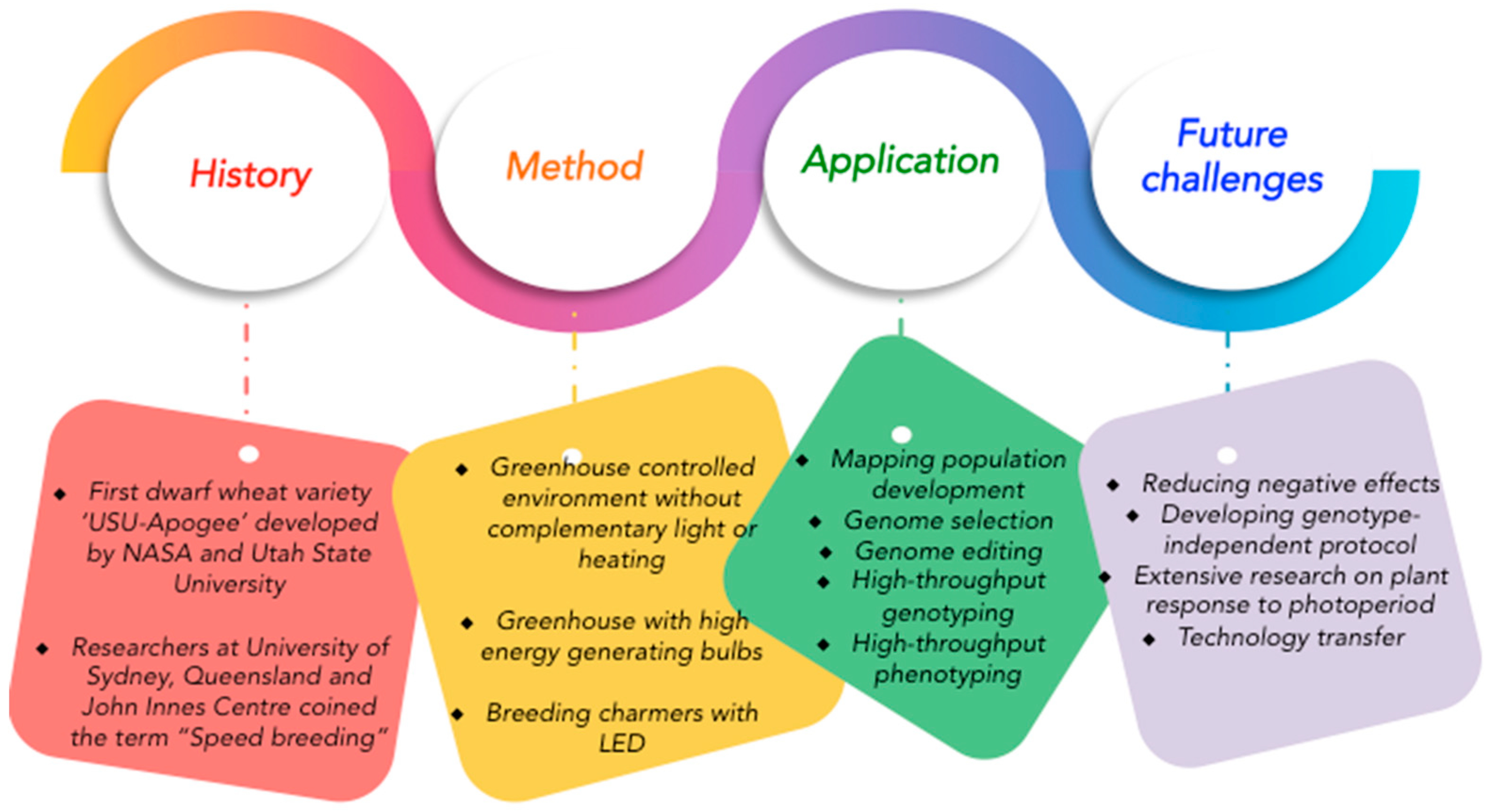
Breeding More Crops in Less Time: A Perspective on Speed Breeding
Abstract
:Simple Summary
Abstract
1. Introduction
| Type of Photoperiod | Family | Species | Generations/Year | Reference |
|---|---|---|---|---|
| Long day | Poaceae | Oat (Avena sativa) | ~7 generations | Liu et al. [26] |
| Barley (Hordeum vulgare) | ~6 generations | Hickey et al. [15] | ||
| Wheat (Triticum aestivum) | 4–6 generations | Mukade et al. [27] | ||
| Fabaceae | Clover (Trifolium subterraneum) | 2.7–6.1 generations | Pazos-Navarro et al. [21] | |
| Lentil (Lens culinaris) | ~8 generations | Mobini et al. [28] | ||
| Chickpea (Cicer arietinum) | ~6 generations | Watson et al. [29]; Atieno et al. [30] | ||
| Pea (Pisum sativum) | 6.8 generations | Ochatt et al. [31]; Ribalta et al. [22] | ||
| 5 generations | Mobini and Warkentin [32] | |||
| Faba bean (Vicia faba) | 7 generations | Mobini et al. [28] | ||
| Narrow-leaf lupin (Lupinus angustifolius) | 5 generations | Croser et al. [14] | ||
| Brassicaceae | Rapeseed (Brassica napus) | ~5 generations | Watson et al. [29] | |
| Linaceae | Flax (Linum usitatissimum) | ~3 generations | Sysoeva et al. [20] | |
| Short day | Poaceae | Rice (Oryza sativa) | ~4–5 generations | Rana et al. [33]; Collard et al. [34] |
| Sorghum (Sorghum bicolor) | 4 generations | Forster et al. [35] | ||
| Fabaceae | Soybean (Glycine max) | ~5 generations | Nagatoshi and Fujita [36]; Jahne et al. [17] | |
| Pigeonpea (Cajanus cajan) | ~4 generations | Saxena et al. [37] | ||
| Bambara groundnut (Vigna subterranea ) | ~4 generations | Ochatt et al. [31] | ||
| Groundnut (Arachis hypogaea) | ~4 generations | O’Connor et al. [38] | ||
| Amaranthaceae | Grain amaranthus (Amaranthus spp.) | ~6 generations | Stetter et al. [39] |
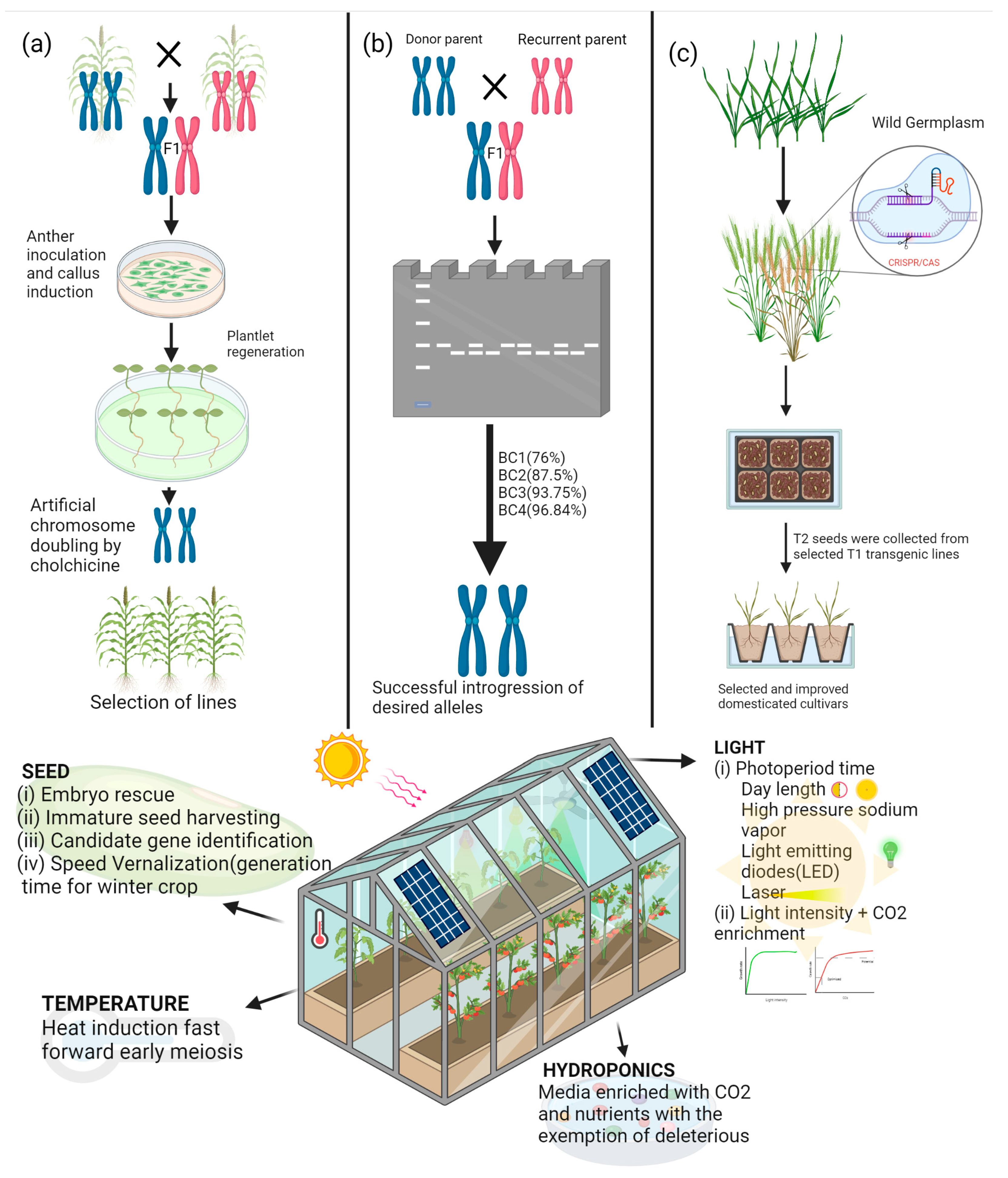
2. Flexible SB Systems for Fast-Tracking Applied and Basic Research
3. SB Applications in Research and Breeding
4. Model Species
4.1. Cereals
4.2. Oilseeds
4.3. Legumes
4.4. Fruit Crops
4.5. Vegetable Crops
5. Opportunities for Combining SB with Modern Breeding and Phenotyping Tools
6. Challenges and Limitations
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IISD. World Population to Reach 9.9 Billion by 2050. Available online: https://sdg.iisd.org/news/world-population-to-reach-9-9-billion-by-2050/ (accessed on 3 February 2022).
- Hussain, B. Modernization in plant breeding approaches for improving biotic stress resistance in crop plants. Turk. J. Agric. For. 2015, 39, 515–530. [Google Scholar] [CrossRef]
