Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Datasets
2.2. Evaluation of the Intrinsic Disorder Predisposition
2.3. PPI Networks
2.4. Disorder-Based Functional Annotations
2.5. CH-CDF Analysis
2.6. Statistical Analysis
2.7. Pathway Analysis
2.8. 3D Model Structures of Main Proteins
3. Results
3.1. Functional Intrinsic Disorder in the Main Proteins
3.1.1. NFH
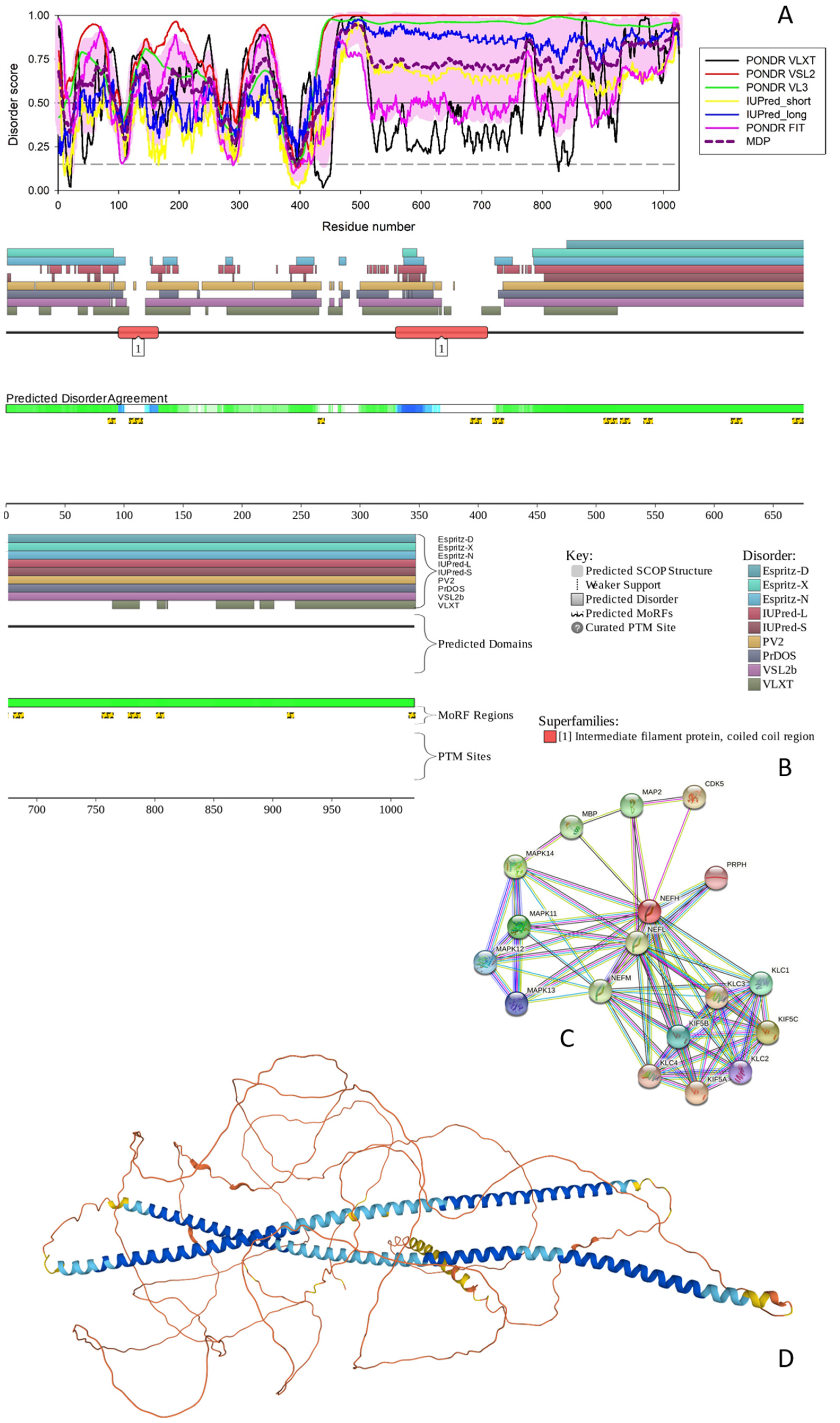
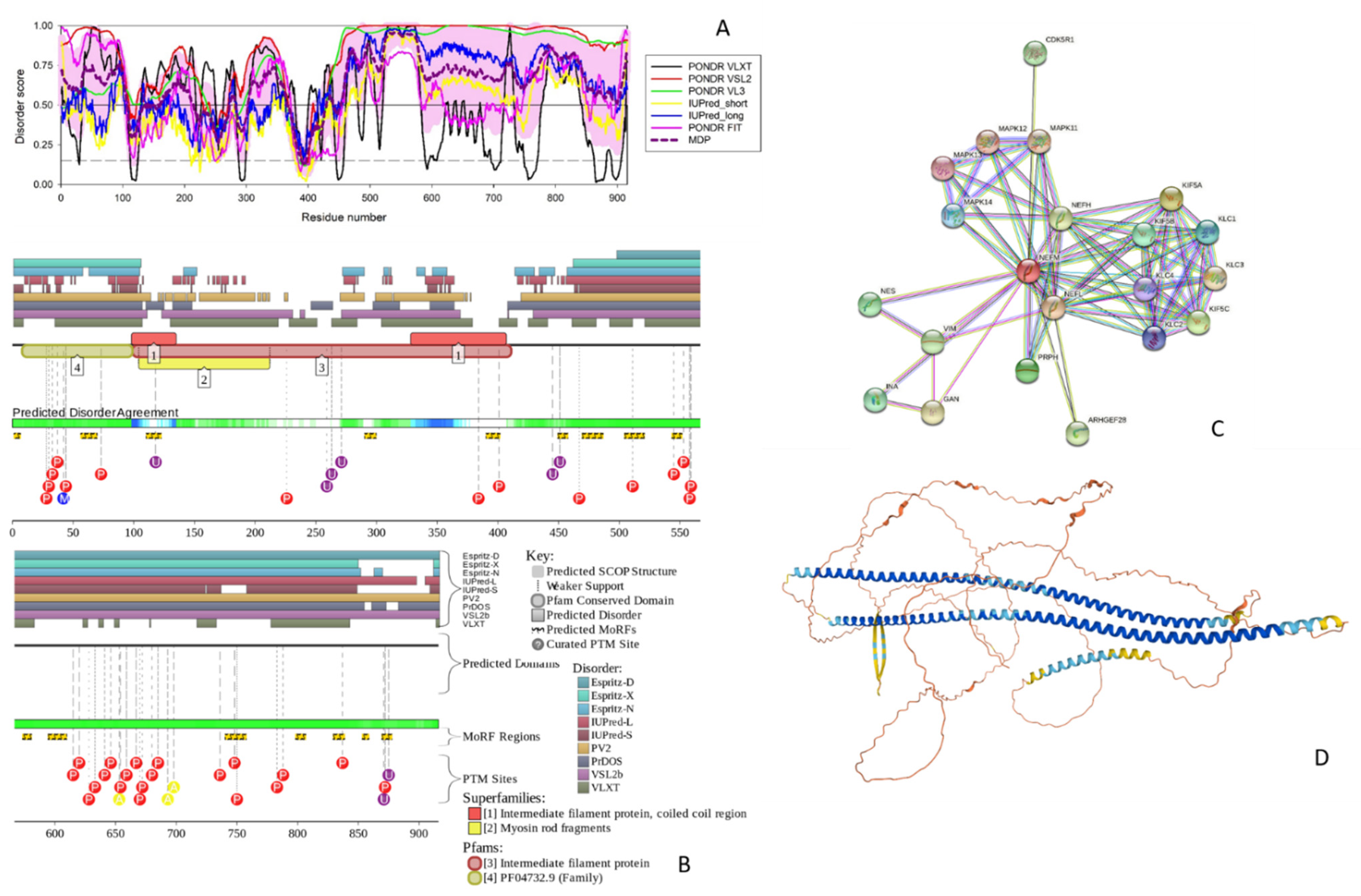


3.1.2. NFM
3.1.3. NFL
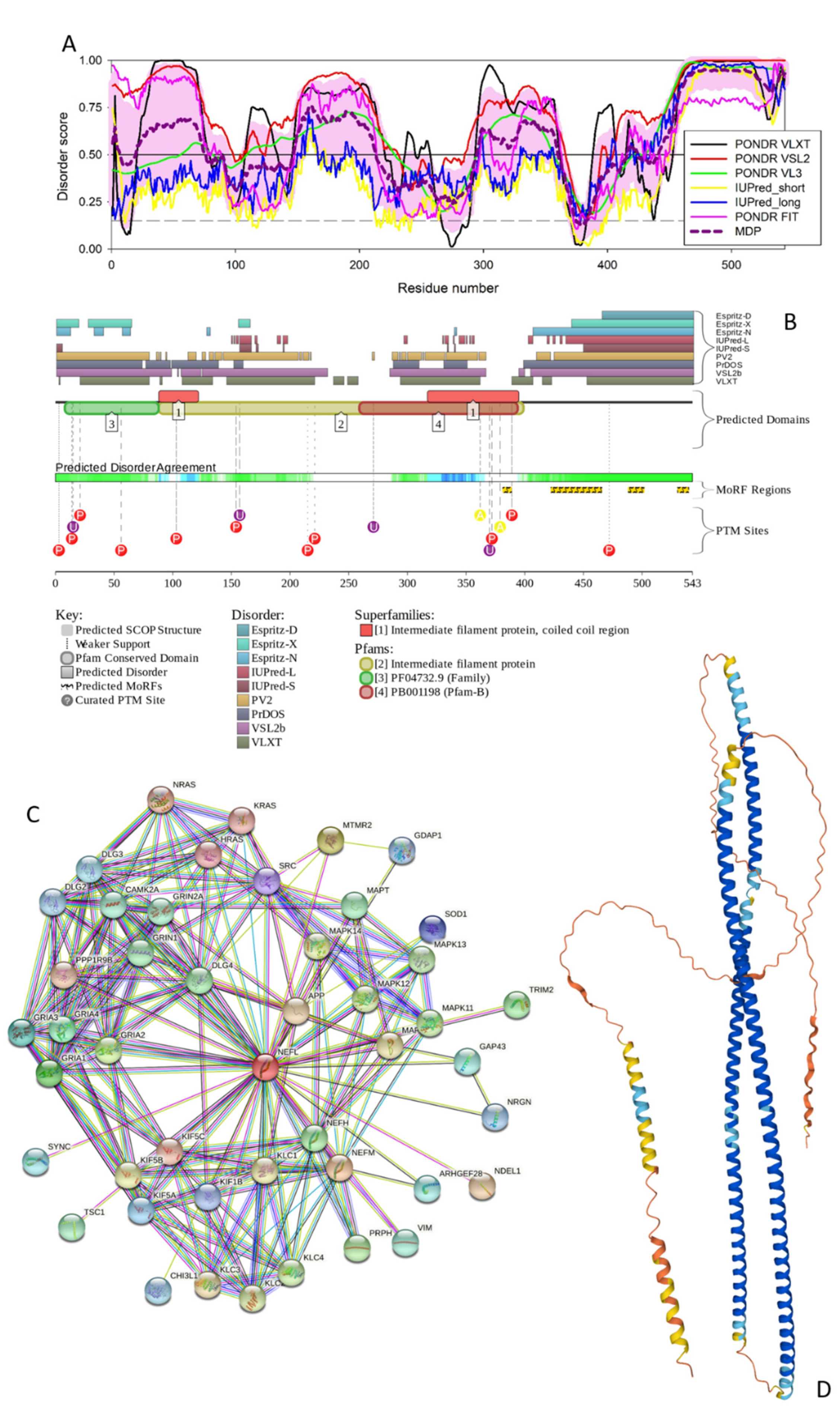
3.1.4. GFAP
3.1.5. MBP
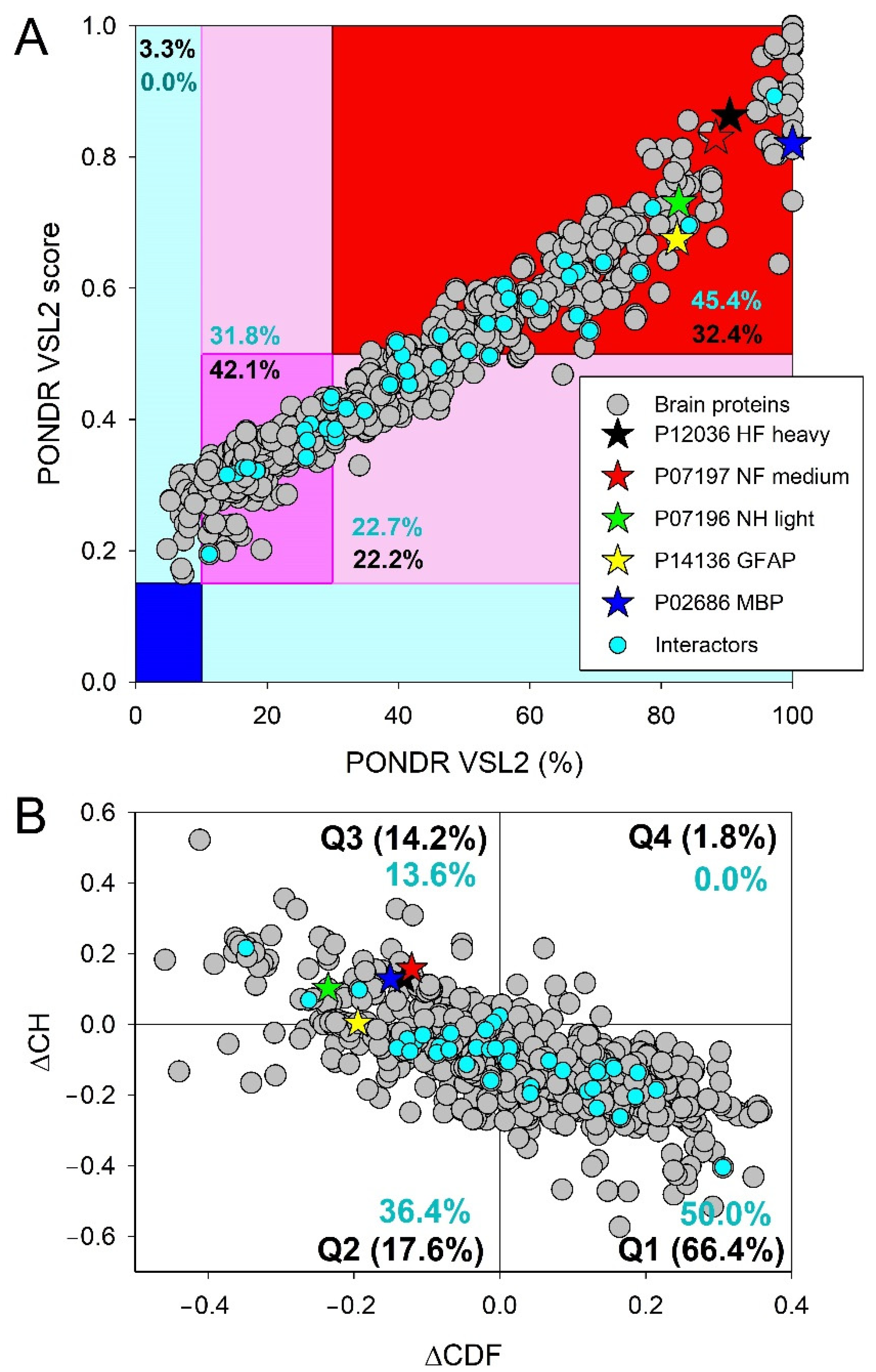
3.1.6. The Main Proteins as Illustrations of the “Stability of Instability” Phenomenon
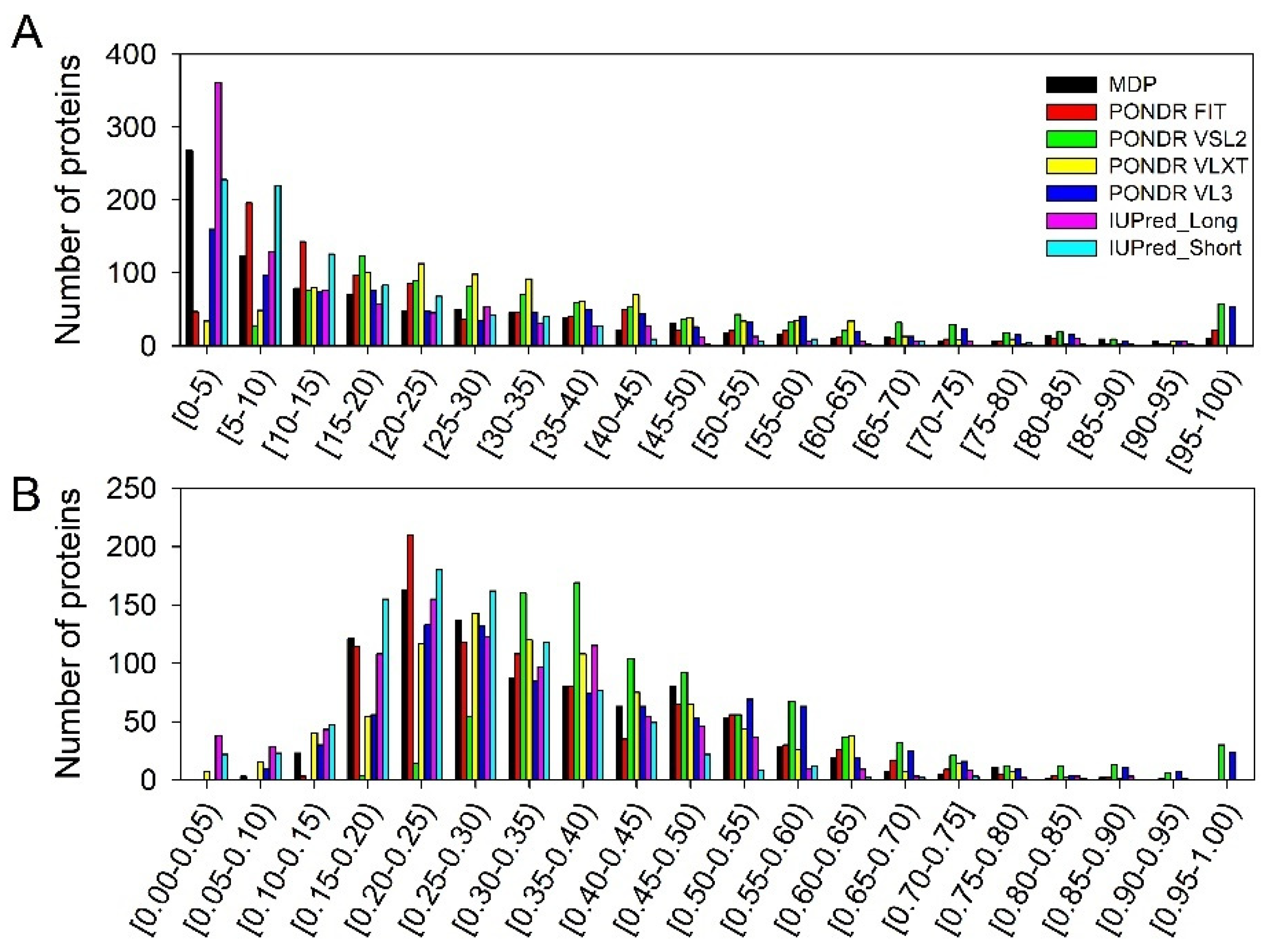
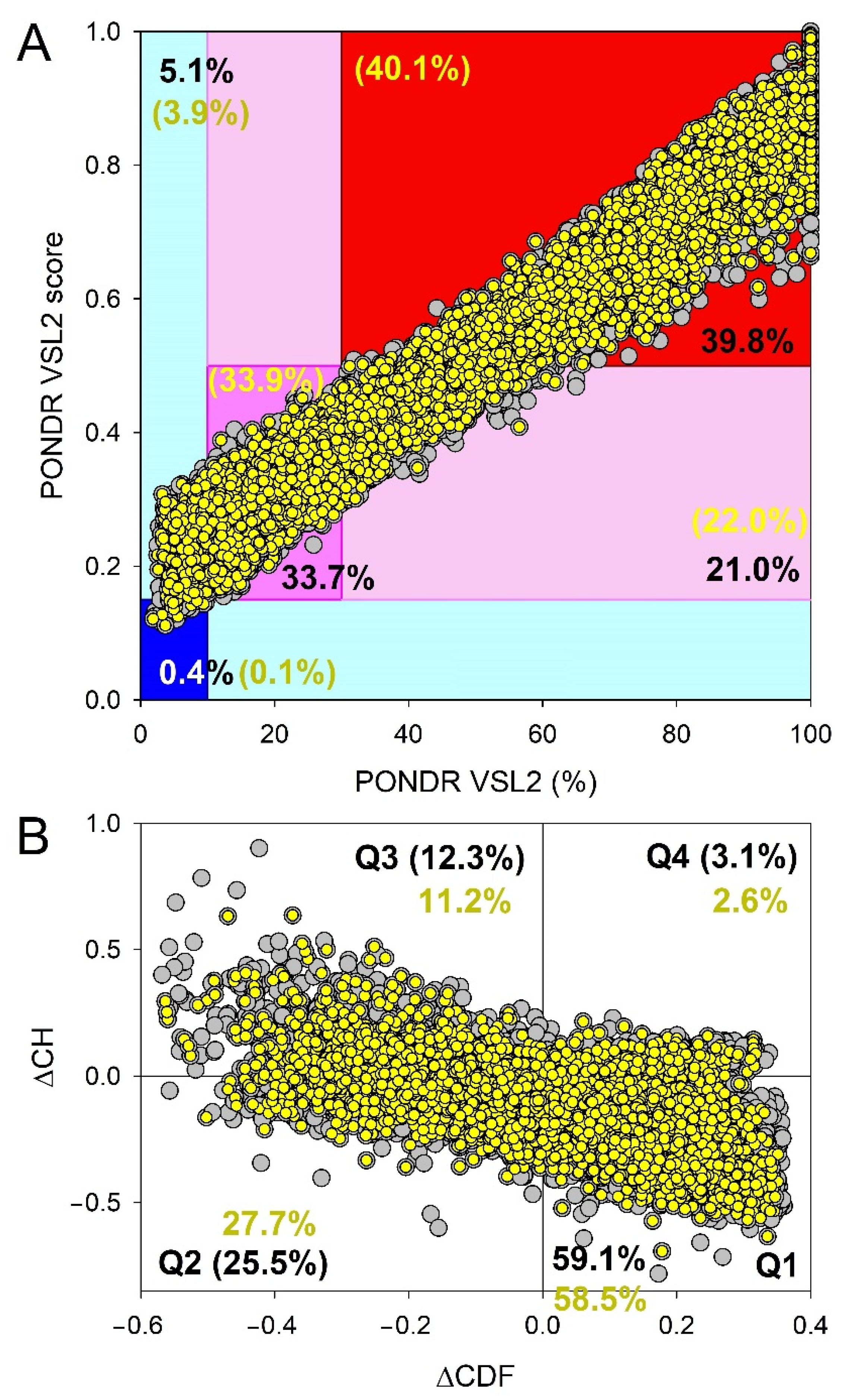
3.2. Global Intrinsic Disorder Analysis of the Heslington Brain Proteins
3.3. Functional Intrinsic Disorder of Proteins Potentially Interacting with Main Proteins
4. Conclusions
- Five main proteins that are assumed to be responsible for the preservation of the Heslington brain (which is at least 2600 years old), NFH, NFM, NFL, GFAP, and MBP are predicted to be highly disordered.
- 44 proteins from the Heslington brain, which are expected to interact with these five main proteins, are also predicted to have high levels of intrinsic disorder.
- Contrarily to the expected substantial (if not complete) elimination of the disordered proteins from the brain found inside a skull buried in a pit in Heslington, Yorkshire, England, many proteins in this Heslington brain are predicted to be highly disordered, with most of these proteins being expected to contain noticeable levels of intrinsic disorder.
- Intrinsic disorder of NFH, NFM, NFL, GFAP, and MBP and their interactors (in combination with other factors, such as the way in which the person was buried) might play a crucial role in preserving the Heslington brain by forming tightly folded brain protein aggregates.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Connor, S.; Ali, E.; Al-Sabah, S.; Anwar, D.; Bergström, E.; Brown, K.A.; Buckberry, J.; Buckley, S.; Collins, M.; Denton, J.; et al. Exceptional preservation of a prehistoric human brain from Heslington, Yorkshire, UK. J. Archaeol. Sci. 2011, 38, 1641–1654. [Google Scholar] [CrossRef]
- Petzold, A.; Lu, C.H.; Groves, M.; Gobom, J.; Zetterberg, H.; Shaw, G.; O’Connor, S. Protein aggregate formation permits millennium-old brain preservation. J. R. Soc. Interface 2020, 17, 20190775. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Rao, M.V.; Veeranna; Nixon, R.A. Neurofilaments and Neurofilament Proteins in Health and Disease. Cold Spring Harb. Perspect. Biol. 2017, 9, a018309. [Google Scholar] [CrossRef] [PubMed]
- Eng, L.F.; Ghirnikar, R.S.; Lee, Y.L. Glial fibrillary acidic protein: GFAP-thirty-one years (1969-2000). Neurochem. Res. 2000, 25, 1439–1451. [Google Scholar] [CrossRef]
- Vermeire, P.J.; Stalmans, G.; Lilina, A.V.; Fiala, J.; Novak, P.; Herrmann, H.; Strelkov, S.V. Molecular Interactions Driving Intermediate Filament Assembly. Cells 2021, 10, 2457. [Google Scholar] [CrossRef]
- Eldirany, S.A.; Lomakin, I.B.; Ho, M.; Bunick, C.G. Recent insight into intermediate filament structure. Curr. Opin. Cell Biol. 2021, 68, 132–143. [Google Scholar] [CrossRef]
