Genome-Wide Analysis of Invertase Gene Family, and Expression Profiling under Abiotic Stress Conditions in Potato
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Database Search and Sequence Retrieval
2.2. Phylogenetic Analysis and Gene Structure Illustration
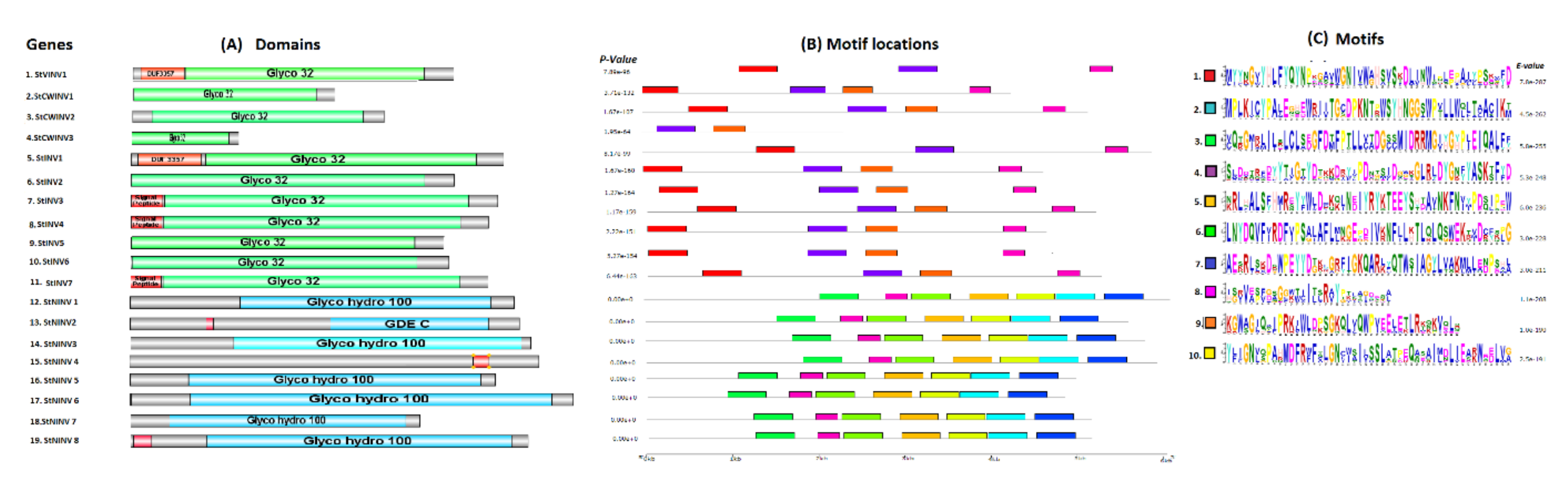
2.3. Functional Motifs and Domain Analysis
2.4. Gene Location, Structural Prediction and 3D Modeling
2.5. Gene Expression Analysis in Different Tissues and Gene Ontology Annotation
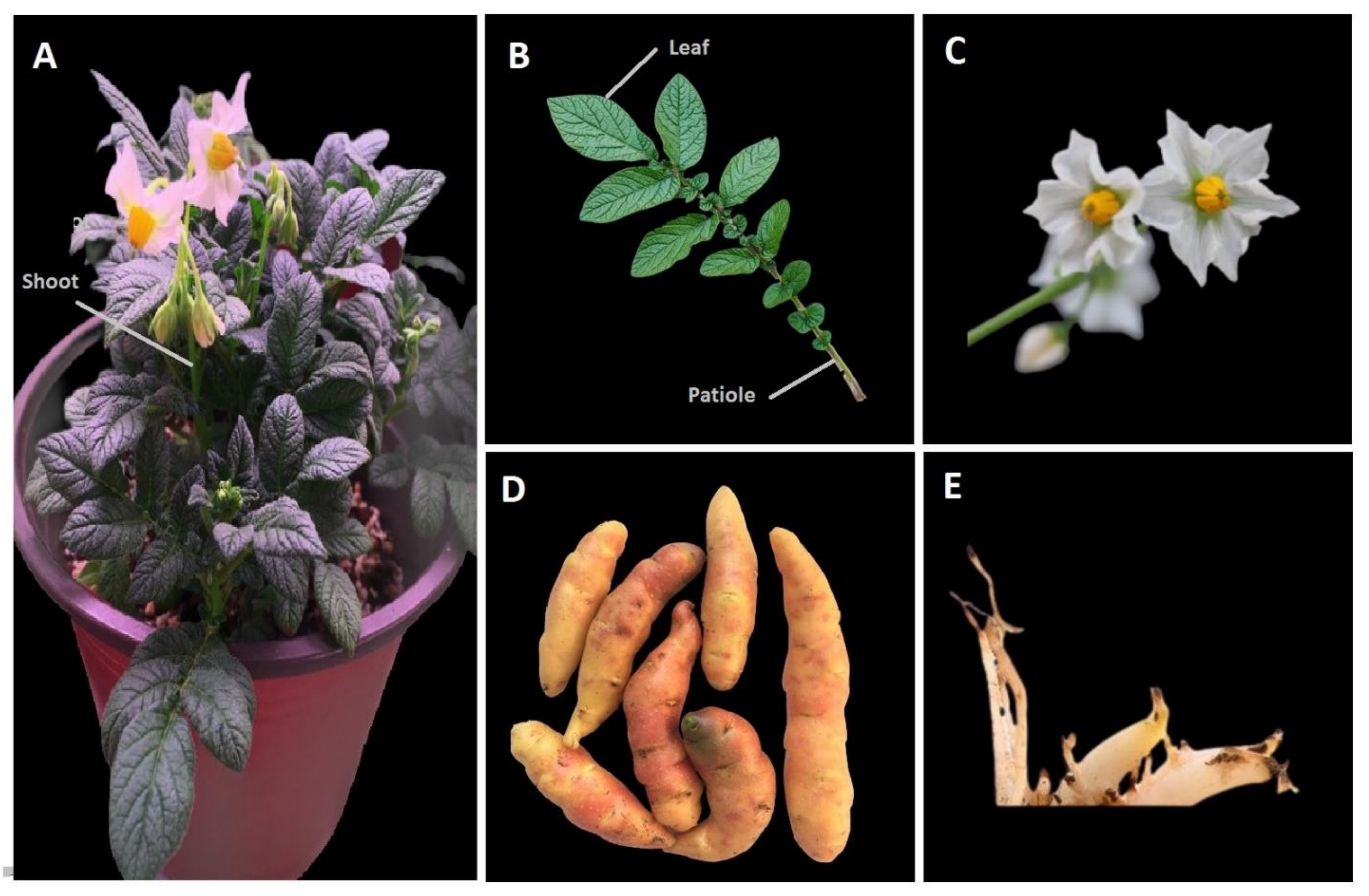
2.6. Plant Materials Propagation Conditions and Treatments
- Growth conditions and Plant material
- B.
