Metabolomics and Chemoinformatics in Agricultural Biotechnology Research: Complementary Probes in Unravelling New Metabolites for Crop Improvement
Abstract
:Simple Summary
Abstract
1. Introduction
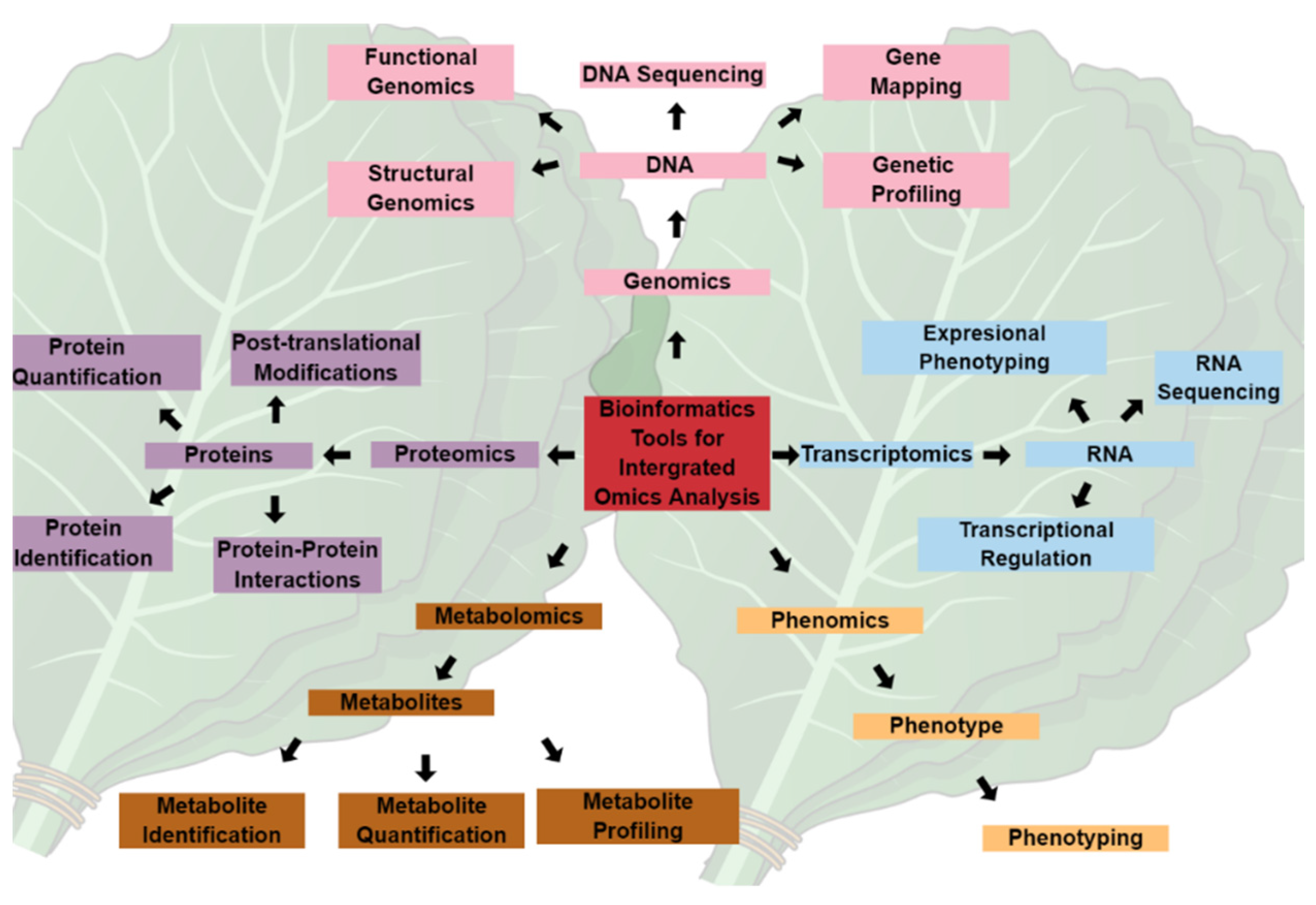
2. Metabolomics and Chemoinformatics Tools as Prospects for Crop Improvement
2.1. Plant Metabolomics
2.2. Analytical Tools and Approaches in Plant Metabolomics Research
2.3. An Overview of the Standardized Workflow for Plant Metabolomics Studies
2.3.1. Sample Preparation: Harvesting and Metabolite Extraction
2.3.2. Chemometrics and Chemoinformatics Tools for Metabolite Annotation and Biomarker Identification
3. Current Applications of Plant Metabolomics in Crop Improvement
3.1. Examples of Metabolomics for the Elucidation of Plant-Growth Promotion
3.2. Elucidation of Plant Response to Biotic and Abiotic Stress
3.2.1. Adaptation to Biotic Stress
3.2.2. Adaptation to (Selected) Abiotic Stress
4. Metabolomics-Assisted Breeding for Crop Improvement
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- United Nations (UN). Growing at a Slower Pace, World Population Is Expected to Reach 9.7 Billion in 2050 and Could Peak at Nearly 11 Billion around 2100. 2019. Available online: https://www.un.org/development/desa/en/news/population/world-population-prospects-2019.html#:~:text=The%20world’s%20population%20is%20expected,United%20Nations%20report%20launched%20today (accessed on 23 December 2021).
- da Cunha Dias, T.A.; Lora, E.E.; Maya, D.M.Y.; del Olmo, O.A. Global potential assessment of available land for bioenergy projects in 2050 within food security limits. Land Use Policy 2021, 105, 105346. [Google Scholar] [CrossRef]
- de Raymond, A.B.; Alpha, A.; Ari, T.B.; Daviron, B.; Nesme, T.; Tétart, G. Systemic risk and food security. Emerging trends and future avenues for research. Glob. Food Secur. 2021, 29, 100547. [Google Scholar] [CrossRef]
- Davis, K.F.; Downs, S.; Gephart, J.A. Towards food supply chain resilience to environmental shocks. Nat. Food 2021, 2, 54–65. [Google Scholar] [CrossRef]
- Etesami, H. Can interaction between silicon and plant growth-promoting rhizobacteria benefit in alleviating abiotic and biotic stresses in crop plants? Agric. Ecosyst. Environ. 2018, 253, 98–112. [Google Scholar] [CrossRef]
- Smyth, S.J.; Webb, S.R.; Phillips, P.W.B. The role of public-private partnerships in improving global food security. Glob. Food Secur. 2021, 31, 100588. [Google Scholar] [CrossRef]
- Singh, B.; Norvell, E.; Wijewardana, C.; Wallace, T.; Chastain, D.; Reddy, K.R. Assessing morphological characteristics of elite cotton lines from different breeding programmes for low temperature and drought tolerance. J. Agron. Crop Sci. 2018, 204, 467–476. [Google Scholar] [CrossRef]
- Martignago, D.; Rico-Medina, A.; Blasco-Escámez, D.; Fontanet-Manzaneque, J.B.; Caño-Delgado, A.I. Drought resistance by engineering plant tissue-specific responses. Front. Plant Sci. 2020, 10, 1676. [Google Scholar] [CrossRef]