- Stewart, M.; Quinlan, R.A.; Moir, R.D. Molecular interactions in paracrystals of a fragment corresponding to the alpha-helical coiled-coil rod portion of glial fibrillary acidic protein: Evidence for an antiparallel packing of molecules and polymorphism related to intermediate filament structure. J. Cell Biol. 1989, 109, 225–234. [Google Scholar] [CrossRef]
- Reeves, S.A.; Helman, L.J.; Allison, A.; Israel, M.A. Molecular cloning and primary structure of human glial fibrillary acidic protein. Proc. Natl. Acad. Sci. USA 1989, 86, 5178–5182. [Google Scholar] [CrossRef]
- Boggs, J.M. Myelin basic protein: A multifunctional protein. Cell. Mol. Life Sci. 2006, 63, 1945–1961. [Google Scholar] [CrossRef]
- Kursula, P. Structural properties of proteins specific to the myelin sheath. Amino Acids 2008, 34, 175–185. [Google Scholar] [CrossRef]
- Martinsen, V.; Kursula, P. Multiple sclerosis and myelin basic protein: Insights into protein disorder and disease. Amino Acids 2022, 54, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Fornasiero, E.F.; Mandad, S.; Wildhagen, H.; Alevra, M.; Rammner, B.; Keihani, S.; Opazo, F.; Urban, I.; Ischebeck, T.; Sakib, M.S.; et al. Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat. Commun. 2018, 9, 4230. [Google Scholar] [CrossRef] [PubMed]
- Toyama, B.H.; Savas, J.N.; Park, S.K.; Harris, M.S.; Ingolia, N.T.; Yates, J.R., 3rd; Hetzer, M.W. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013, 154, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Didonna, A.; Opal, P. The role of neurofilament aggregation in neurodegeneration: Lessons from rare inherited neurological disorders. Mol. Neurodegener. 2019, 14, 19. [Google Scholar] [CrossRef]
- Sihag, R.K.; Inagaki, M.; Yamaguchi, T.; Shea, T.B.; Pant, H.C. Role of phosphorylation on the structural dynamics and function of types III and IV intermediate filaments. Exp. Cell Res. 2007, 313, 2098–2109. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Midic, U.; Xie, H.; Xue, B.; Vucetic, S.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genom. 2009, 10 (Suppl. 1), S7. [Google Scholar] [CrossRef]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Erdos, G.; Dosztanyi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 2005, 61 (Suppl. 7), 176–182. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Dayhoff, G.W.I.; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein Sci. 2022, in press.
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Uversky, V.N. Analyzing IDPs in interactomes. In Intrinsically Disordered Proteins; Kragelund, B.B., Skriver, K., Eds.; Humana: New York, NY, USA, 2020; pp. 895–945. [Google Scholar]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Alonso-Lopez, D.; Campos-Laborie, F.J.; Gutierrez, M.A.; Lambourne, L.; Calderwood, M.A.; Vidal, M.; De Las Rivas, J. APID database: Redefining protein-protein interaction experimental evidences and binary interactomes. Database 2019, 2019, baz005. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol. Biosyst. 2008, 4, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 2011, 77, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Oldfield, C.J.; Van, Y.Y.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder and induced pluripotent stem cells. Mol. Biosyst. 2012, 8, 134–150. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac. Symp. Biocomput. 2012, 128–139. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003, 4, P3. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mukai, H.; Toshimori, M.; Shibata, H.; Kitagawa, M.; Shimakawa, M.; Miyahara, M.; Sunakawa, H.; Ono, Y. PKN associates and phosphorylates the head-rod domain of neurofilament protein. J. Biol. Chem. 1996, 271, 9816–9822. [Google Scholar] [CrossRef]
