EXOC6 (Exocyst Complex Component 6) Is Associated with the Risk of Type 2 Diabetes and Pancreatic β-Cell Dysfunction
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression Data from Human Pancreatic Islets
2.2. Screening for T2D-Associated Genetic Variants in Exoc6/6b
2.3. Maintaining of INS-1 (832/13) Cells
2.4. siRNA Silencing and Insulin Secretion
2.5. Cell Viability Assay
2.6. Apoptosis Assay
2.7. Glucose Uptake Assay
2.8. Intracellular Reactive Oxygen Species (ROS)
2.9. Quantitative PCR (qPCR)
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
3.1. Expression of EXOC6 and EXOC6B in Human Pancreatic Islets
3.2. Associated Genetic Variants in EXOC6/6B with the Risk of T2D
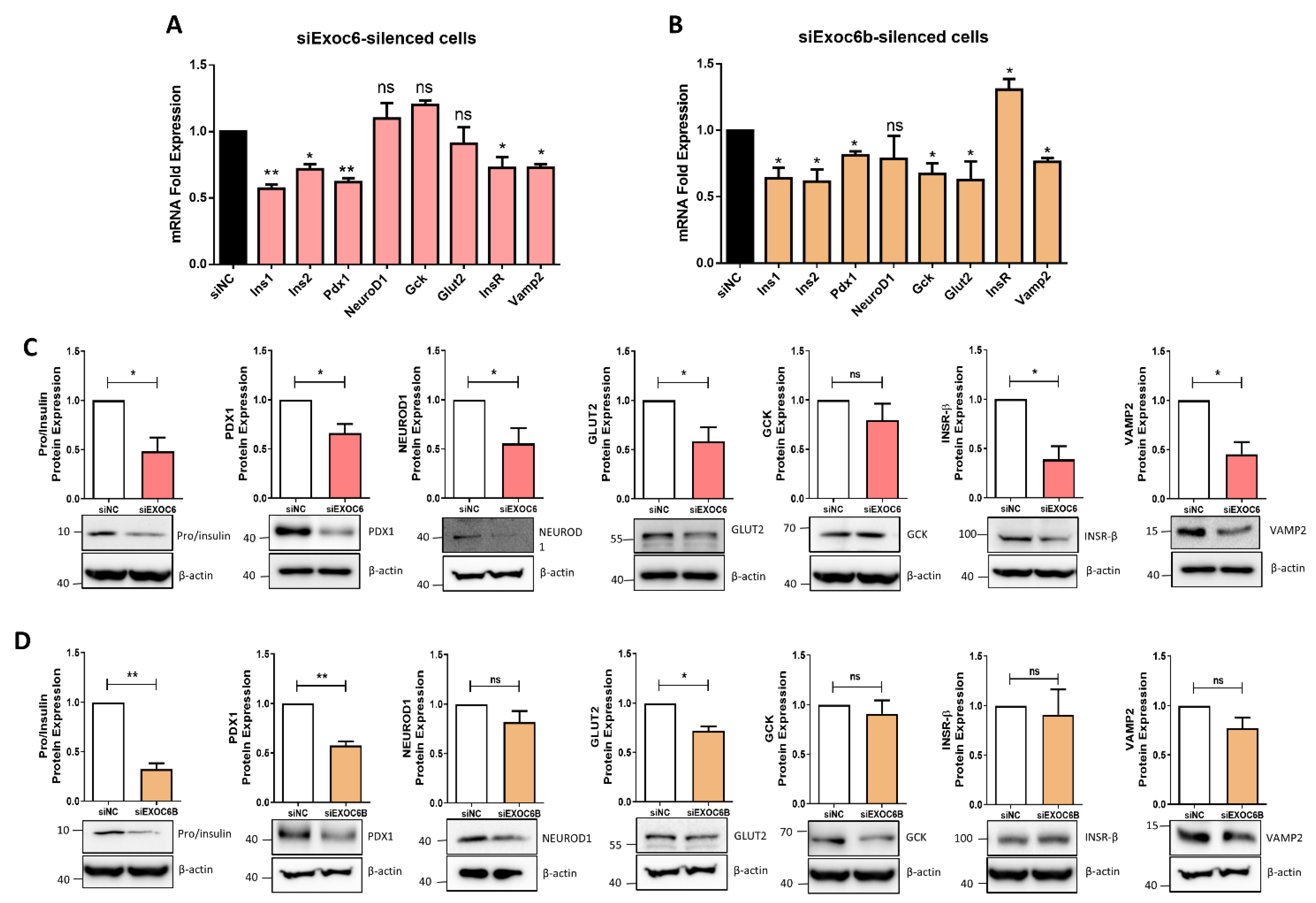
3.3. Expression Silencing of Exoc6/6B Cells Reduces Insulin Secretion and Influence Cell Functions in INS-1 Cells
3.4. Silencing of Exoc6/6b Modulates the Expression of genes Involved in β Cell Function
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scully, T. Diabetes in numbers. Nature 2012, 485, S2–S3. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.; Ramracheya, R.; Rorsman, P. Autocrine regulation of insulin secretion. Diabetes Obes. Metab. 2012, 14, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Ruiter, M.; Buijs, R.M.; Kalsbeek, A. Hormones and the autonomic nervous system are involved in suprachiasmatic nucleus modulation of glucose homeostasis. Curr. Diabetes Rev. 2006, 2, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O. Glucose Metabolism. In Stroke Revisited: Diabetes in Stroke; Springer: Berlin/Heidelberg, Germany, 2021; pp. 3–13. [Google Scholar]
- Zawalich, W.S.; Rasmussen, H. Control of insulin secretion: A model involving Ca2+, cAMP and diacylglycerol. Mol. Cell. Endocrinol. 1990, 70, 119–137. [Google Scholar] [CrossRef]
- Van de Bunt, M.; Gloyn, A. A tale of two glucose transporters: How GLUT2 re-emerged as a contender for glucose transport into the human beta cell. Diabetologia 2012, 55, 2312–2315. [Google Scholar] [CrossRef] [PubMed]
- Jahn, R.; Südhof, T.C. Membrane fusion and exocytosis. Annu. Rev. Biochem. 1999, 68, 863–911. [Google Scholar] [CrossRef] [PubMed]
- TerBush, D.R.; Maurice, T.; Roth, D.; Novick, P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996, 15, 6483–6494. [Google Scholar] [CrossRef]
- Whyte, J.R.; Munro, S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002, 115, 2627–2637. [Google Scholar] [CrossRef]
- Ewart, M.-A.; Clarke, M.; Kane, S.; Chamberlain, L.H.; Gould, G.W. Evidence for a role of the exocyst in insulin-stimulated Glut4 trafficking in 3T3-L1 adipocytes. J. Biol. Chem. 2005, 280, 3812–3816. [Google Scholar] [CrossRef]
- Lyons, P.D. Insulin stimulates the phosphorylation of the exocyst protein Sec8 in adipocytes. Biosci. Rep. 2009, 29, 229–235. [Google Scholar] [CrossRef][Green Version]