- Lin, Z.; Cogan, N.O.I.; Pembleton, L.W.; Spangenberg, G.C.; Forster, J.W.; Hayes, B.J.; Daetwyler, H.D. Genetic Gain and Inbreeding from Genomic Selection in a Simulated Commercial Breeding Program for Perennial Ryegrass. Plant Genome. 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moose, S.P.; Mumm, R.H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohra, A.; Jha, U.C.; Godwin, I.D.; Varshney, R.K. Genomic interventions for sustainable agriculture. Plant Biotechnol. J. 2020, 18, 2388–2405. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Singh, V.K.; Bohra, A.; Kumar, A.; Reif, J.C.; Varshney, R.K. Genomics and breeding innovations for enhancing genetic gain for climate resilience and nutrition traits. Theor. Appl. Genet. 2021, 134, 1829–1843. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing Future Crops: Genomics-Assisted Breeding Comes of Age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Cobb, J.N.; Juma, R.U.; Biswas, P.S.; Arbelaez, J.D.; Rutkoski, J.; Atlin, G.; Hagen, T.; Quinn, M.; Ng, E.H. Enhancing the rate of genetic gain in public-sector plant breeding programs: Lessons from the breeder’s equation. Theor. Appl. Genet. 2019, 132, 627–645. [Google Scholar] [CrossRef] [Green Version]
- Brim, C.A. A Modified Pedigree Method of Selection in Soybeans 1. Crop Sci. 1966, 6, 220. [Google Scholar] [CrossRef] [Green Version]
- Goulden, C.H. Problems in Plant Selection; Cambridge University Press: Cambridge, UK, 1939; pp. 132–133. [Google Scholar]
- Borlaug, N. Wheat Breeding and Its Impact on World Food Supply; Finlay, K.W., Shephard, K.W., Eds.; Australian Academy of Sciences: Canberra, Australia, 1968; pp. 1–36. [Google Scholar]
- Ghosh, S.; Watson, A.; Gonzalez-Navarro, O.E.; Ramirez-Gonzalez, R.H.; Yanes, L.; Mendoza-Suárez, M.; Simmonds, J.; Wells, R.; Rayner, T.; Green, P.; et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 2018, 13, 2944–2963. [Google Scholar] [CrossRef] [Green Version]
- Mobini, S.H.; Lulsdorf, M.; Warkentin, T.D.; Vandenberg, A. Low red: Far-red light ratio causes faster in vitro flowering in lentil. Can. J. Plant Sci. 2016, 96, 908–918. [Google Scholar] [CrossRef] [Green Version]
- Croser, J.S.; Pazos-Navarro, M.; Bennett, R.G.; Tschirren, S.; Edwards, K.; Erskine, W.; Creasy, R.; Ribalta, F.M. Time to flowering of temperate pulses in vivo and generation turnover in vivo–in vitro of narrow-leaf lupin accelerated by low red to far-red ratio and high intensity in the far-red region. Plant Cell Tissue Organ Cult. 2016, 127, 591–599. [Google Scholar] [CrossRef]
- Hickey, L.T.; Germán, S.E.; Pereyra, S.A.; Diaz, J.E.; Ziems, L.A.; Fowler, R.A.; Platz, G.J.; Franckowiak, J.D.; Dieters, M.J. Speed breeding for multiple disease resistance in barley. Euphytica 2017, 213, 64. [Google Scholar] [CrossRef]
- Cazzola, F.; Bermejo, C.J.; Guindon, M.F.; Cointry, E. Speed breeding in pea (Pisum sativum L.), an efficient and simple system to accelerate breeding programs. Euphytica 2020, 216, 178. [Google Scholar] [CrossRef]
- Jähne, F.; Hahn, V.; Würschum, T.; Leiser, W.L. Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 2020, 133, 2335–2342. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, N.E. Microchemical and morphological studies of effect of light on plants. Bot. Gaz. 1926, 81, 173–195. [Google Scholar] [CrossRef]
- Wheeler, R.M. A historical background of plant lighting: An introduction to the workshop. Hortic. Sci. 2008, 43, 1942–1943. [Google Scholar] [CrossRef] [Green Version]