- Andreeva, A.; Howorth, D.; Brenner, S.E.; Hubbard, T.J.; Chothia, C.; Murzin, A.G. SCOP database in 2004: Refinements integrate structure and sequence family data. Nucleic Acids Res. 2004, 32, D226–D229. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995, 247, 536–540. [Google Scholar] [CrossRef] [PubMed]
- de Lima Morais, D.A.; Fang, H.; Rackham, O.J.; Wilson, D.; Pethica, R.; Chothia, C.; Gough, J. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 2011, 39, D427–D434. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef]
- Janmey, P.A.; Leterrier, J.-F.; Herrmann, H. Assembly and structure of neurofilaments. Curr. Opin. Colloid Interface Sci. 2003, 8, 40–47. [Google Scholar] [CrossRef]
- Carter, J.; Gragerov, A.; Konvicka, K.; Elder, G.; Weinstein, H.; Lazzarini, R.A. Neurofilament (NF) assembly; divergent characteristics of human and rodent NF-L subunits. J. Biol. Chem. 1998, 273, 5101–5108. [Google Scholar] [CrossRef]
- Carpenter, D.A.; Ip, W. Neurofilament triplet protein interactions: Evidence for the preferred formation of NF-L-containing dimers and a putative function for the end domains. J. Cell Sci. 1996, 109 Pt 10, 2493–2498. [Google Scholar] [CrossRef]
- Peysselon, F.; Xue, B.; Uversky, V.N.; Ricard-Blum, S. Intrinsic disorder of the extracellular matrix. Mol. Biosyst. 2011, 7, 3353–3365. [Google Scholar] [CrossRef]
- Szappanos, B.; Suveges, D.; Nyitray, L.; Perczel, A.; Gaspari, Z. Folded-unfolded cross-predictions and protein evolution: The case study of coiled-coils. FEBS Lett. 2010, 584, 1623–1627. [Google Scholar] [CrossRef] [PubMed]
- Alderson, T.R.; Pritišanac, I.; Moses, A.M.; Forman-Kay, J.D. Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2. bioRxiv 2022. [Google Scholar] [CrossRef]
- He, J.; Turzo, S.B.A.; Seffernick, J.T.; Kim, S.S.; Lindert, S. Prediction of Intrinsic Disorder Using Rosetta ResidueDisorder and AlphaFold2. J. Phys. Chem. B 2022, 126, 8439–8446. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.B.; Perminov, A.; Bekele, S.; Kedziora, G.; Farajollahi, S.; Varaljay, V.; Hinkle, K.; Molinero, V.; Meister, K.; Hung, C.; et al. AlphaFold2 models indicate that protein sequence determines both structure and dynamics. Sci. Rep. 2022, 12, 10696. [Google Scholar] [CrossRef]
- Laurents, D.V. AlphaFold 2 and NMR Spectroscopy: Partners to Understand Protein Structure, Dynamics and Function. Front. Mol. Biosci. 2022, 9, 906437. [Google Scholar] [CrossRef]
- Ruff, K.M.; Pappu, R.V. AlphaFold and Implications for Intrinsically Disordered Proteins. J. Mol. Biol. 2021, 433, 167208. [Google Scholar] [CrossRef]
- Heins, S.; Wong, P.C.; Muller, S.; Goldie, K.; Cleveland, D.W.; Aebi, U. The rod domain of NF-L determines neurofilament architecture, whereas the end domains specify filament assembly and network formation. J. Cell Biol. 1993, 123, 1517–1533. [Google Scholar] [CrossRef]
