genetic Mapping Combined with Transcriptome Profiling Reveals That Both Target-Site Resistance and Increased Detoxification Confer Resistance to the Pyrethroid Bifenthrin in the Spider Mite Tetranychus urticae. Biology. 2022; 11(11):1630. https://doi.org/10.3390/biology11111630
Chicago/Turabian StyleDe Beer, Berdien, Marilou Vandenhole, Christine Njiru, Pieter Spanoghe, Wannes Dermauw, and Thomas Van Leeuwen. 2022. "High-Resolution Genetic Mapping Combined with Transcriptome Profiling Reveals That Both Target-Site Resistance and Increased Detoxification Confer Resistance to the Pyrethroid Bifenthrin in the Spider Mite Tetranychus urticae" Biology 11, no. 11: 1630. https://doi.org/10.3390/biology11111630
APA StyleDe Beer, B., Vandenhole, M., Njiru, C., Spanoghe, P., Dermauw, W., & Van Leeuwen, T. (2022). High-Resolution Genetic Mapping Combined with Transcriptome Profiling Reveals That Both Target-Site Resistance and Increased Detoxification Confer Resistance to the Pyrethroid Bifenthrin in the Spider Mite Tetranychus urticae. Biology, 11(11), 1630. https://doi.org/10.3390/biology11111630