- Sano, H.; Peck, G.R.; Blachon, S.; Lienhard, G.E. A potential link between insulin signaling and GLUT4 translocation: Association of Rab10-GTP with the exocyst subunit Exoc6/6b. Biochem. Biophys. Res. Commun. 2015, 465, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, I.; Ackerman IV, W.; Vandre, D.; Robinson, J. Exocyst complex protein expression in the human placenta. Placenta 2014, 35, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Ashrafzadeh, P. Exploring Cellular Dynamics: From Vesicle Tethering to Cell Migration. Ph.D. Thesis, Uppsala University, Uppsala, Sweden, 2016. [Google Scholar]
- Zou, W.; Yadav, S.; DeVault, L.; Jan, Y.N.; Sherwood, D.R. RAB-10-dependent membrane transport is required for dendrite arborization. PLoS Genet. 2015, 11, e1005484. [Google Scholar] [CrossRef] [PubMed]
- Van Bergen, N.J.; Ahmed, S.M.; Collins, F.; Cowley, M.; Vetro, A.; Dale, R.C.; Hock, D.H.; de Caestecker, C.; Menezes, M.; Massey, S. Mutations in the exocyst component EXOC2 cause severe defects in human brain development. J. Exp. Med. 2020, 217, e20192040. [Google Scholar] [CrossRef]
- Naegeli, K.M.; Hastie, E.; Garde, A.; Wang, Z.; Keeley, D.P.; Gordon, K.L.; Pani, A.M.; Kelley, L.C.; Morrissey, M.A.; Chi, Q. Cell invasion in vivo via rapid exocytosis of a transient lysosome-derived membrane domain. Dev. Cell 2017, 43, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Walch-Solimena, C.; Collins, R.N.; Novick, P.J. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 1997, 137, 1495–1509. [Google Scholar] [CrossRef]
- Tsuboi, T.; Ravier, M.A.; Xie, H.; Ewart, M.-A.; Gould, G.W.; Baldwin, S.A.; Rutter, G.A. Mammalian exocyst complex is required for the docking step of insulinvesicle exocytosis. J. Biol. Chem. 2005, 280, 25565–25570. [Google Scholar] [CrossRef]
- Lopez, J.A.; Kwan, E.P.; Xie, L.; He, Y.; James, D.E.; Gaisano, H.Y. The RalA GTPase is a central regulator of insulin exocytosis from pancreatic islet beta cells. J. Biol. Chem. 2008, 283, 17939–17945. [Google Scholar] [CrossRef]
- Ljubicic, S.; Bezzi, P.; Vitale, N.; Regazzi, R. The GTPase RalA regulates different steps of the secretory process in pancreatic β-cells. PLoS ONE 2009, 4, e7770. [Google Scholar] [CrossRef][Green Version]
- Xie, L.; Zhu, D.; Kang, Y.; Liang, T.; He, Y.; Gaisano, H.Y. Exocyst sec5 regulates exocytosis of newcomer insulin granules underlying biphasic insulin secretion. PLoS ONE 2013, 8, e67561. [Google Scholar] [CrossRef]
- Baumann, K. Keeping insulin secretion in check. Nat. Rev. Mol. Cell Biol. 2016, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, B.A.; Young, M.; Carter, L.; Pang, A.P.; Corley, M.J.; Fogelgren, B.; Polgar, N. The exocyst complex regulates insulin-stimulated glucose uptake of skeletal muscle cells. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E957–E972. [Google Scholar] [CrossRef] [PubMed]
- Gyanwali, G.C. The Role of Host Exocytosis in Internalin A (InlA)-Mediated Entry of Listeria Monocytogenes into Human Cells. Ph.D. Thesis, University of Otago, Dunedin, New Zealand, 2021. [Google Scholar]