- Uceda-Castro, R.; van Asperen, J.V.; Vennin, C.; Sluijs, J.A.; van Bodegraven, E.J.; Margarido, A.S.; Robe, P.A.J.; van Rheenen, J.; Hol, E.M. GFAP splice variants fine-tune glioma cell invasion and tumour dynamics by modulating migration persistence. Sci. Rep. 2022, 12, 424. [Google Scholar] [CrossRef]
- Bignami, A.; Dahl, D. Astrocyte-specific protein and neuroglial differentiation. An immunofluorescence study with antibodies to the glial fibrillary acidic protein. J. Comp. Neurol. 1974, 153, 27–38. [Google Scholar] [CrossRef]
- Bignami, A.; Eng, L.F.; Dahl, D.; Uyeda, C.T. Localization of the glial fibrillary acidic protein in astrocytes by immunofluorescence. Brain Res. 1972, 43, 429–435. [Google Scholar] [CrossRef]
- van Asperen, J.V.; Robe, P.; Hol, E.M. GFAP Alternative Splicing and the Relevance for Disease—A Focus on Diffuse Gliomas. ASN Neuro 2022, 14, 17590914221102065. [Google Scholar] [CrossRef] [PubMed]
- Geisler, N.; Weber, K. Amino acid sequence data on glial fibrillary acidic protein (GFA); implications for the subdivision of intermediate filaments into epithelial and non-epithelial members. EMBO J. 1983, 2, 2059–2063. [Google Scholar] [CrossRef] [PubMed]
- Middeldorp, J.; Boer, K.; Sluijs, J.A.; De Filippis, L.; Encha-Razavi, F.; Vescovi, A.L.; Swaab, D.F.; Aronica, E.; Hol, E.M. GFAPdelta in radial glia and subventricular zone progenitors in the developing human cortex. Development 2010, 137, 313–321. [Google Scholar] [CrossRef] [PubMed]
- van Bodegraven, E.J.; van Asperen, J.V.; Robe, P.A.J.; Hol, E.M. Importance of GFAP isoform-specific analyses in astrocytoma. Glia 2019, 67, 1417–1433. [Google Scholar] [CrossRef] [PubMed]
- van Bodegraven, E.J.; van Asperen, J.V.; Sluijs, J.A.; van Deursen, C.B.J.; van Strien, M.E.; Stassen, O.; Robe, P.A.J.; Hol, E.M. GFAP alternative splicing regulates glioma cell-ECM interaction in a DUSP4-dependent manner. FASEB J. 2019, 33, 12941–12959. [Google Scholar] [CrossRef]
- Kawajiri, A.; Yasui, Y.; Goto, H.; Tatsuka, M.; Takahashi, M.; Nagata, K.; Inagaki, M. Functional significance of the specific sites phosphorylated in desmin at cleavage furrow: Aurora-B may phosphorylate and regulate type III intermediate filaments during cytokinesis coordinatedly with Rho-kinase. Mol. Biol. Cell 2003, 14, 1489–1500. [Google Scholar] [CrossRef]
- Kosako, H.; Amano, M.; Yanagida, M.; Tanabe, K.; Nishi, Y.; Kaibuchi, K.; Inagaki, M. Phosphorylation of glial fibrillary acidic protein at the same sites by cleavage furrow kinase and Rho-associated kinase. J. Biol. Chem. 1997, 272, 10333–10336. [Google Scholar] [CrossRef]
- Jin, Z.; Fu, Z.; Yang, J.; Troncosco, J.; Everett, A.D.; Van Eyk, J.E. Identification and characterization of citrulline-modified brain proteins by combining HCD and CID fragmentation. Proteomics 2013, 13, 2682–2691. [Google Scholar] [CrossRef]
- Etienne-Manneville, S. Cytoplasmic Intermediate Filaments in Cell Biology. Annu. Rev. Cell Dev. Biol. 2018, 34, 1–28. [Google Scholar] [CrossRef]
- Harauz, G.; Ishiyama, N.; Hill, C.M.; Bates, I.R.; Libich, D.S.; Fares, C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron 2004, 35, 503–542. [Google Scholar] [CrossRef]
- Harauz, G.; Ladizhansky, V.; Boggs, J.M. Structural polymorphism and multifunctionality of myelin basic protein. Biochemistry 2009, 48, 8094–8104. [Google Scholar] [CrossRef] [PubMed]