- Scossa, F.; Alseekh, S.; Fernie, A.R. Integrating multi-omics data for crop improvement. J. Plant Physiol. 2021, 257, 153352. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet. Res. Int. 2015, 2015, 431487. [Google Scholar] [CrossRef] [Green Version]
- Shamshad, M.; Sharma, A. The usage of genomic selection strategy in plant breeding. In Next Generation Plant Breed; Çiftçi, Y.O., Ed.; IntechOpen: London, UK, 2018; Chapter 5; pp. 93–108. [Google Scholar]
- Lassoued, R.; Phillips, P.W.; Smyth, S.J.; Hesseln, H. Estimating the cost of regulating genome edited crops: Expert judgment and overconfidence. GM Crops Food 2019, 10, 44–62. [Google Scholar] [CrossRef] [Green Version]
- Billet, K.; Houillé, B.; Dugé de Bernonville, T.; Besseau, S.; Oudin, A.; Courdavault, V.; Delanoue, G.; Guérin, L.; Clastre, M.; Giglioli-Guivarc’h, N.; et al. Field-based metabolomics of Vitis vinifera L. stems provides new insights for genotype discrimination and polyphenol metabolism structuring. Front. Plant Sci. 2018, 9, 798. [Google Scholar] [CrossRef]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [Green Version]
- Deborde, C.; Moing, A.; Roch, L.; Jacob, D.; Rolin, D.; Giraudeau, P. Plant metabolism as studied by NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 102, 61–97. [Google Scholar] [CrossRef]
- Gao, Y.; Chen, Y.; Wang, Z.; Xu, J.; Yue, X.; He, J.; Abliz, Z. Development of simultaneous targeted metabolite quantification and untargeted metabolomics strategy using dual-column liquid chromatography coupled with tandem mass spectrometry. Anal. Chim. Acta. 2018, 1037, 369–379. [Google Scholar] [CrossRef]
- Kokova, D.; Mayboroda, O.A. Twenty years on: Metabolomics in helminth research. Trends Parasitol. 2019, 35, 282–288. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal signalling related to defence priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef] [Green Version]
- Burgess, K.; Rankin, N.; Weidt, S. Handbook of Pharmacogenomics and Stratified Medicine. In Metabolomics; Academic Press: Oxford, UK, 2014; Chapter 10; pp. 181–205. [Google Scholar]
- Gowda, G.A.; Alvarado, L.Z.; Raftery, D. Metabolomics (Chp 5). In Nutrition in the Prevention and Treatment of Disease, 4th ed.; Academic Press: London, UK, 2017; pp. 103–122. [Google Scholar]
- Srinivasan, T.S.; Kannan, R.R. Single-cell-type metabolomics for crop improvement. In Single-Cell Omics: Application in Biomedicine and Agriculture, 2nd ed.; Barh, D., Azevedo, V., Eds.; Academic Press: London, UK, 2019; Chapter 16; pp. 315–339. [Google Scholar]
- Beckles, D.M.; Roessner, U. Plant metabolomics: Applications and opportunities for agricultural biotechnology. In Plant Biotechnology and Agriculture: Prospects for the 21st Century; Altman, A., Hasegawa, M., Eds.; Academic Press: London, UK, 2012; pp. 67–81. [Google Scholar]
- Razzaq, A.; Sadia, B.; Raza, A.; Khalid Hameed, M.; Saleem, F. Metabolomics: A way forward for crop improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef] [Green Version]