- Escrevente, C.; Bento-Lopes, L.; Ramalho, J.S.; Barral, D.C. A novel Rab11-Rab3a cascade required for lysosome exocytosis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Tanaka, T.; Goto, K.; Iino, M. Diverse functions and signal transduction of the exocyst complex in tumor cells. J. Cell. Physiol. 2017, 232, 939–957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-M.; Ellis, S.; Sriratana, A.; Mitchell, C.A.; Rowe, T. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J. Biol. Chem. 2004, 279, 43027–43034. [Google Scholar] [CrossRef]
- Hachim, M.Y.; Hachim, I.Y.; Al Heialy, S.; Taneera, J. Cellular exocytosis gene (EXOC6/6B): A potential molecular link for the susceptibility and mortality of COVID-19 in diabetic patients. bioRxiv 2020. [Google Scholar] [CrossRef]
- Fadista, J.; Vikman, P.; Laakso, E.O.; Mollet, I.G.; Esguerra, J.L.; Taneera, J.; Storm, P.; Osmark, P.; Ladenvall, C.; Prasad, R.B. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 13924–13929. [Google Scholar] [CrossRef]
- Taneera, J.; Fadista, J.; Ahlqvist, E.; Atac, D.; Ottosson-Laakso, E.; Wollheim, C.B.; Groop, L. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum. Mol. Genet. 2015, 24, 1945–1955. [Google Scholar] [CrossRef]
- Alonso, L.; Piron, A.; Moran, I.; Guindo-Martinez, M.; Bonas-Guarch, S.; Atla, G.; Miguel-Escalada, I.; Royo, R.; Puiggros, M.; Garcia-Hurtado, X. TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep. 2021, 37, 109807. [Google Scholar] [CrossRef]
- Taneera, J.; Dhaiban, S.; Hachim, M.; Mohammed, A.K.; Mukhopadhyay, D.; Bajbouj, K.; Hamoudi, R.; Salehi, A.; Hamad, M. Reduced expression of Chl1 gene impairs insulin secretion by down-regulating the expression of key molecules of β-cell function. Exp. Clin. Endocrinol. Diabetes 2021, 129, 864–872. [Google Scholar] [CrossRef]
- Hectors, T.L.; Vanparys, C.; Pereira-Fernandes, A.; Martens, G.A.; Blust, R. Evaluation of the INS-1 832/13 cell line as a beta-cell based screening system to assess pollutant effects on beta-cell function. PLoS ONE 2013, 8, e60030. [Google Scholar] [CrossRef] [PubMed]
- Taneera, J.; Mohammed, I.; Mohammed, A.K.; Hachim, M.; Dhaiban, S.; Malek, A.; Dunér, P.; Elemam, N.M.; Sulaiman, N.; Hamad, M. Orphan G-protein coupled receptor 183 (GPR183) potentiates insulin secretion and prevents glucotoxicity-induced β-cell dysfunction. Mol. Cell. Endocrinol. 2020, 499, 110592. [Google Scholar] [CrossRef] [PubMed]
- El-Huneidi, W.; Anjum, S.; Mohammed, A.K.; Unnikannan, H.; Saeed, R.; Bajbouj, K.; Abu-Gharbieh, E.; Taneera, J. Copine 3 “CPNE3” is a novel regulator for insulin secretion and glucose uptake in pancreatic β-cells. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Lobo, G.; Fulmer, D.; Guo, L.; Dang, Y.; Su, Y.; Ilatovskaya, D.V.; Nihalani, D.; Rohrer, B.; Body, S.C. The exocyst acting through the primary cilium is necessary for renal ciliogenesis, cystogenesis, and tubulogenesis. J. Biol. Chem. 2019, 294, 6710–6718. [Google Scholar] [CrossRef]
- McClure-Begley, T.D.; Klymkowsky, M.W. Nuclear roles for cilia-associated proteins. Cilia 2017, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, L.E.; Tang, K.; Kats, E.S.; Schutz-Geschwender, A.; Lipschutz, J.H.; Mostov, K.E. ERK and MMPs sequentially regulate distinct stages of epithelial tubule development. Dev. Cell 2004, 7, 21–32. [Google Scholar] [CrossRef]