- Givogri, M.I.; Bongarzone, E.R.; Schonmann, V.; Campagnoni, A.T. Expression and regulation of golli products of myelin basic protein gene during in vitro development of oligodendrocytes. J. Neurosci. Res. 2001, 66, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Vassall, K.A.; Bamm, V.V.; Harauz, G. MyelStones: The executive roles of myelin basic protein in myelin assembly and destabilization in multiple sclerosis. Biochem. J. 2015, 472, 17–32. [Google Scholar] [CrossRef] [PubMed]
- de Ferra, F.; Engh, H.; Hudson, L.; Kamholz, J.; Puckett, C.; Molineaux, S.; Lazzarini, R.A. Alternative splicing accounts for the four forms of myelin basic protein. Cell 1985, 43, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Kamholz, J.; Toffenetti, J.; Lazzarini, R.A. Organization and expression of the human myelin basic protein gene. J. Neurosci. Res. 1988, 21, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Barbarese, E.; Carson, J.H.; Braun, P.E. Accumulation of the four myelin basic proteins in mouse brain during development. J. Neurochem. 1978, 31, 779–782. [Google Scholar] [CrossRef]
- Kim, J.; Mastronardi, F.; Wood, D.; Lubman, D.; Zand, R.; Moscarello, M. Multiple sclerosis: An important role for post-translational modifications of myelin basic protein in pathogenesis. Mol. Cell. Proteom. 2003, 2, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Moscarello, M.A. Myelin Basic Protein, The “Executive” Molecule of the Myelin Membrane. In Cell Biology and Pathology of Myelin: Evolving Biological Concepts and Therapeutic Approaches; Juurlink, B.H.J., Devon, R.M., Doucette, J.R., Nazarali, A.J., Schreyer, D.J., Verge, V.M.K., Eds.; Plenum Press: New York, NY, USA, 1997; pp. 13–25. [Google Scholar]
- Chao, L.P.; Einstein, E.R. Physical properties of the bovine encephalitogenic protein; molecular weight and conformation. J. Neurochem. 1970, 17, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Majava, V.; Wang, C.; Myllykoski, M.; Kangas, S.M.; Kang, S.U.; Hayashi, N.; Baumgartel, P.; Heape, A.M.; Lubec, G.; Kursula, P. Structural analysis of the complex between calmodulin and full-length myelin basic protein, an intrinsically disordered molecule. Amino Acids 2010, 39, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Krigbaum, W.R.; Hsu, T.S. Molecular conformation of bovine A1 basic protein, a coiling macromolecule in aqueous solution. Biochemistry 1975, 14, 2542–2546. [Google Scholar] [CrossRef] [PubMed]
- Libich, D.S.; Ahmed, M.A.; Zhong, L.; Bamm, V.V.; Ladizhansky, V.; Harauz, G. Fuzzy complexes of myelin basic protein: NMR spectroscopic investigations of a polymorphic organizational linker of the central nervous system. Biochem. Cell Biol. 2010, 88, 143–155. [Google Scholar] [CrossRef]
- Sedzik, J.; Kirschner, D.A. Is myelin basic protein crystallizable? Neurochem. Res. 1992, 17, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Beniac, D.R.; Luckevich, M.D.; Czarnota, G.J.; Tompkins, T.A.; Ridsdale, R.A.; Ottensmeyer, F.P.; Moscarello, M.A.; Harauz, G. Three-dimensional structure of myelin basic protein. I. Reconstruction via angular reconstitution of randomly oriented single particles. J. Biol. Chem. 1997, 272, 4261–4268. [Google Scholar] [CrossRef] [PubMed]
- Golds, E.E.; Braun, P.E. Protein associations and basic protein conformation in the myelin membrane. The use of difluorodinitrobenzene as a cross-linking reagent. J. Biol. Chem. 1978, 253, 8162–8170. [Google Scholar] [CrossRef] [PubMed]
