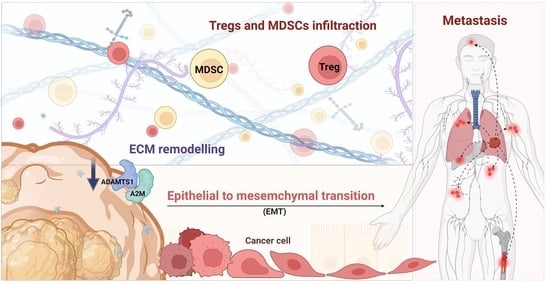
Downregulated ADAMTS1 Incorporating A2M Contributes to Tumorigenesis and Alters Tumor Immune Microenvironment in Lung Adenocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Bioinformatics
2.3. NGS and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Immunoblot
2.5. Characterization of the Tumor Immune Microenvironment
2.6. ADAMTS1 Knockdown
2.7. Wound Healing Analysis
2.8. Animal Model
2.9. Statistical Analysis
3. Results
3.1. Lung Tumors Expressed Lower Levels of ADAMTS1
3.2. The Phenotypic Association and Survival Significance of ADAMTS1
3.3. Protein Interaction Network of ADAMTS1
3.4. The Impact of A2M in Patients with LUAD
3.5. GSEA Analysis of ADAMTS1
3.6. The Role of ADAMTS1 in the Tumor Microenvironment
3.7. Knockdown of ATAMDS1 Promotes Cancer Migration, EMT, and Metastasis In Vitro and In Vivo Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
- The CancerSEA website (http://biocc.hrbmu.edu.cn/CancerSEA/).
- The GSCAL website (http://bioinfo.life.hust.edu.cn/GSCA).
- The KM plotter database (http://kmplot.com/analysis/).
- The Oncomine database: http://www.oncomine.org.
- The Pathway Commons (https://www.pathwaycommons.org/).
- TIMER (http://timer.cistrome.org/).
- The UALCAN website (http://ualcan.path.uab.edu/).
Acknowledgments
Conflicts of Interest
Appendix A
| Factors | Coefficient | Standard Error | p-Value | 95% Confidential Interval |
|---|---|---|---|---|
| A2M | 0.713 | 0.044 | *** | 0.626 to 0.8 |
| ACAN | 0.096 | 0.025 | *** | 0.047 to 0.145 |
| HPX | −0.040 | 0.023 | 0.076 | −0.084 to 0.004 |
| IFNA21 | 0.227 | 0.151 | 0.134 | −0.07 to 0.524 |
| MAZ | −0.139 | 0.056 | * | −0.249 to −0.028 |
| PLA2G10 | −0.111 | 0.022 | *** | −0.154 to −0.068 |
| PYHIN1 | −0.026 | 0.035 | 0.458 | −0.094 to 0.042 |
| SPI1 | −0.082 | 0.046 | 0.076 | −0.172 to 0.008 |
| ZIC2 | −0.051 | 0.017 | ** | −0.084 to −0.018 |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Guo, H.; Li, H.; Zhu, L.; Feng, J.; Huang, X.; Baak, J.P.A. “How Long Have I Got?” in Stage IV NSCLC Patients With at Least 3 Months Up to 10 Years Survival, Accuracy of Long-, Intermediate-, and Short-Term Survival Prediction Is Not Good Enough to Answer This Question. Front. Oncol. 2021, 11, 761042. [Google Scholar] [CrossRef] [PubMed]
- Henke, E.; Nandigama, R.; Ergün, S. Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Front. Mol. Biosci. 2020, 6, 160. [Google Scholar] [CrossRef] [Green Version]
- Dinca, S.C.; Greiner, D.; Weidenfeld, K.; Bond, L.; Barkan, D.; Jorcyk, C.L. Novel mechanism for OSM-promoted extracellular matrix remodeling in breast cancer: LOXL2 upregulation and subsequent ECM alignment. Breast Cancer Res. BCR 2021, 23, 56. [Google Scholar] [CrossRef]