- Kafina, M.D.; Paw, B.H. Intracellular iron and heme trafficking and metabolism in developing erythroblasts. Metallomics 2017, 9, 1193–1203. [Google Scholar] [CrossRef]
- Updegraff, B.L.; Zhou, X.; Guo, Y.; Padanad, M.S.; Chen, P.-H.; Yang, C.; Sudderth, J.; Rodriguez-Tirado, C.; Girard, L.; Minna, J.D. Transmembrane protease TMPRSS11B promotes lung cancer growth by enhancing lactate export and glycolytic metabolism. Cell Rep. 2018, 25, 2223–2233.e6. [Google Scholar] [CrossRef]
- Martin-Urdiroz, M.; Deeks, M.J.; Horton, C.G.; Dawe, H.R.; Jourdain, I. The exocyst complex in health and disease. Front. Cell Dev. Biol. 2016, 4, 24. [Google Scholar] [CrossRef]
- Dietl, P.; Haller, T.; Mair, N.; Frick, M. Mechanisms of surfactant exocytosis in alveolar type II cells in vitro and in vivo. Physiology 2001, 16, 239–243. [Google Scholar] [CrossRef]
- Sladek, R.; Rocheleau, G.; Rung, J.; Dina, C.; Shen, L.; Serre, D.; Boutin, P.; Vincent, D.; Belisle, A.; Hadjadj, S. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007, 445, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Lu, F.; Dong, M.; Lin, Y.; Li, H.; Chen, J.; Shen, C.; Jin, G.; Hu, Z.; Shen, H. Genetic variants of IDE-KIF11-HHEX at 10q23. 33 associated with type 2 diabetes risk: A fine-mapping study in Chinese population. PLoS ONE 2012, 7, e35060. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Qian, Y.; Lu, F.; Dong, M.; Lin, Y.; Li, H.; Shen, C.; Dai, J.; Jiang, Y.; Jin, G. Genetic variants at 10q23. 33 are associated with plasma lipid levels in a Chinese population. J. Biomed. Res. 2014, 28, 53. [Google Scholar]
- Liu, R.-K.; Lin, X.; Wang, Z.; Greenbaum, J.; Qiu, C.; Zeng, C.-P.; Zhu, Y.-Y.; Shen, J.; Deng, H.-W. Identification of novel functional CpG-SNPs associated with Type 2 diabetes and birth weight. Aging 2021, 13, 10619. [Google Scholar] [CrossRef]
- Sugawara, T.; Kano, F.; Murata, M. Rab2A is a pivotal switch protein that promotes either secretion or ER-associated degradation of (pro) insulin in insulin-secreting cells. Sci. Rep. 2014, 4, 1–14. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Z.; Yang, Y.; Li, Q.; Zeng, R.; Kang, J.; Wu, J. Rab1A mediates proinsulin to insulin conversion in β-cells by maintaining Golgi stability through interactions with golgin-84. Protein Cell 2016, 7, 692–696. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thorens, B.; Guillam, M.-T.; Beermann, F.; Burcelin, R.; Jaquet, M. Transgenic reexpression of GLUT1 or GLUT2 in pancreatic β cells rescues GLUT2-null mice from early death and restores normal glucose-stimulated insulin secretion. J. Biol. Chem. 2000, 275, 23751–23758. [Google Scholar] [CrossRef]
- Taneera, J.; Mussa, B.; Saber-Ayad, M.; Dhaiban, S.; Aljaibeji, H.; Sulaiman, N. Maturity-onset diabetes of the young: An overview with focus on the middle east. Curr. Mol. Med. 2017, 17, 549–562. [Google Scholar] [CrossRef]