- Smith, R. Sedimentation analysis of the self-association of bovine myelin basic protein. Biochemistry 1980, 19, 1826–1831. [Google Scholar] [CrossRef]
- Smith, R. Self-association of myelin basic protein: Enhancement by detergents and lipids. Biochemistry 1982, 21, 2697–2701. [Google Scholar] [CrossRef] [PubMed]
- Moskaitis, J.E.; Shriver, L.C.; Campagnoni, A.T. The association of myelin basic protein with itself and other proteins. Neurochem. Res. 1987, 12, 409–417. [Google Scholar] [CrossRef]
- Edwards, A.M.; Ross, N.W.; Ulmer, J.B.; Braun, P.E. Interaction of myelin basic protein and proteolipid protein. J. Neurosci. Res. 1989, 22, 97–102. [Google Scholar] [CrossRef]
- Smith, R. The basic protein of CNS myelin: Its structure and ligand binding. J. Neurochem. 1992, 59, 1589–1608. [Google Scholar] [CrossRef]
- Smith, R. Non-covalent cross-linking of lipid bilayers by myelin basic protein: A possible role in myelin formation. Biochim. Biophys. Acta 1977, 470, 170–184. [Google Scholar] [CrossRef]
- Han, H.; Myllykoski, M.; Ruskamo, S.; Wang, C.; Kursula, P. Myelin-specific proteins: A structurally diverse group of membrane-interacting molecules. Biofactors 2013, 39, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Anthony, J.S.; Moscarello, M.A. A conformation change induced in the basic encephalitogen by lipids. Biochim. Biophys. Acta 1971, 243, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Smith, R. The secondary structure of myelin basic protein extracted by deoxycholate. Biochim. Biophys. Acta 1977, 491, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Keniry, M.A.; Smith, R. Dependence on lipid structure of the coil-to-helix transition of bovine myelin basic protein. Biochim. Biophys. Acta 1981, 668, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Mendz, G.L.; Moore, W.J.; Carnegie, P.R.; Proton, N.M.R. Evidence for secondary and tertiary structure in myelin basic proteins. Biochem. Biophys. Res. Commun. 1982, 105, 1333–1340. [Google Scholar] [CrossRef]
- Stuart, B.H. A Fourier transform infrared spectroscopic study of the secondary structure of myelin basic protein in reconstituted myelin. Biochem. Mol. Biol. Int. 1996, 38, 839–845. [Google Scholar]
- Vanlangenakker, N.; Vanden Berghe, T.; Krysko, D.V.; Festjens, N.; Vandenabeele, P. Molecular mechanisms and pathophysiology of necrotic cell death. Curr. Mol. Med. 2008, 8, 207–220. [Google Scholar] [CrossRef]
- Trojanowski, J.Q.; Schmidt, M.L.; Shin, R.W.; Bramblett, G.T.; Rao, D.; Lee, V.M. Altered tau and neurofilament proteins in neuro-degenerative diseases: Diagnostic implications for Alzheimer’s disease and Lewy body dementias. Brain Pathol. 1993, 3, 45–54. [Google Scholar] [CrossRef]
- Julien, J.P.; Couillard-Despres, S.; Meier, J. Transgenic mice in the study of ALS: The role of neurofilaments. Brain Pathol. 1998, 8, 759–769. [Google Scholar] [CrossRef]
- Julien, J.P.; Mushynski, W.E. Neurofilaments in health and disease. Prog. Nucleic Acid Res. Mol. Biol. 1998, 61, 1–23. [Google Scholar] [CrossRef]