- Alawiye, T.T.; Babalola, O.O. Metabolomics: Current application and prospects in crop production. Biologia 2021, 7, 227–239. [Google Scholar] [CrossRef]
- Albrecht, U.; Fiehn, O.; Bowman, K.D. Metabolic variations in different citrus rootstock cultivars associated with different responses to Huanglongbing. Plant Physiol. Biochem. 2016, 107, 33–44. [Google Scholar] [CrossRef]
- Sun, W.; Chen, Z.; Hong, J.; Shi, J. Promoting human nutrition and health through plant metabolomics: Current status and challenges. Biology 2020, 10, 20. [Google Scholar] [CrossRef]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 4. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Babalola, O.O.; Du Toit, L. Metabolomic applications for understanding complex tripartite plant-microbe interactions: Strategies and perspectives. Biotechnol. Rep. 2020, 25, e00425. [Google Scholar] [CrossRef]
- Kang, Z.; Babar, M.A.; Khan, N.; Guo, J.; Khan, J.; Islam, S.; Shrestha, S.; Shahir, D. Comparative metabolomic profiling in the roots and leaves in contrasting genotypes reveals complex mechanisms involved in post-anthesis drought tolerance in wheat. PLoS ONE 2019, 14, e0213502. [Google Scholar] [CrossRef]
- Nephali, L.; Piater, L.A.; Dubery, I.A.; Patterson, V.; Huyser, J.; Burgess, K.; Tugizimana, F. Biostimulants for plant growth and mitigation of abiotic stresses: A metabolomics perspective. Metabolites 2020, 10, 505. [Google Scholar] [CrossRef]
- Boiteau, R.M.; Hoyt, D.W.; Nicora, C.D.; Kinmonth-Schultz, H.A.; Ward, J.K.; Bingol, K. Structure elucidation of unknown metabolites in metabolomics by combined NMR and MS/MS prediction. Metabolites 2018, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Cuperlovic-Culf, M.; Vaughan, M.M.; Vermillion, K.; Surendra, A.; Teresi, J.; McCormick, S.P. Effects of atmospheric CO2 level on the metabolic response of resistant and susceptible wheat to Fusarium graminearum infection. Mol. Plant-Microbe Interact. 2019, 32, 379–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, A.K.; Carroll, A.J.; Estavillo, G.M.; Rebetzke, G.J.; Pogson, B.J. Wheat drought tolerance in the field is predicted by amino acid responses to glasshouse-imposed drought. J. Exp. Bot. 2019, 70, 4931–4948. [Google Scholar] [CrossRef]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic profiling of the host response of tomato (Solanum lycopersicum) following infection by Ralstonia solanacearum. Int. J. Mol. Sci. 2019, 20, 3945. [Google Scholar] [CrossRef] [Green Version]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolomic evaluation of tissue-specific defense responses in tomato plants modulated by PGPR-priming against Phytophthora capsici infection. Plants 2021, 10, 1530. [Google Scholar] [CrossRef]
- Nephali, L.; Moodley, V.; Piater, L.; Steenkamp, P.; Buthelezi, N.; Dubery, I.; Burgess, K.; Huyser, J.; Tugizimana, F. A metabolomic landscape of maize plants treated with a microbial biostimulant under well-watered and drought conditions. Front. Plant Sci. 2021, 12, 676632. [Google Scholar] [CrossRef] [PubMed]