- Sysoeva, M.I.; Markovskaya, E.F.; Shibaeva, T.G. Plants under Continuous Light: A Review. Plant Stress 2010, 4, 5–17. [Google Scholar]
- Pazos-Navarro, M.; Castello, M.; Bennett, R.G.; Nichols, P.; Croser, J. In vitro-assisted single-seed descent for breeding-cycle compression in subterranean clover (Trifolium subterraneum L.). Crop Pasture Sci. 2017, 68, 958. [Google Scholar] [CrossRef]
- Ribalta, F.M.; Pazos-Navarro, M.; Nelson, K.; Edwards, K.; Ross, J.J.; Bennett, R.; Munday, C.; Erskine, W.; Ochatt, S.J.; Croser, J. Precocious floral initiation and identification of exact timing of embryo physiological maturity facilitate germination of immature seeds to truncate the lifecycle of pea. Plant Growth Regul. 2017, 81, 345–353. [Google Scholar] [CrossRef]
- Ballare, C.L.; Scopel, A.L.; Stapleton, A.E.; Yanovsky, M.J. Solar Ultraviolet-B Radiation Affects Seedling Emergence, DNA Integrity, Plant Morphology, Growth Rate, and Attractiveness to Herbivore Insects in Datura ferox. Plant Physiol. 1996, 112, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Basu, C.; Meinhardt-Wollweber, M.; Roth, B. LEDs for energy efficient greenhouse lighting. Renew. Sustain. Energy Rev. 2015, 49, 139–147. [Google Scholar] [CrossRef] [Green Version]
- Christopher, J.; Richard, C.; Chenu, K.; Christopher, M.; Borrell, A.; Hickey, L. Integrating Rapid Phenotyping and Speed Breeding to Improve Stay-Green and Root Adaptation of Wheat in Changing, Water-Limited, Australian Environments. Procedia Environ. Sci. 2015, 29, 175–176. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zwer, P.; Wang, H.; Liu, C.; Lu, Z.; Wang, Y.; Yan, G. A fast generation cycling system for oat and triticale breeding. Plant Breed. 2016, 135, 574–579. [Google Scholar] [CrossRef]
- Mukade, K. New Procedures for Accelerating Generation Advancement in Wheat Breeding. JARQ 1974, 8, 1–5. [Google Scholar]
- Mobini, S.H.; Lulsdorf, M.; Warkentin, T.; Vandenberg, A. Plant growth regulators improve in vitro flowering and rapid generation advancement in lentil and faba bean. Vitr. Cell. Dev. Biol.-Plant 2014, 51, 71–79. [Google Scholar] [CrossRef]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Hatta, M.A.M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Atieno, J.; Li, Y.; Langridge, P.; Dowling, K.; Brien, C.; Berger, B.; Varshney, R.; Sutton, T. Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci. Rep. 2017, 7, 1300. [Google Scholar] [CrossRef] [Green Version]
- Ochatt, S.J.; Sangwan, R.S.; Marget, P.; Assoumou Ndong, Y.; Rancillac, M.; Perney, P. New Approaches towards the Shortening of Generation Cycles for Faster Breeding of Protein Legumes. Plant Breed. 2002, 121, 436–440. [Google Scholar] [CrossRef]
- Mobini, S.H.; Warkentin, T.D. A simple and efficient method of in vivo rapid generation technology in pea (Pisum sativum L.). Vitr. Cell. Dev. Biol.-Plant 2016, 52, 530–536. [Google Scholar] [CrossRef]