- Galvin, J.E.; Nakamura, M.; McIntosh, T.K.; Saatman, K.E.; Sampathu, D.; Raghupathi, R.; Lee, V.M.; Trojanowski, J.Q. Neurofilament-rich intraneuronal inclusions exacerbate neurodegenerative sequelae of brain trauma in NFH/LacZ transgenic mice. Exp. Neurol. 2000, 165, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Gotow, T. Neurofilaments in health and disease. Med. Electron Microsc. 2000, 33, 173–199. [Google Scholar] [CrossRef] [PubMed]
- Perrone Capano, C.; Pernas-Alonso, R.; di Porzio, U. Neurofilament homeostasis and motoneurone degeneration. Bioessays 2001, 23, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Eyer, J.; Peterson, A. Neurofilament-deficient axons and perikaryal aggregates in viable transgenic mice expressing a neurofilament-beta-galactosidase fusion protein. Neuron 1994, 12, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Tu, P.H.; Robinson, K.A.; de Snoo, F.; Eyer, J.; Peterson, A.; Lee, V.M.; Trojanowski, J.Q. Selective degeneration of Purkinje cells with Lewy body-like inclusions in aged NFHLACZ transgenic mice. J. Neurosci. 1997, 17, 1064–1074. [Google Scholar] [CrossRef][Green Version]
- Quinlan, R.A.; Brenner, M.; Goldman, J.E.; Messing, A. GFAP and its role in Alexander disease. Exp. Cell Res. 2007, 313, 2077–2087. [Google Scholar] [CrossRef]
- Li, R.; Messing, A.; Goldman, J.E.; Brenner, M. GFAP mutations in Alexander disease. Int. J. Dev. Neurosci. 2002, 20, 259–268. [Google Scholar] [CrossRef]
- Hagemann, T.L. Alexander disease: Models, mechanisms, and medicine. Curr. Opin. Neurobiol. 2022, 72, 140–147. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem Sci 1999, 24, 329–332. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E. The “Correctly Folded” state of proteins: Is it a metastable state? Angew. Chem. Int. Ed. Engl. 2002, 41, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Makin, O.S.; Atkins, E.; Sikorski, P.; Johansson, J.; Serpell, L.C. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. USA 2005, 102, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Meersman, F.; Dobson, C.M. Probing the pressure-temperature stability of amyloid fibrils provides new insights into their molecular properties. Biochim. Biophys. Acta 2006, 1764, 452–460. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Sambashivan, S.; Nelson, R.; Ivanova, M.I.; Sievers, S.A.; Apostol, M.I.; Thompson, M.J.; Balbirnie, M.; Wiltzius, J.J.; McFarlane, H.T.; et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 2007, 447, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Shammas, S.L.; Knowles, T.P.; Baldwin, A.J.; Macphee, C.E.; Welland, M.E.; Dobson, C.M.; Devlin, G.L. Perturbation of the stability of amyloid fibrils through alteration of electrostatic interactions. Biophys. J. 2011, 100, 2783–2791. [Google Scholar] [CrossRef]
- Radzicka, A.; Wolfenden, R. Rates of uncatalyzed peptide bond hydrolysis in neutral solution and the transition state affinities of proteases. J. Am. Chem. Soc. 1996, 118, 6105–6109. [Google Scholar] [CrossRef]
- Goodwin, R.J.; Dungworth, J.C.; Cobb, S.R.; Pitt, A.R. Time-dependent evolution of tissue markers by MALDI-MS imaging. Proteomics 2008, 8, 3801–3808. [Google Scholar] [CrossRef]
- Vass, A.A. Beyond the grave-understanding human decomposition. Microbiol. Today 2001, 28, 190–193. [Google Scholar]
- Schepers, G. The fossil brain. S. Afr. Archaeol. Bull. 1949, 4, 71–82. [Google Scholar] [CrossRef]