- Othibeng, K.; Nephali, L.; Ramabulana, A.T.; Steenkamp, P.; Petras, D.; Kang, K.B.; Opperman, H.; Huyser, J.; Tugizimana, F. A metabolic choreography of maize plants treated with a humic substance-based biostimulant under normal and starved conditions. Metabolites 2021, 11, 403. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, H.; Hardy, N.; Beckmann, M.; Draper, J.; Smith, A.R.; Taylor, J.; Fiehn, O.; Goodacre, R.; Bino, R.J.; Hall, R.; et al. A proposed framework for the description of plant metabolomics experiments and their results. Nat. Biotechnol. 2004, 22, 1601–1606. [Google Scholar] [CrossRef]
- Fiehn, O.; Robertson, D.; Griffin, J.; van der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Fernie, A.R.; Aharoni, A.; Willmitzer, L.; Stitt, M.; Tohge, T.; Kopka, J.; Carroll, A.J.; Saito, K.; Fraser, P.D.; DeLuca, V. Recommendations for reporting metabolite data. Plant Cell 2011, 23, 2477–2482. [Google Scholar] [CrossRef] [Green Version]
- Hamany Djande, C.Y.; Pretorius, C.; Tugizimana, F.; Piater, L.A.; Dubery, I.A. Metabolomics: A tool for cultivar phenotyping and investigation of grain crops. Agronomy 2020, 10, 831. [Google Scholar] [CrossRef]
- Martins, M.; Caldana, C.; Wolf, L.D.; Abreu, L.G.F.D. The importance of experimental design, quality assurance, and control in plant metabolomics experiments. In Plant Metabolomics; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2018; pp. 3–17. [Google Scholar]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2010, 21, 4–13. [Google Scholar] [CrossRef]
- Riet, K.B.; Ndlovu, N.; Piater, L.A.; Dubery, I.A. Simultaneous analysis of defense-related phytohormones in Arabidopsis thaliana responding to fungal infection. Appl. Plant Sci. 2016, 4, 1600013. [Google Scholar] [CrossRef]
- Pétriacq, P.; Williams, A.; Cotton, A.; McFarlane, A.E.; Rolfe, S.A.; Ton, J. Metabolite profiling of non-sterile rhizosphere soil. Plant J. 2017, 92, 147–162. [Google Scholar] [CrossRef]
- Altemimi, A.; Lakhssassi, N.; Baharlouei, A.; Watson, D.G.; Lightfoot, D.A. Phytochemicals: Extraction, isolation, and identification of bioactive compounds from plant extracts. Plants 2017, 6, 42. [Google Scholar] [CrossRef]
- Gong, Z.G.; Hu, J.; Wu, X.; Xu, Y.J. The recent developments in sample preparation for mass spectrometry-based metabolomics. Crit. Rev. Anal. Chem. 2017, 4, 325–331. [Google Scholar] [CrossRef]
- Gasteiger, J. Chemoinformatics: Achievements and challenges, a personal view. Molecules 2016, 21, 151. [Google Scholar] [CrossRef] [Green Version]
- Jónsdóttir, S.O.; Jørgensen, F.S.; Brunak, S. Prediction methods and databases within chemoinformatics: Emphasis on drugs and drug candidates. Bioinformatics 2005, 21, 2145–2160. [Google Scholar] [CrossRef]
- Iwaniak, A.; Hrynkiewicz, M.; Bucholska, J.; Minkiewicz, P.; Darewicz, M. Understanding the nature of bitter-taste di- and tripeptides derived from food proteins based on chemometric analysis. J. Food Biochem. 2019, 43, e12500. [Google Scholar] [CrossRef] [Green Version]