- qRT-PCR analysis
3. Results and Discussion
3.1. Genome Wide Analysis of Invertase Genes
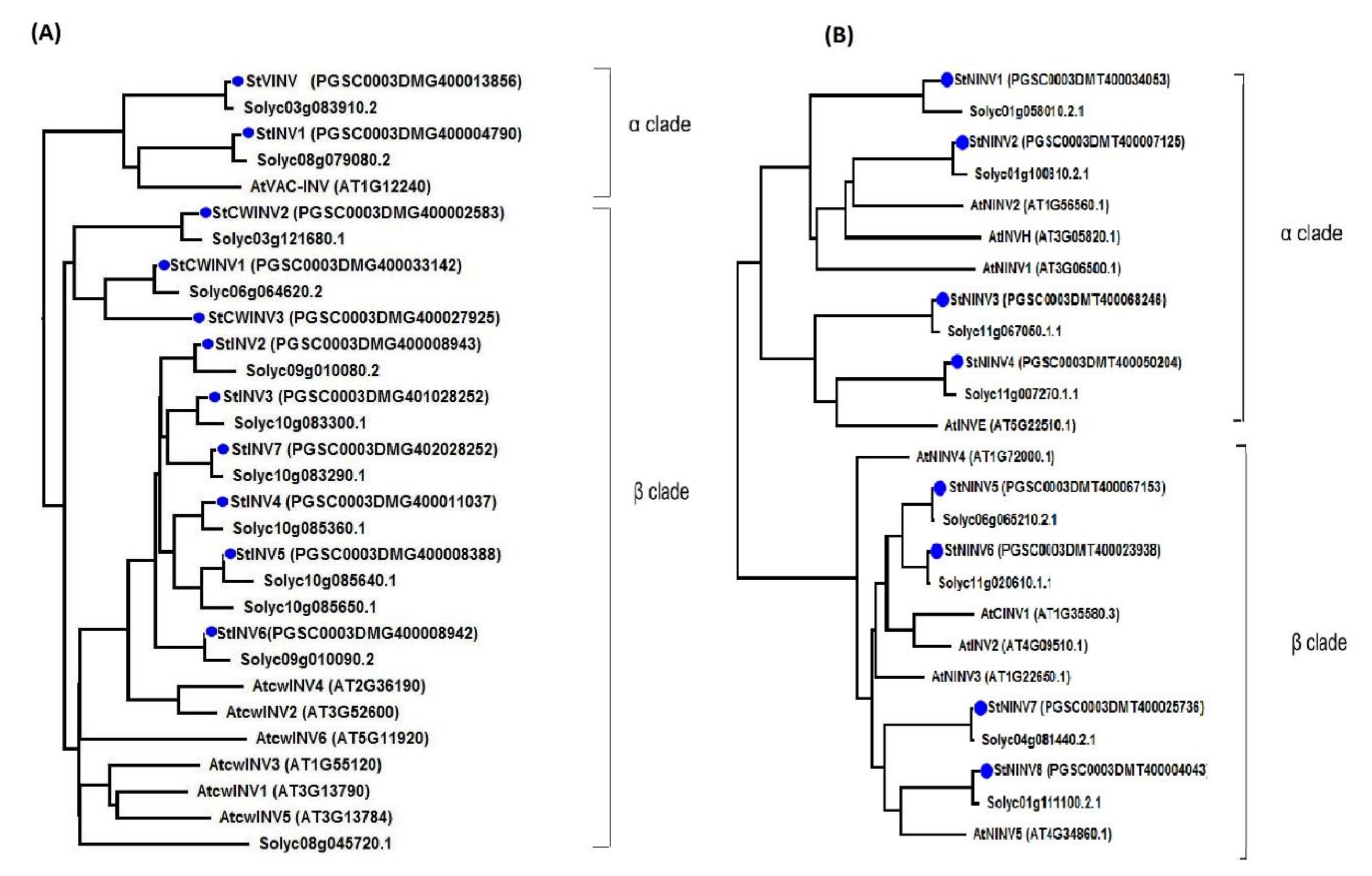
3.2. Phylogenetic Analysis and Gene Structure Illustration
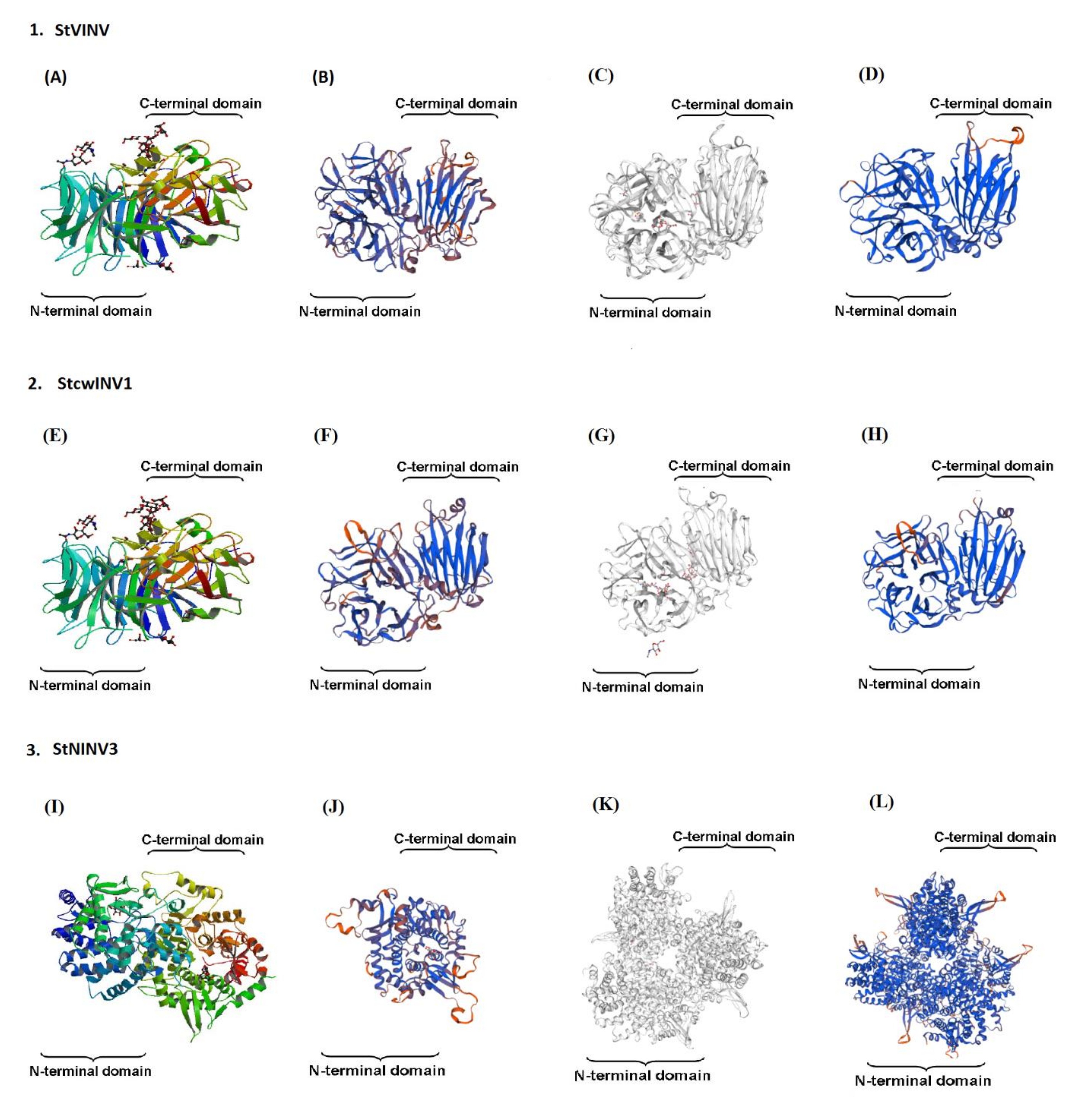
3.3. Gene Structure Prediction and 3-Dimensional Modeling
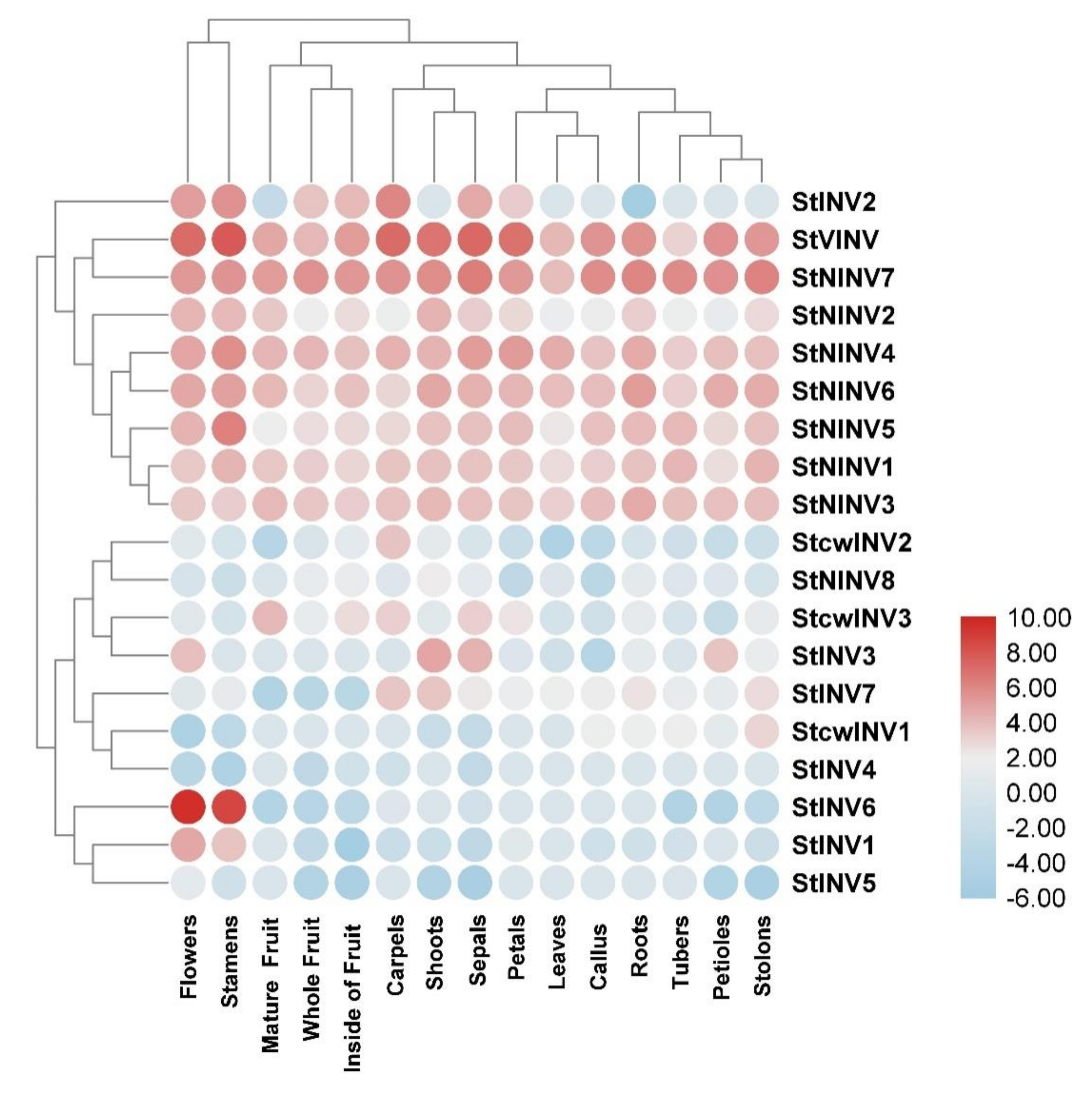
3.4. Gene Expression Pattern in Selected Tissues
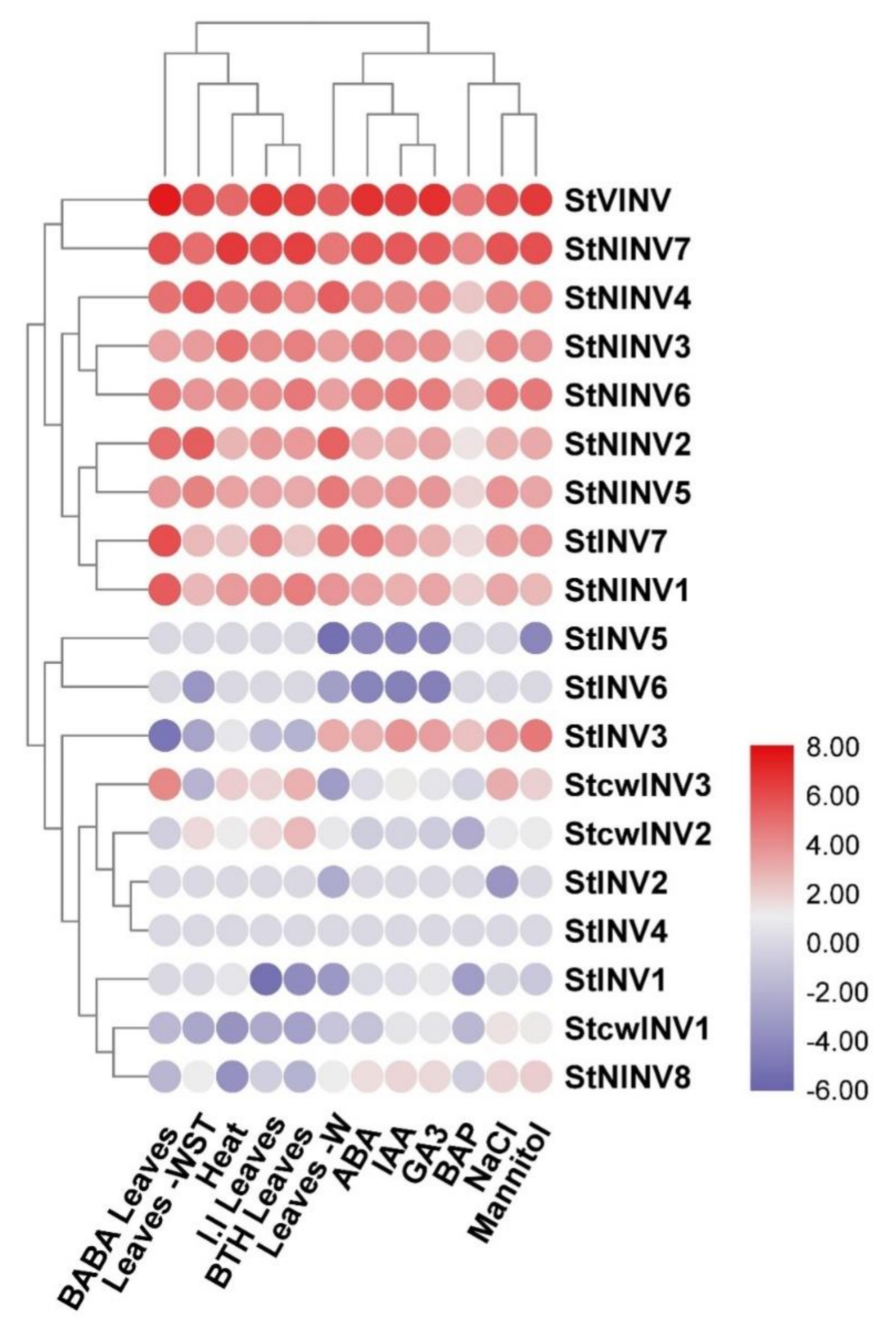
3.5. Gene Expression Analysis under Different Biotic and Abiotic Stresses
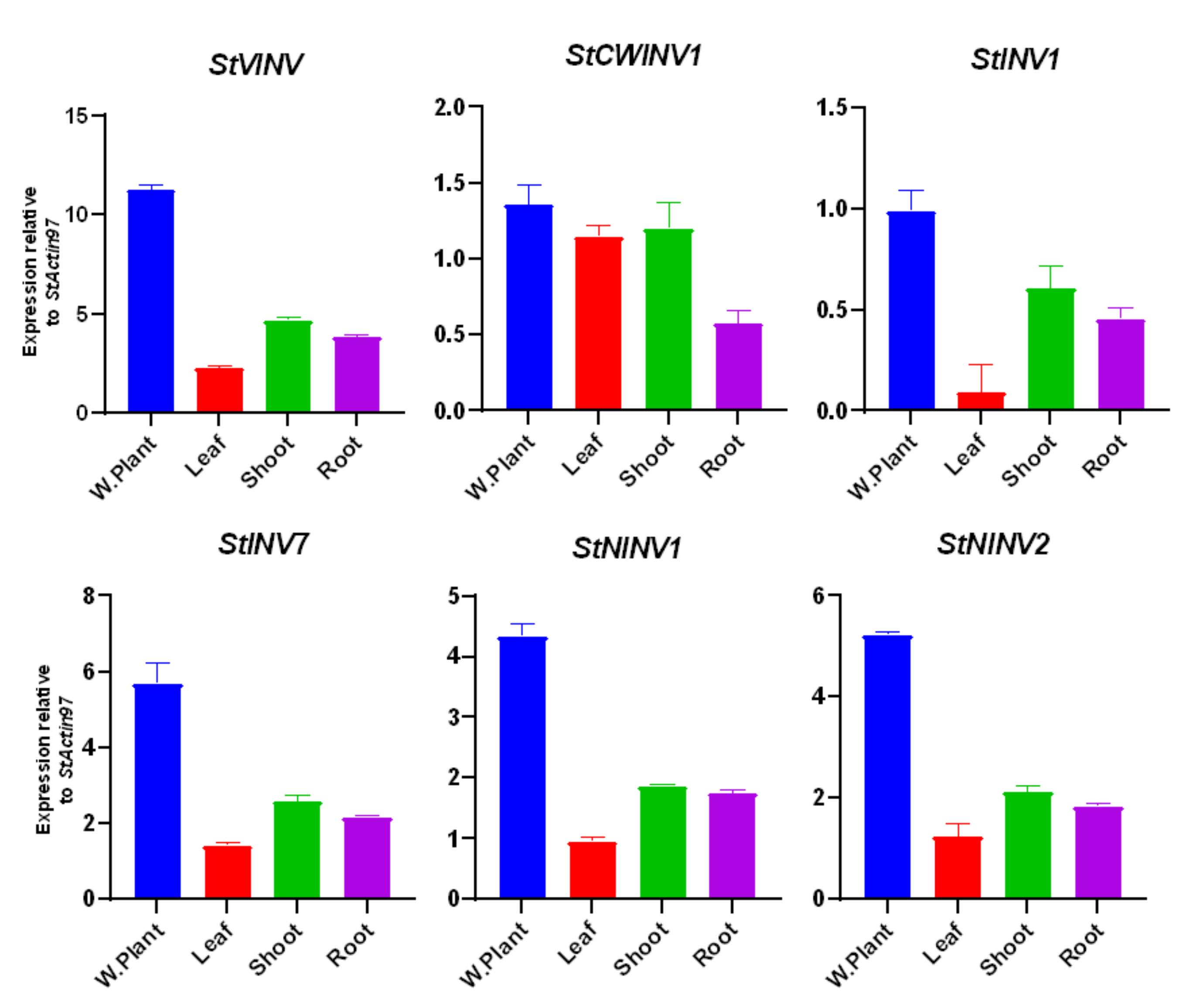
3.6. Validation of Invertase Gene Expression in Different Tissues
3.7. Validation of Invertase Gene Expression under Abiotic Stress Conditions
3.8. GO Annotation of Solanum Tuberosum Invertase Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roitsch, T.; Gonzalez, M.C. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Lammens, W.; Le Roy, K.; Van Laere, A.; Rabijns, A.; Van den Ende, W. Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose. J. Mol. Biol. 2008, 377, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.B.; Qin, Y.L.; Qi, Z.Q.; Niu, Y.; Liu, Z.J.; Liu, W.X.; He, H.; Cao, Z.M.; Yang, Y. Genome-Wide Analysis, Expression Profile, and Characterization of the Acid Invertase Gene Family in Pepper. Int. J. Mol. Sci. 2018, 20, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, Y.-L.; Jin, Y.; Yang, Y.-J.; Li, G.-J.; Boyer, J.S. Sugar Input, Metabolism, and Signaling Mediated by Invertase: Roles in Development, Yield Potential, and Response to Drought and Heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Draffehn, A.M.; Meller, S.; Li, L.; Gebhardt, C. Natural diversity of potato (Solanum tuberosum) invertases. BMC Plant Biol. 2010, 10, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Richael, C.; Chamberlain, P.; Busse, J.S.; Bussan, A.J.; Jiang, J.; Bethke, P.C. Vacuolar invertase gene silencing in potato (Solanum tuberosum L.) improves processing quality by decreasing the frequency of sugar-end defects. PLoS ONE 2014, 9, e93381. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, K.; Su, X.; Rao, P.; An, X. Genome-Wide Identification of the Invertase Gene Family in Populus. PLoS ONE 2015, 10, e0138540. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Geng, M.-T.; Wu, X.-H.; Liu, J.; Li, R.-M.; Hu, X.-W.; Guo, J.-C. Genome-wide identification, 3D modeling, expression and enzymatic activity analysis of cell wall invertase gene family from cassava (Manihot esculenta Crantz). Int. J. Mol. Sci. 2014, 15, 7313–7331. [Google Scholar] [CrossRef] [Green Version]
- Sherson, S.M.; Alford, H.L.; Forbes, S.M.; Wallace, G.; Smith, S.M. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 2003, 54, 525–531. [Google Scholar] [CrossRef] [Green Version]
- Bhaskar, P.B.; Wu, L.; Busse, J.S.; Whitty, B.R.; Hamernik, A.J.; Jansky, S.H.; Buell, C.R.; Bethke, P.C.; Jiang, J. Suppression of the vacuolar invertase gene prevents cold-induced sweetening in potato. Plant Physiol. 2010, 154, 939–948. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Gong, H.; He, Q.; Zeng, Z.; Busse, J.S.; Jin, W.; Bethke, P.C.; Jiang, J. Silencing of vacuolar invertase and asparagine synthetase genes and its impact on acrylamide formation of fried potato products. Plant Biotechnol. J. 2016, 14, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Brain, P.; Sagan, D. Acquiring Genomes: A Theory of the Origin of Species; Argulis, L.M., Sagan, D., Eds.; Basic Books: New York, NY, USA, 2008; p. 1. [Google Scholar]
- Ji, X.; Van den Ende, W.; Van Laere, A.; Cheng, S.; Bennett, J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef] [PubMed]