- Uversky, V.N. Paradoxes and wonders of intrinsic disorder: Stability of instability. Intrinsically Disord. Proteins 2017, 5, e1327757. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Kimzey, A.L.; Masselon, C.D.; Bruce, J.E.; Garner, E.C.; Brown, C.J.; Dunker, A.K.; Smith, R.D.; Ackerman, E.J. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 2001, 10, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Fontana, A.; Zambonin, M.; Polverino de Laureto, P.; De Filippis, V.; Clementi, A.; Scaramella, E. Probing the conformational state of apomyoglobin by limited proteolysis. J. Mol. Biol. 1997, 266, 223–230. [Google Scholar] [CrossRef]
- Fontana, A.; Polverino de Laureto, P.; De Phillips, V. Molecular aspects of proteolysis of globular proteins. In Protein Stability and Stabilization; van den Tweel, W., Harder, A., Buitelear, M., Eds.; Elsevier Science: Amsterdam, The Netherlands, 1993; pp. 101–110. [Google Scholar]
- Fontana, A.; Fassina, G.; Vita, C.; Dalzoppo, D.; Zamai, M.; Zambonin, M. Correlation between sites of limited proteolysis and segmental mobility in thermolysin. Biochemistry 1986, 25, 1847–1851. [Google Scholar] [CrossRef]
- Gummesson, S.; Hallgren, F.; Kjellström, A. Keep your head high: Skulls on stakes and cranial trauma in Mesolithic Sweden. Antiquity 2018, 92, 74–90. [Google Scholar] [CrossRef]
- Drake, J.L.; Whitelegge, J.P.; Jacobs, D.K. First sequencing of ancient coral skeletal proteins. Sci. Rep. 2020, 10, 19407. [Google Scholar] [CrossRef]

| Term | Genes | p-Value | Count |
|---|---|---|---|
| hsa04915:Estrogen signaling pathway | KRT33B, HSPA8, KRT19, KRT18, KRT39, KRT27, KRT15, KRT13, CALM1, KRT31, CALM2, CTSD | 1.78 × 10−11 | 12 |
| hsa05150:Staphylococcus aureus infection | KRT33B, KRT19, KRT18, KRT39, KRT27, KRT15, KRT13, KRT31 | 2.35 × 10−7 | 8 |
| hsa05022:Pathways of neurodegeneration—multiple diseases | PSMD14, HSPA5, NEFL, CTNNB1, NEFM, CAPN1, CALM1, CALM2, NEFH, RAB5A | 2.59 × 10−4 | 10 |
| hsa05010:Alzheimer disease | PSMD14, CTNNB1, CAPN1, IDE, CALM1, CALM2, RTN4 | 0.00804 | 7 |
| hsa05152:Tuberculosis | LAMP1, CALM1, CALM2, CTSD, RAB5A | 0.00943 | 5 |
| hsa04114:Oocyte meiosis | CALM1, CALM2, YWHAZ, YWHAG | 0.02269 | 4 |
| hsa05014:Amyotrophic lateral sclerosis | PSMD14, HSPA5, NEFL, NEFM, NEFH, RAB5A | 0.02575 | 6 |
| hsa05418:Fluid shear stress and atherosclerosis | CTNNB1, TXN, CALM1, CALM2 | 0.02646 | 4 |
| hsa05012:Parkinson disease | PSMD14, HSPA5, TXN, CALM1, CALM2 | 0.03424 | 5 |
| hsa04141:Protein processing in endoplasmic reticulum | HSPA8, HSPA5, CAPN1, CRYAB | 0.04472 | 4 |
| hsa05417:Lipid and atherosclerosis | HSPA8, HSPA5, CALM1, CALM2 | 0.07770 | 4 |
| hsa04916:Melanogenesis | CTNNB1, CALM1, CALM2 | 0.08006 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammed, A.S.; Uversky, V.N. Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain. Biology 2022, 11, 1704. https://doi.org/10.3390/biology11121704
Mohammed AS, Uversky VN. Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain. Biology. 2022; 11(12):1704. https://doi.org/10.3390/biology11121704
Chicago/Turabian StyleMohammed, Aaron S., and Vladimir N. Uversky. 2022. "Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain" Biology 11, no. 12: 1704. https://doi.org/10.3390/biology11121704
APA StyleMohammed, A. S., & Uversky, V. N. (2022). Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain. Biology, 11(12), 1704. https://doi.org/10.3390/biology11121704