- Rana, M.M.; Takamatsu, T.; Baslam, M.; Kaneko, K.; Itoh, K.; Harada, N.; Sugiyama, T.; Ohnishi, T.; Kinoshita, T.; Takagi, H.; et al. Salt Tolerance Improvement in Rice through Efficient SNP Marker-Assisted Selection Coupled with Speed-Breeding. Int. J. Mol. Sci. 2019, 20, 2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collard, B.C.Y.; Beredo, J.C.; Lenaerts, B.; Mendoza, R.; Santelices, R.; Lopena, V.; Verdeprado, H.; Raghavan, C.; Gregorio, G.B.; Vial, L.; et al. Revisiting rice breeding methods—Evaluating the use of rapid generation advance (RGA) for routine rice breeding. Plant Prod. Sci. 2017, 20, 337–352. [Google Scholar] [CrossRef] [Green Version]
- Forster, B.P. Accelerated plant breeding. CAB Rev. 2014, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Nagatoshi, Y.; Fujita, Y. Accelerating Soybean Breeding in a CO2-Supplemented Growth Chamber. Plant Cell Physiol. 2019, 60, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, K.; Saxena, R.K.; Varshney, R.K. Use of immature seed germination and single seed descent for rapid genetic gains in pigeonpea. Plant Breed. 2017, 136, 954–957. [Google Scholar] [CrossRef] [Green Version]
- O'Connor, D.J.; Wright, G.C.; Dieters, M.J.; George, D.L.; Hunter, M.N.; Tatnell, J.R.; Fleischfresser, D.B. Development and Application of Speed Breeding Technologies in a Commercial Peanut Breeding Program. Peanut Sci. 2013, 40, 107–114. [Google Scholar] [CrossRef]
- Stetter, M.G.; Zeitler, L.; Steinhaus, A.; Kroener, K.; Biljecki, M.; Schmid, K.J. Crossing Methods and Cultivation Conditions for Rapid Production of Segregating Populations in Three Grain Amaranth Species. Front. Plant Sci. 2016, 7, 816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croser, J.; Mao, D.; Dron, N.; Michelmore, S.; McMurray, L.; Preston, C.; Bruce, D.; Ogbonnaya, F.C.; Ribalta, F.M.; Hayes, J.; et al. Evidence for the Application of Emerging Technologies to Accelerate Crop Improvement—A Collaborative Pipeline to Introgress Herbicide Tolerance Into Chickpea. Front. Plant Sci. 2021, 12, 779122. [Google Scholar] [CrossRef] [PubMed]
- Lulsdorf, M.M.; Banniza, S. Rapid generation cycling of an F2 population derived from a cross between Lens culinaris Medik. and Lens ervoides (Brign.) Grande after aphanomyces root rot selection. Plant Breed. 2018, 137, 486–491. [Google Scholar] [CrossRef]
- Gosal, S.S.; Pathak, D.; Wani, S.H.; Vij, S.; Pathak, M. Accelerated Breeding of Plants: Methods and Applications. In Accelerated Plant Breeding, Volume 1; Gosal, S.S., Wani, S.H., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–29. ISBN 978-3-030-41865-6. [Google Scholar]
- Khoo, K.H.P.; Sheedy, J.G.; Taylor, J.D.; Croser, J.S.; Hayes, J.E.; Sutton, T.; Thompson, J.P.; Mather, D.E. A QTL on the Ca7 chromosome of chickpea affects resistance to the root-lesion nematode Pratylenchus thornei. Mol. Breed. 2021, 41, 78. [Google Scholar] [CrossRef]