- Vargas, W.; Cumino, A.; Salerno, G.L. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta 2003, 216, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Dahal, K.; Li, X.-Q.; Tai, H.; Creelman, A.; Bizimungu, B. Improving Potato Stress Tolerance and Tuber Yield Under a Climate Change Scenario—A Current Overview. Front. Plant Sci. 2019, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Wiberley-Bradford, A.; Busse, J.; Jiang, J.; Bethke, P. Sugar metabolism, chip color, invertase activity, and gene expression during long-term cold storage of potato (Solanum tuberosum) tubers from wild-type and vacuolar invertase silencing lines of Katahdin. BMC Res. Notes 2014, 7, 801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef] [Green Version]
- Kumar, J.; Das, S.; Teoh, S.L. Dietary Acrylamide and the Risks of Developing Cancer: Facts to Ponder. Front. Nutr. 2018, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. 1), S162–S173. [Google Scholar] [CrossRef]
- World Wide Web Consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2018, 47, D520–D528. [Google Scholar] [CrossRef] [Green Version]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [Green Version]
- Filloux, C.; Cédric, M.; Romain, P.; Lionel, F.; Christophe, K.; Dominique, R.; Abderrahman, M.; Daniel, P. An integrative method to normalize RNA-Seq data. BMC Bioinform. 2014, 15, 188. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Shi, A.; Mou, B. Genome-wide identification and expression analysis of the CBF/DREB1 gene family in lettuce. Sci. Rep. 2020, 10, 5733. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Colunga, S.; López-González, C.; Morales-Elías, N.; Massange-Sánchez, J.; Trachsel, S.; Tiessen, A. Genome-wide analysis of the invertase gene family from maize. Plant Mol. Biol. 2018, 97, 385–406. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.Y.; Vorst, O.; Fiers, M.W.; Stiekema, W.J.; Nap, J.P. In plants, highly expressed genes are the least compact. Trends Genet. TIG 2006, 22, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Lammens, W.; Le Roy, K.; Yuan, S.; Vergauwen, R.; Rabijns, A.; Van Laere, A.; Strelkov, S.V.; Van den Ende, W. Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose. Plant J. 2012, 70, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Hu, H.X.; Cai, K.; Xia, L.Y.; Yang, F.; Jiang, Y.L.; Chen, Y.; Zhou, C.Z. Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB. FEBS Lett. 2018, 592, 1589–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Cai, K.; Hu, H.X.; Jiang, Y.L.; Yang, F.; Hu, P.F.; Cao, D.D.; Li, W.F.; Chen, Y.; Zhou, C.Z. Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase. J. Biol. Chem. 2016, 291, 25667–25677. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.-Z.; Pang, F.-H.; Cai, W.-J.; Chen, X.-D.; Zhao, M.-Z.; Yu, H.-M. Genome-wide analysis of the invertase genes in strawberry (Fragaria×ananassa). J. Integr. Agric. 2021, 20, 2652–2665. [Google Scholar] [CrossRef]
- Eom, S.H.; Rim, Y.; Hyun, T.K. Genome-wide identification and evolutionary analysis of neutral/alkaline invertases in Brassica rapa. Biotechnol. Biotechnol. Equip. 2019, 33, 1158–1163. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Xi, H.; Chen, X.; Zhao, Y.; Yao, J.; Si, J.; Zhang, L. Genome-wide identification and expression pattern of alkaline/neutral invertase gene family in Dendrobium catenatum. Biotechnol. Biotechnol. Equip. 2021, 35, 527–537. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, Y.; Ding, S.; Zhang, Q.; Chen, Y.; Zhang, J. Molecular cloning, structure, phylogeny and expression analysis of the invertase gene family in sugarcane. BMC Plant Biol. 2017, 17, 109. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.N.; Tanveer, M.; Abbas, A.; Fahad, S.; Baloch, M.S.; Ahmad, M.I.; Saud, S.; Song, Y. Targeting salt stress coping mechanisms for stress tolerance in Brassica: A research perspective. Plant Physiol. Biochem. 2021, 158, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.M.; Raveendran, M.; Oane, R.; Ismail, A.; Lafitte, R.; Bruskiewich, R.; Cheng, S.H.; Bennett, J. Tissue-Specific Expression and Drought Responsiveness of Cell-Wall Invertase Genes of Rice at Flowering. Plant Mol. Biol. 2005, 59, 945–964. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yang, K.; Li, G.; Li, Y.; Gao, Z. Identification and Expression Analyses of Invertase Genes in Moso Bamboo Reveal Their Potential Drought Stress Functions. Front Genet. 2021, 12, 696300. [Google Scholar] [CrossRef] [PubMed]
- Morales, F.; Ancín, M.; Fakhet, D.; González-Torralba, J.; Gámez, A.L.; Seminario, A.; Soba, D.; Ben Mariem, S.; Garriga, M.; Aranjuelo, I. Photosynthetic Metabolism under Stressful Growth Conditions as a Bases for Crop Breeding and Yield Improvement. Plants 2020, 9, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, F.; Zeng, F.; Shen, Q.; Abbas, A.; Cheng, J.; Jiang, W.; Chen, G.; Shah, A.N.; Holford, P.; Tanveer, M.; et al. Molecular evolution and functional modification of plant miRNAs with CRISPR. Trends Plant Sci. 2022; in press. [Google Scholar] [CrossRef]
- Nishanth, M.J.; Sheshadri, S.A.; Rathore, S.S.; Srinidhi, S.; Simon, B. Expression analysis of Cell wall invertase under abiotic stress conditions influencing specialized metabolism in Catharanthus roseus. Sci. Rep. 2018, 8, 15059. [Google Scholar] [CrossRef] [Green Version]