- Wang, J.; Gu, W.; Chen, C. Knocking down insulin receptor in pancreatic beta cell lines with lentiviral-small hairpin RNA reduces glucose-stimulated insulin secretion via decreasing the gene expression of insulin, GLUT2 and Pdx1. Int. J. Mol. Sci. 2018, 19, 985. [Google Scholar] [CrossRef]
- Suriben, R.; Kaihara, K.A.; Paolino, M.; Reichelt, M.; Kummerfeld, S.K.; Modrusan, Z.; Dugger, D.L.; Newton, K.; Sagolla, M.; Webster, J.D. β-cell insulin secretion requires the ubiquitin ligase COP1. Cell 2015, 163, 1457–1467. [Google Scholar] [CrossRef]



| S. No. | Genes/Symbol | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|---|
| 1. | Exoc1 | TGTCAAGATGAGCCACCACG | GGCGATGCTCTCAGGTTCAC |
| 2. | Exoc2 | ACTCCCTGCAGTCGTTGAAG | CCTGGGTTTTAGGCTGCTGA |
| 3. | Exoc3 | CAGCTGCGCGGATGTGTA | GTTGCAACAGCCTCCAGGTC |
| 4. | Exoc4 | TGCAAACCTGGAGCCAGAAAT | CGAAGGAGACACTGTTTGGC |
| 5. | Exco5 | ACCGAAGGTTCCAAGATGCT | ACATCTCCAACACTGGCAGG |
| 6. | Exoc7 | CCATTGGGGCCAAAGCTCTA | AACGGTGCCATCTTTAGGCA |
| 7. | Exoc8 | TCGAAGGGCAGTGTCTCAAC | CAATTCGAAGCTGGCGGATG |
| 8. | Hprt | TTGTGTCATCAGCGAAAGTGG | CACAGGACTAGAACGTCTGCT |
| 9. | Vamp2 | TGGTGGACATCATGAGGGTG | GCTTGGCTGCACTTGTTTCA |
| DIAGRAM DIAMANTE | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Reference Allele | Alternate Allele | Type | Sample Size | Effect Allele | MAF | OR | SE | p-Value |
| rs947591 | C | A | Upstream | 231420 | A | 0.48 | 1.07 | 0.0064 | 2.4 × 10−25 |
| rs2488071 | A | G | Upstream | 231420 | A | 0.46 | 1.048 | 0.0064 | 9.1 × 10−21 |
| rs2488073 | A | G | Upstream | 231420 | A | 0.48 | 1.047 | 0.0064 | 2.5 × 10−20 |
| DIAGRAM 1000G | |||||||||
| rs947591 | C | A | Upstream | A | - | 1.09 | 0.013 | 7.4 × 10−13 | |
| rs2488073 | A | G | Upstream | A | - | 0.917 | 0.013 | 8.5 × 10−12 | |
| rs2488071 | A | G | Upstream | A | - | 0.919 | 0.013 | 3.1 × 10−11 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulaiman, N.; Yaseen Hachim, M.; Khalique, A.; Mohammed, A.K.; Al Heialy, S.; Taneera, J. EXOC6 (Exocyst Complex Component 6) Is Associated with the Risk of Type 2 Diabetes and Pancreatic β-Cell Dysfunction. Biology 2022, 11, 388. https://doi.org/10.3390/biology11030388
Sulaiman N, Yaseen Hachim M, Khalique A, Mohammed AK, Al Heialy S, Taneera J. EXOC6 (Exocyst Complex Component 6) Is Associated with the Risk of Type 2 Diabetes and Pancreatic β-Cell Dysfunction. Biology. 2022; 11(3):388. https://doi.org/10.3390/biology11030388
Chicago/Turabian StyleSulaiman, Nabil, Mahmood Yaseen Hachim, Anila Khalique, Abdul Khader Mohammed, Saba Al Heialy, and Jalal Taneera. 2022. "EXOC6 (Exocyst Complex Component 6) Is Associated with the Risk of Type 2 Diabetes and Pancreatic β-Cell Dysfunction" Biology 11, no. 3: 388. https://doi.org/10.3390/biology11030388
APA StyleSulaiman, N., Yaseen Hachim, M., Khalique, A., Mohammed, A. K., Al Heialy, S., & Taneera, J. (2022). EXOC6 (Exocyst Complex Component 6) Is Associated with the Risk of Type 2 Diabetes and Pancreatic β-Cell Dysfunction. Biology, 11(3), 388. https://doi.org/10.3390/biology11030388