- Dadu, R.H.R.; Bar, I.; Ford, R.; Sambasivam, P.; Croser, J.; Ribalta, F.; Kaur, S.; Sudheesh, S.; Gupta, D. Lens orientalis Contributes Quantitative Trait Loci and Candidate Genes Associated with Ascochyta Blight Resistance in Lentil. Front. Plant Sci. 2021, 12, 703283. [Google Scholar] [CrossRef]
- Taylor, C.M.; Garg, G.; Berger, J.D.; Ribalta, F.M.; Croser, J.S.; Singh, K.B.; Cowling, W.A.; Kamphuis, L.G.; Nelson, M.N. A Trimethylguanosine Synthase1-like (TGS1) homologue is implicated in vernalisation and flowering time control. Theor. Appl. Genet. 2021, 134, 3411–3426. [Google Scholar] [CrossRef]
- Zaman, S.U.; Malik, A.I.; Kaur, P.; Ribalta, F.M.; Erskine, W. Waterlogging Tolerance at Germination in Field Pea: Variability, Genetic Control, and Indirect Selection. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Zhang, P.; Liu, H.; Lu, Z.; Yan, G. A fully in vitro protocol towards large scale production of recombinant inbred lines in wheat (Triticum aestivum L.). Plant Cell Tissue Organ Cult. 2017, 128, 655–661. [Google Scholar] [CrossRef]
- Ferrie, A.M.R. Doubled haploid production in nutraceutical species: A review. Euphytica 2007, 158, 347–357. [Google Scholar] [CrossRef]
- Ortiz, R.; Trethowan, R.; Ferrara, G.O.; Iwanaga, M.; Dodds, J.H.; Crouch, J.H.; Crossa, J.; Braun, H.-J. High yield potential, shuttle breeding, genetic diversity, and a new international wheat improvement strategy. Euphytica 2007, 157, 365–384. [Google Scholar] [CrossRef]
- Aldwinckle, H.S. Flowering of apple seedlings 16–20 months after germination. Hortic. Sci. 1975, 10, 124–126. [Google Scholar]
- Van Nocker, S.; Gardiner, S.E. Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 2014, 1, 14022. [Google Scholar] [CrossRef] [Green Version]
- De Pauw, R.M.; Clarke, J.M. Acceleration of generation advancement in spring wheat. Euphytica 1976, 25, 415–418. [Google Scholar] [CrossRef]
- Robertson, L.D.; Curtis, B.C. Germination of Immature Kernels of Winter Wheat. Crop Sci. 1967, 7, 269–270. [Google Scholar] [CrossRef]
- Tanaka, J.; Hayashi, T.; Iwata, H. A practical, rapid generation-advancement system for rice breeding using simplified biotron breeding system. Breed. Sci. 2016, 66, 542–551. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, T.; Yoshino, M.; Yamakawa, H.; Kinoshita, T. The Biotron Breeding System: A Rapid and Reliable Procedure for Genetic Studies and Breeding in Rice. Plant Cell Physiol. 2011, 52, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Alahmad, S.; Dinglasan, E.; Leung, K.M.; Riaz, A.; Derbal, N.; Voss-Fels, K.P.; Able, J.A.; Bassi, F.M.; Christopher, J.; Hickey, L.T. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods 2018, 14, 36. [Google Scholar] [CrossRef]
- Zhang, Z.; Wei, W.; Zhu, H.; Challa, G.S.; Bi, C.; Trick, H.N.; Li, W. W3 Is a New Wax Locus That Is Essential for Biosynthesis of β-Diketone, Development of Glaucousness, and Reduction of Cuticle Permeability in Common Wheat. PLoS ONE 2015, 10, e0140524. [Google Scholar] [CrossRef] [Green Version]