- Gowda, G.A.; Raftery, D. Recent advances in NMR-Based metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef]
- Pretorius, C.J.; Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomics for biomarker discovery: Key signatory metabolic profiles for the identification and discrimination of oat cultivars. Metabolites 2021, 11, 165. [Google Scholar] [CrossRef]
- Bartel, J.; Krumsiek, J.; Theis, F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013, 4, e201301009. [Google Scholar] [CrossRef] [Green Version]
- Alonso, A.; Marsal, S.; Julià, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef] [Green Version]
- Steuer, A.E.; Brockbals, L.; Kraemer, T. Metabolomic strategies in biomarker research-new approach for indirect identification of drug consumption and sample manipulation in clinical and forensic toxicology? Front. Chem. 2019, 7, 319. [Google Scholar] [CrossRef]
- Chele, K.H.; Tinte, M.M.; Piater, L.A.; Dubery, I.A.; Tugizimana, F. Soil salinity, a serious environmental issue and plant responses: A metabolomics perspective. Metabolites 2021, 11, 724. [Google Scholar] [CrossRef]
- Benton, H.P.; Wong, D.M.; Trauger, S.A.; Siuzdak, G. XCMS2: Processing tandem mass spectrometry data for metabolite identification and structural characterization. Anal. Chem. 2008, 80, 6382–6389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polettini, A.E.; Kutzler, J.; Sauer, C.; Bleicher, S.; Schultis, W. LC-QTOF-MS presumptive identification of synthetic cannabinoids without reference chromatographic retention/mass spectral information. I. reversed-phase retention time QSPR prediction as an aid to identification of new/unknown compounds. J. Anal. Toxicol. 2021, 45, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Coene, K.; Kluijtmans, L.; van der Heeft, E.; Engelke, U.; de Boer, S.; Hoegen, B.; Kwast, H.; van de Vorst, M.; Huigen, M.; Keularts, I.; et al. Next-generation metabolic screening: Targeted and untargeted metabolomics for the diagnosis of inborn errors of metabolism in individual patients. J. Inherit. Metab. Dis. 2018, 41, 337–353. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Shen, M.; Li, J.; Dong, Y.; Liu, H.; Peng, J.; Hu, Y.; Sun, Y. Profiling of plant growth-promoting metabolites by phosphate-solubilizing bacteria in maize rhizosphere. Plants 2012, 10, 1071. [Google Scholar] [CrossRef]
- Buswell, W.; Schwarzenbacher, R.E.; Luna, E.; Sellwood, M.; Chen, B.; Flors, V.; Pétriacq, P.; Ton, J. Chemical priming of immunity without costs to plant growth. New Phytol. 2018, 218, 1205–1216. [Google Scholar] [CrossRef] [Green Version]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Ann. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef] [Green Version]
- Westman, S.M.; Kloth, K.J.; Hanson, J.; Ohlsson, A.B.; Albrectsen, B.R. Defence priming in Arabidopsis—A Meta-Analysis. Sci. Rep. 2019, 9, 13309. [Google Scholar] [CrossRef]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef] [Green Version]