- Albacete, A.; Cantero-Navarro, E.; Großkinsky, D.K.; Arias, C.L.; Balibrea, M.E.; Bru, R.; Fragner, L.; Ghanem, M.E.; González, M.d.l.C.; Hernández, J.A.; et al. Ectopic overexpression of the cell wall invertase gene CIN1 leads to dehydration avoidance in tomato. J. Exp. Bot. 2014, 66, 863–878. [Google Scholar] [CrossRef]
- Abbas, A.; Shah, A.N.; Tanveer, M.; Ahmed, W.; Shah, A.A.; Fiaz, S.; Waqas, M.M.; Ullah, S. MiRNA fine tuning for crop improvement: Using advance computational models and biotechnological tools. Mol. Biol. Rep. 2022, 1–14, Online ahead of print. [Google Scholar] [CrossRef]
- Shah, A.N.; Tanveer, M.; Abbas, A.; Yildirim, M.; Shah, A.A.; Ahmad, M.I.; Wang, Z.; Sun, W.; Song, Y. Combating Dual Challenges in Maize Under High Planting Density: Stem Lodging and Kernel Abortion. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
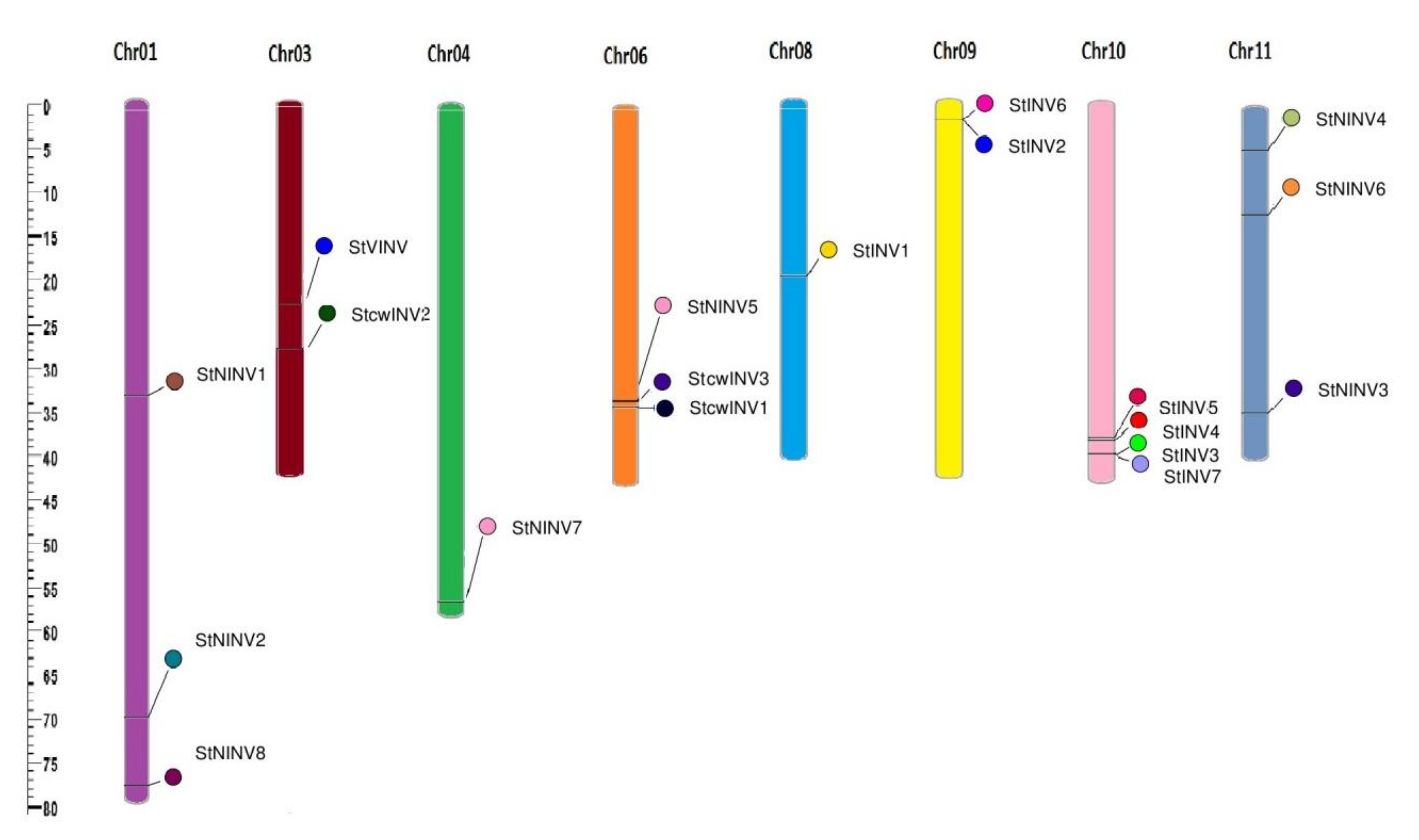



| GENE ID | NAMES | START | END | CHROMOSOME | ORIENTATION |
|---|---|---|---|---|---|
| PGSC0003DMG400013856 | StVINV | 39255144 | 39259094 | 3 | Forward |
| PGSC0003DMG400002583 | StcwINV1 | 61076945 | 61079609 | 6 | Forward |
| PGSC0003DMG400027925 | StcwINV2 | 27921873 | 27923870 | 3 | Forward |
| PGSC0003DMG400033142 | StcwINV3 | 47626455 | 47633517 | 6 | Forward |
| PGSC0003DMG400004790 | StINV1 | 52701644 | 52705608 | 8 | Forward |
| PGSC0003DMG400008943 | StINV2 | 2474975 | 2477132 | 9 | Reverse |
| PGSC0003DMG401028252 | StINV3 | 55840813 | 55843004 | 10 | Forward |
| PGSC0003DMG400011037 | StINV4 | 53706687 | 53710614 | 10 | Reverse |
| PGSC0003DMG400008388 | StINV5 | 53235938 | 53238048 | 10 | Reverse |
| PGSC0003DMG400008942 | StINV6 | 2469738 | 2471543 | 9 | Reverse |
| PGSC0003DMG402028252 | StINV7 | 55851974 | 55856628 | 10 | Forward |
| PGSC0003DMG400013088 | StNINV1 | 37067454 | 37072142 | 1 | Reverse |