- Rizal, G.; Karki, S.; Alcasid, M.; Montecillo, F.; Acebron, K.; Larazo, N.; Garcia, R.; Slamet-Loedin, I.H.; Quick, W.P. Shortening the Breeding Cycle of Sorghum, a Model Crop for Research. Crop Sci. 2014, 54, 520–529. [Google Scholar] [CrossRef]
- Burris, J.S. Effect of Seed Maturation and Plant Population on Soybean Seed Quality. Agron. J. 1973, 65, 440–441. [Google Scholar] [CrossRef]
- Roumet, P.; Morin, F. Germination of Immature Soybean Seeds to Shorten Reproductive Cycle Duration. Crop Sci. 1997, 37, 521–525. [Google Scholar] [CrossRef]
- Dagustu, N.; Bayram, G.; Sincik, M.; Bayraktaroglu, M. The Short Breeding Cycle Protocol Effective on Diverse Genotypes of Sunflower (Helianthus annuus L.). Turkish J. Field Crop. 2012, 17, 124–128. [Google Scholar]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Espósito, M.A.; Almirón, P.; Gatti, I.; Cravero, V.P.; Anido, F.S.L.; Cointry, E.L. Methodology A rapid method to increase the number of F1 plants in pea (Pisum sativum) breeding programs. Genet. Mol. Res. 2012, 11, 2729–2732. [Google Scholar] [CrossRef]
- Samineni, S.; Sen, M.; Sajja, S.B.; Gaur, P.M. Rapid generation advance (RGA) in chickpea to produce up to seven generations per year and enable speed breeding. Crop J. 2020, 8, 164–169. [Google Scholar] [CrossRef]
- Parmar, S.; Deshmukh, D.B.; Kumar, R.; Manohar, S.S.; Joshi, P.; Sharma, V.; Chaudhari, S.; Variath, M.T.; Gangurde, S.S.; Bohar, R.; et al. Single Seed-Based High-Throughput Genotyping and Rapid Generation Advancement for Accelerated Groundnut Genetics and Breeding Research. Agronomy 2021, 11, 1226. [Google Scholar] [CrossRef]
- Baier, K.; Maynard, C.; Powell, W.A. Early Flowering in Chestnut Species Induced under High Intensity, High Dose Light in Growth Chambers. J. Am. Chestnut Found. 2012, 26, 8–10. [Google Scholar]
- Flachowsky, H.; Le Roux, P.-M.; Peil, A.; Patocchi, A.; Richter, K.; Hanke, M.-V. Application of a high-speed breeding technology to apple (Malus × domestica) based on transgenic early flowering plants and marker-assisted selection. New Phytol. 2011, 192, 364–377. [Google Scholar] [CrossRef]
- Vira, B.; Wildburger, C.; Mansourian, S.; International Union of Forestry Research Organizations (Eds.) Forests, Trees and Landscapes for Food Security and Nutrition: A Global Assessment Report; IUFRO World Series; IUFRO: Vienna, Austria, 2015; ISBN 978-3-902762-40-5. [Google Scholar]
- Souza, L.S.; Diniz, R.P.; Neves, R.; Alves, A.A.C.; de Oliveira, E.J. Grafting as a strategy to increase flowering of cassava. Sci. Hortic. 2018, 240, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Demers, D.-A.; Dorais, M.; Wien, C.H.; Gosselin, A. Effects of supplemental light duration on greenhouse tomato (Lycopersicon esculentum Mill.) plants and fruit yields. Sci. Hortic. 1998, 74, 295–306. [Google Scholar] [CrossRef]
- Bhattaraj, S.P.; de la Pena, R.C.; Midmore, D.J.; Palchamy, K. In vitro culture of immature seed for rapid generation advancement in tomato. Euphytica 2009, 167, 23–30. [Google Scholar] [CrossRef]