- Toba, H.; Ikemoto, M.J.; Kobara, M.; Nakata, T. Secreted protein acidic and rich in cysteine (SPARC) and a disintegrin and metalloproteinase with thrombospondin type 1 motif (ADAMTS1) increments by the renin-angiotensin system induce renal fibrosis in deoxycorticosterone acetate-salt hypertensive rats. Eur. J. Pharmacol. 2022, 914, 174681. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.R.; Santamaria, S.; Koch, C.D.; Ahnström, J.; Apte, S.S. Identification of novel ADAMTS1, ADAMTS4 and ADAMTS5 cleavage sites in versican using a label-free quantitative proteomics approach. J. Proteom. 2021, 249, 104358. [Google Scholar] [CrossRef] [PubMed]
- de Arao Tan, I.; Ricciardelli, C.; Russell, D.L. The metalloproteinase ADAMTS1: A comprehensive review of its role in tumorigenic and metastatic pathways. Int. J. Cancer 2013, 133, 2263–2276. [Google Scholar] [CrossRef] [PubMed]
- Kuno, K.; Terashima, Y.; Matsushima, K. ADAMTS-1 is an active metalloproteinase associated with the extracellular matrix. J. Biol. Chem. 1999, 274, 18821–18826. [Google Scholar] [CrossRef] [Green Version]
- Tokmak, A.; Ozaksit, G.; Sarikaya, E.; Kuru-Pekcan, M.; Kosem, A. Decreased ADAMTS-1, -9 and -20 levels in women with endometrial polyps: A possible link between extracellular matrix proteases and endometrial pathologies. J. Obstet. Gynaecol. J. Inst. Obstet. Gynaecol. 2019, 39, 845–850. [Google Scholar] [CrossRef]
- Chen, S.Z.; Ning, L.F.; Xu, X.; Jiang, W.Y.; Xing, C.; Jia, W.P.; Chen, X.L.; Tang, Q.Q.; Huang, H.Y. The miR-181d-regulated metalloproteinase Adamts1 enzymatically impairs adipogenesis via ECM remodeling. Cell Death Differ. 2016, 23, 1778–1791. [Google Scholar] [CrossRef] [Green Version]
- Freitas, V.; Bussador Do Amaral, J.; Silva, T.; Santos, E.; Mangone, F.; Pinheiro, J.; Jaeger, R.; Nagai, M.; Machado-Santelli, G. Decreased expression of ADAMTS-1 in human breast tumors stimulates migration and invasion. Mol. Cancer 2013, 12, 2. [Google Scholar] [CrossRef]
- Thorseth, M.-L.; Carretta, M.; Jensen, C.; Mølgaard, K.; Jürgensen, H.J.; Engelholm, L.H.; Behrendt, N.; Willumsen, N.; Madsen, D.H. Uncovering mediators of collagen degradation in the tumor microenvironment. Matrix Biol. Plus 2022, 13, 100101. [Google Scholar] [CrossRef] [PubMed]
- Niland, S.; Riscanevo, A.X.; Eble, J.A. Matrix Metalloproteinases Shape the Tumor Microenvironment in Cancer Progression. Int. J. Mol. Sci. 2021, 23, 146. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet. Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2018, 47, D900–D908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.J.; Hu, F.F.; Xia, M.X.; Han, L.; Zhang, Q.; Guo, A.Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Rodchenkov, I.; Babur, O.; Luna, A.; Aksoy, B.A.; Wong, J.V.; Fong, D.; Franz, M.; Siper, M.C.; Cheung, M.; Wrana, M.; et al. Pathway Commons 2019 Update: Integration, analysis and exploration of pathway data. Nucleic Acids Res. 2019, 48, D489–D497. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Wang, S.; Dong, Q.Z.; Shen, Z.; Wang, W.; Tao, S.; Gu, C.; Liu, J.; Xie, Y.; et al. Prospero-related homeobox 1 drives angiogenesis of hepatocellular carcinoma through selectively activating interleukin-8 expression. Hepatology 2017, 66, 1894–1909. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-P.; Wu, K.-J. Hypoxia-regulated target genes implicated in tumor metastasis. J. Biomed. Sci. 2012, 19, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Baena, F.J.; Redondo-García, S.; Peris-Torres, C.; Martino-Echarri, E.; Fernández-Rodríguez, R.; Plaza-Calonge, M.D.C.; Anderson, P.; Rodríguez-Manzaneque, J.C. ADAMTS1 protease is required for a balanced immune cell repertoire and tumour inflammatory response. Sci. Rep. 2018, 8, 13103. [Google Scholar] [CrossRef] [PubMed]
- Tan, I.A.; Frewin, K.; Ricciardelli, C.; Russell, D.L. ADAMTS1 Promotes Adhesion to Extracellular Matrix Proteins and Predicts Prognosis in Early Stage Breast Cancer Patients. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2019, 52, 1553–1568. [Google Scholar]
- Noriega-Guerra, H.; Cruz, M.C.; Ribeiro, P.R.L.; Strnadel, J.; Wang, H.; Klemke, R.L.; Jaeger, R.G.; Freitas, V.M. ADAMTS-1 disrupts HGF/c-MET signaling and HGF-stimulated cellular processes in fibrosarcoma. Exp. Cell Res. 2018, 363, 271–282. [Google Scholar] [CrossRef]
- Wang, B.; Chen, S.; Zhao, J.Q.; Xiang, B.L.; Gu, X.; Zou, F.; Zhang, Z.H. ADAMTS-1 inhibits angiogenesis via the PI3K/Akt-eNOS-VEGF pathway in lung cancer cells. Transl. Cancer Res. 2019, 8, 2725–2735. [Google Scholar] [CrossRef]
- de Assis Lima, M.; da Silva, S.V.; Serrano-Garrido, O.; Hülsemann, M.; Santos-Neres, L.; Rodríguez-Manzaneque, J.C.; Hodgson, L.; Freitas, V.M. Metalloprotease ADAMTS-1 decreases cell migration and invasion modulating the spatiotemporal dynamics of Cdc42 activity. Cell. Signal. 2021, 77, 109827. [Google Scholar] [CrossRef]
- Liu, Y.J.; Xu, Y.; Yu, Q. Full-length ADAMTS-1 and the ADAMTS-1 fragments display pro- and antimetastatic activity, respectively. Oncogene 2006, 25, 2452–2467. [Google Scholar] [CrossRef] [Green Version]
- Hsu, Y.L.; Hung, J.Y.; Chang, W.A.; Jian, S.F.; Lin, Y.S.; Pan, Y.C.; Wu, C.Y.; Kuo, P.L. Hypoxic Lung-Cancer-Derived Extracellular Vesicle MicroRNA-103a Increases the Oncogenic Effects of Macrophages by Targeting PTEN. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 568–581. [Google Scholar] [CrossRef] [Green Version]
- Tsai, Y.M.; Wu, K.L.; Chang, Y.Y.; Chang, W.A.; Huang, Y.C.; Jian, S.F.; Tsai, P.H.; Lin, Y.S.; Chong, I.W.; Hung, J.Y.; et al. Loss of miR-145-5p Causes Ceruloplasmin Interference with PHD-Iron Axis and HIF-2α Stabilization in Lung Adenocarcinoma-Mediated Angiogenesis. Int. J. Mol. Sci. 2020, 21, 5081. [Google Scholar] [CrossRef]
- Arandjelovic, S.; Freed, T.A.; Gonias, S.L. Growth factor-binding sequence in human alpha2-macroglobulin targets the receptor-binding site in transforming growth factor-beta. Biochemistry 2003, 42, 6121–6127. [Google Scholar] [CrossRef]