| PGSC0003DMG400002756 | StNINV2 | 78430399 | 78435789 | 1 | Reverse |
| PGSC0003DMG400026530 | StNINV3 | 39908750 | 39913516 | 11 | Forward |
| PGSC0003DMG400019494 | StNINV4 | 5048165 | 5052692 | 11 | Forward |
| PGSC0003DMG400026107 | StNINV5 | 46662131 | 46665477 | 6 | Forward |
| PGSC0003DMG400009257 | StNINV6 | 13677099 | 13683141 | 11 | Forward |
| PGSC0003DMG400009936 | StNINV7 | 70804924 | 70808440 | 4 | Reverse |
| PGSC0003DMG400001596 | StNINV8 | 87160865 | 87164427 | 1 | Reverse |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, A.; Shah, A.N.; Shah, A.A.; Nadeem, M.A.; Alsaleh, A.; Javed, T.; Alotaibi, S.S.; Abdelsalam, N.R. Genome-Wide Analysis of Invertase Gene Family, and Expression Profiling under Abiotic Stress Conditions in Potato. Biology 2022, 11, 539. https://doi.org/10.3390/biology11040539
Abbas A, Shah AN, Shah AA, Nadeem MA, Alsaleh A, Javed T, Alotaibi SS, Abdelsalam NR. Genome-Wide Analysis of Invertase Gene Family, and Expression Profiling under Abiotic Stress Conditions in Potato. Biology. 2022; 11(4):539. https://doi.org/10.3390/biology11040539
Chicago/Turabian StyleAbbas, Asad, Adnan Noor Shah, Anis Ali Shah, Muhammad Azhar Nadeem, Ahmad Alsaleh, Talha Javed, Saqer S. Alotaibi, and Nader R. Abdelsalam. 2022. "Genome-Wide Analysis of Invertase Gene Family, and Expression Profiling under Abiotic Stress Conditions in Potato" Biology 11, no. 4: 539. https://doi.org/10.3390/biology11040539
APA StyleAbbas, A., Shah, A. N., Shah, A. A., Nadeem, M. A., Alsaleh, A., Javed, T., Alotaibi, S. S., & Abdelsalam, N. R. (2022). Genome-Wide Analysis of Invertase Gene Family, and Expression Profiling under Abiotic Stress Conditions in Potato. Biology, 11(4), 539. https://doi.org/10.3390/biology11040539