- Manzur, J.; Oliva-Alarcón, M.; Rodríguez-Burruezo, A. In vitro germination of immature embryos for accelerating generation advancement in peppers (Capsicum annuum L.). Sci. Hortic. 2014, 170, 203–210. [Google Scholar] [CrossRef]
- Geboloğlu, N.; Bozmaz, S.; Aydin, M.; Cakmak, P. The role of growth regulators, embryo age and genotypes on immature embryo germination and rapid generation advancement in tomato (Lycopersicon esculentum Mill.). Afr. J. Biotechnol. 2011, 10, 4895–4900. [Google Scholar]
- Borovsky, Y.; Mohan, V.; Shabtai, S.; Paran, I. CaFT-LIKE is a flowering promoter in pepper and functions as florigen in tomato. Plant Sci. 2020, 301, 110678. [Google Scholar] [CrossRef]
- Velez-Ramirez, A.I.; Van Ieperen, W.; Vreugdenhil, D.; Van Poppel, P.M.J.A.; Heuvelink, E.; Millenaar, F.F. A single locus confers tolerance to continuous light and allows substantial yield increase in tomato. Nat. Commun. 2014, 5, 4549. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Wu, J.-J.; Tang, T.; Liu, K.-D.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef] [Green Version]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-Free Genome Editing of Brassica oleracea and B. rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 2018, 9, 1594. [Google Scholar] [CrossRef]
- Bao, A.; Zhang, C.; Huang, Y.; Chen, H.; Zhou, X.; Cao, D. Genome editing technology and application in soybean improvement. Oil Crop Sci. 2020, 5, 31–40. [Google Scholar] [CrossRef]
- Angeles-Shim, R.B.; Reyes, V.P.; del Valle, M.M.; Lapis, R.S.; Shim, J.; Sunohara, H.; Jena, K.K.; Ashikari, M.; Doi, K. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast. Rice Sci. 2020, 27, 113–123. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Herzog, E.; Dreisigacker, S.; Sukurmaran, S.; Watson, A.; Frisch, M.; Hayes, B.J.; Hickey, L.T. Speed GS to accelerate genetic gain in spring wheat. In Applications of Genetic and Genomic Research in Cereals, 1st ed.; Miedaner, T., Korzun, V., Eds.; Woodhead Publishing: Cambridge, MA, USA, 2018. [Google Scholar] [CrossRef]
- Watson, A.; Hickey, L.T.; Christopher, J.; Rutkoski, J.; Poland, J.; Hayes, B.J. Multivariate Genomic Selection and Potential of Rapid Indirect Selection with Speed Breeding in Spring Wheat. Crop Sci. 2019, 59, 1945–1959. [Google Scholar] [CrossRef] [Green Version]
- Al Tamimi, N.; Brien, C.; Oakey, H.; Berger, B.; Saade, S.; Ho, Y.S.; Schmöckel, S.M.; Tester, M.; Negrao, S. Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nat. Commun. 2016, 7, 13342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, C.; Hickey, L.; Fletcher, S.; Chenu, K.; Borrell, A.; Christopher, J. High-throughput Phenotyping of Wheat Seminal Root Traits in a Breeding Context. Procedia Environ. Sci. 2015, 29, 102–103. [Google Scholar] [CrossRef] [Green Version]
- Walter, J.; Edwards, J.; Cai, J.; McDonald, G.; Miklavcic, S.; Kuchel, H. High-Throughput Field Imaging and Basic Image Analysis in a Wheat Breeding Programme. Front. Plant Sci. 2019, 10, 449. [Google Scholar] [CrossRef]