- Lindner, I.; Hemdan, N.Y.; Buchold, M.; Huse, K.; Bigl, M.; Oerlecke, I.; Ricken, A.; Gaunitz, F.; Sack, U.; Naumann, A.; et al. Alpha2-macroglobulin inhibits the malignant properties of astrocytoma cells by impeding beta-catenin signaling. Cancer Res. 2010, 70, 277–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurz, S.; Thieme, R.; Amberg, R.; Groth, M.; Jahnke, H.G.; Pieroh, P.; Horn, L.C.; Kolb, M.; Huse, K.; Platzer, M.; et al. The anti-tumorigenic activity of A2M-A lesson from the naked mole-rat. PLoS ONE 2017, 12, e0189514. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.B.; Chua, M.L.K.; Yeong, J.P.S.; Tan, S.J.; Lim, W.-T.; Lim, C.T. Pan-cancer analysis connects tumor matrisome to immune response. npj Precis. Oncol. 2019, 3, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Principe, D.R.; Chiec, L.; Mohindra, N.A.; Munshi, H.G. Regulatory T-Cells as an Emerging Barrier to Immune Checkpoint Inhibition in Lung Cancer. Front. Oncol. 2021, 11, 684098. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Espinosa, S.; Morales, X.; Senent, Y.; Alignani, D.; Tavira, B.; Macaya, I.; Ruiz, B.; Moreno, H.; Remírez, A.; Sainz, C.; et al. Complement C5a induces the formation of neutrophil extracellular traps by myeloid-derived suppressor cells to promote metastasis. Cancer Lett. 2022, 529, 70–84. [Google Scholar] [CrossRef]
- Genova, C.; Dellepiane, C.; Carrega, P.; Sommariva, S.; Ferlazzo, G.; Pronzato, P.; Gangemi, R.; Filaci, G.; Coco, S.; Croce, M. Therapeutic Implications of Tumor Microenvironment in Lung Cancer: Focus on Immune Checkpoint Blockade. Front. Immunol. 2021, 12, 799455. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-C.; Chang, C.-Y.; Huang, Y.-C.; Wu, K.-L.; Chiang, H.-H.; Chang, Y.-Y.; Liu, L.-X.; Hung, J.-Y.; Hsu, Y.-L.; Wu, Y.-Y.; et al. Downregulated ADAMTS1 Incorporating A2M Contributes to Tumorigenesis and Alters Tumor Immune Microenvironment in Lung Adenocarcinoma. Biology 2022, 11, 760. https://doi.org/10.3390/biology11050760
Lee H-C, Chang C-Y, Huang Y-C, Wu K-L, Chiang H-H, Chang Y-Y, Liu L-X, Hung J-Y, Hsu Y-L, Wu Y-Y, et al. Downregulated ADAMTS1 Incorporating A2M Contributes to Tumorigenesis and Alters Tumor Immune Microenvironment in Lung Adenocarcinoma. Biology. 2022; 11(5):760. https://doi.org/10.3390/biology11050760
Chicago/Turabian StyleLee, Hsiao-Chen, Chao-Yuan Chang, Yung-Chi Huang, Kuan-Li Wu, Hung-Hsing Chiang, Yung-Yun Chang, Lian-Xiu Liu, Jen-Yu Hung, Ya-Ling Hsu, Yu-Yuan Wu, and et al. 2022. "Downregulated ADAMTS1 Incorporating A2M Contributes to Tumorigenesis and Alters Tumor Immune Microenvironment in Lung Adenocarcinoma" Biology 11, no. 5: 760. https://doi.org/10.3390/biology11050760
APA StyleLee, H.-C., Chang, C.-Y., Huang, Y.-C., Wu, K.-L., Chiang, H.-H., Chang, Y.-Y., Liu, L.-X., Hung, J.-Y., Hsu, Y.-L., Wu, Y.-Y., & Tsai, Y.-M. (2022). Downregulated ADAMTS1 Incorporating A2M Contributes to Tumorigenesis and Alters Tumor Immune Microenvironment in Lung Adenocarcinoma. Biology, 11(5), 760. https://doi.org/10.3390/biology11050760