- Phyu, P.; Islam, M.R.; Cruz, P.C.S.; Collard, B.C.Y.; Kato, Y. Use of NDVI for indirect selection of high yield in tropical rice breeding. Euphytica 2020, 216, 74. [Google Scholar] [CrossRef]
- Samantara, K.; Reyes, V.P.; Agrawal, N.; Mohapatra, S.R.; Jena, K.K. Advances and trends on the utilization of multi-parent advanced generation intercross (MAGIC) for crop improvement. Euphytica 2021, 217, 189. [Google Scholar] [CrossRef]
- Kitony, J.K.; Sunohara, H.; Tasaki, M.; Mori, J.-I.; Shimazu, A.; Reyes, V.; Yasui, H.; Yamagata, Y.; Yoshimura, A.; Yamasaki, M.; et al. Development of an Aus-Derived Nested Association Mapping (Aus-NAM) Population in Rice. Plants 2021, 10, 1255. [Google Scholar] [CrossRef] [PubMed]
- Wanga, M.A.; Shimelis, H.; Mark, J.M.; Laing, D. Opportunities and challenges of speed breeding: A review. Plant Breed. 2021, 140, 185–194. [Google Scholar] [CrossRef]
- Barrios, P.M.G.; Bhatta, M.; Halley, M.; Sandro, P.; Gutiérrez, L. Speed breeding and early panicle harvest accelerates oat (Avena sativa L.) breeding cycles. Crop Sci. 2020, 61, 320–330. [Google Scholar] [CrossRef]
- Sharma, A.; Jones, J.B.; White, F.F. Recent advances in developing disease resistance in plants. F1000Research 2019, 8, 1934. [Google Scholar] [CrossRef] [Green Version]
- Bennett, R.; Ribalta, F.M.; Pazos-Navarro, M.; Leonforte, A.; Croser, J.S. Discrimination of boron tolerance in Pisum sativum L. genotypes using a rapid, high-throughput hydroponic screen and precociously germinated seed grown under far-red enriched light. Plant Methods 2017, 13, 70. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.A.; Chitikineni, A.; Lam, H.-M.; Hickey, L.T.; Croser, J.S.; Bayer, P.E.; et al. Fast-forward breeding for a food-secure world. Trends Genet. 2021, 37, 1124–1136. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samantara, K.; Bohra, A.; Mohapatra, S.R.; Prihatini, R.; Asibe, F.; Singh, L.; Reyes, V.P.; Tiwari, A.; Maurya, A.K.; Croser, J.S.; et al. Breeding More Crops in Less Time: A Perspective on Speed Breeding. Biology 2022, 11, 275. https://doi.org/10.3390/biology11020275
Samantara K, Bohra A, Mohapatra SR, Prihatini R, Asibe F, Singh L, Reyes VP, Tiwari A, Maurya AK, Croser JS, et al. Breeding More Crops in Less Time: A Perspective on Speed Breeding. Biology. 2022; 11(2):275. https://doi.org/10.3390/biology11020275
Chicago/Turabian StyleSamantara, Kajal, Abhishek Bohra, Sourav Ranjan Mohapatra, Riry Prihatini, Flora Asibe, Lokendra Singh, Vincent P. Reyes, Abha Tiwari, Alok Kumar Maurya, Janine S. Croser, and et al. 2022. "Breeding More Crops in Less Time: A Perspective on Speed Breeding" Biology 11, no. 2: 275. https://doi.org/10.3390/biology11020275
APA StyleSamantara, K., Bohra, A., Mohapatra, S. R., Prihatini, R., Asibe, F., Singh, L., Reyes, V. P., Tiwari, A., Maurya, A. K., Croser, J. S., Wani, S. H., Siddique, K. H. M., & Varshney, R. K. (2022). Breeding More Crops in Less Time: A Perspective on Speed Breeding. Biology, 11(2), 275. https://doi.org/10.3390/biology11020275