- Fraiture, M.; Brunner, F. Killing two birds with one stone: Trans-kingdom suppression of PAMP/MAMP-induced immunity by T3E from enteropathogenic bacteria. Front. Microbiol. 2014, 5, 320. [Google Scholar] [CrossRef]
- Malik, N.A.A.; Kumar, I.S.; Nadarajah, K. Elicitor and receptor molecules: Orchestrators of plant defense and immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI continuum in phytophthora-plant interactions. Front. Plant Sci. 2020, 11, 593905. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Concurrent metabolic profiling and quantification of aromatic amino acids and phytohormones in Solanum lycopersicum plants responding to Phytophthora capsici. Metabolites 2020, 10, 466. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Piater, L.A.; Dubery, I.A.; Tugizimana, F.; Mhlongo, M.I. Rhizosphere tripartite interactions and PGPR-mediated metabolic reprogramming towards ISR and plant priming: A metabolomics review. Biology 2022, 11, 346. [Google Scholar] [CrossRef]
- Jones, O.A.; Maguire, M.L.; Griffin, J.L.; Jung, Y.H.; Shibato, J.; Rakwal, R.; Agrawal, G.K.; Jwa, N.S. Using metabolic profiling to assess plant-pathogen interactions: An example using rice (Oryza sativa) and the blast pathogen Magnaporthe grisea. Eur. J. Plant Pathol. 2011, 129, 539–554. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.; Tzin, V.; Romeis, J.; Peng, Y.; Li, Y. Combined transcriptome and metabolome analyses to understand the dynamic responses of rice plants to attack by the rice stem borer Chilo suppressalis (Lepidoptera: Crambidae). BMC Plant Biol. 2016, 16, 259. [Google Scholar] [CrossRef] [Green Version]
- Shavit, R.; Batyrshina, Z.S.; Dotan, N.; Tzin, V. Cereal aphids differently affect benzoxazinoid levels in durum wheat. PLoS ONE 2018, 13, e0208103. [Google Scholar] [CrossRef]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 2016, 17, 767. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Bedia, C.; Cardoso, P.; Dalmau, N.; Garreta-Lara, E.; Gómez-Canela, C.; Gorrochategui, E.; Navarro-Reig, M.; Ortiz-Villanueva, E.; Puig-Castellví, F.; Tauler, R. Applications of metabolomics analysis in environmental research. In Comprehensive Analytical Chemistry; Jaumot, J., Bedia, C., Tauler, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Chapter 19; pp. 533–582. [Google Scholar]
- Hein, J.A.; Sherrard, M.E.; Manfredi, K.P.; Abebe, T. The fifth leaf and spike organs of barley (Hordeum vulgare L.) display different physiological and metabolic responses to drought stress. BMC Plant Biol. 2016, 16, 248. [Google Scholar] [CrossRef] [Green Version]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef]
- Skirycz, A.; Inzé, D. More from less: Plant growth under limited water. Curr. Opin. Biotechnol. 2010, 21, 197–203. [Google Scholar] [CrossRef]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and proteomics reveal drought-stress responses of leaf tissues from spring-wheat. Sci. Rep. 2018, 8, 5710. [Google Scholar] [CrossRef] [Green Version]
- Marček, T.; Hamow, K.Á.; Végh, B.; Janda, T.; Darko, E. Metabolic response to drought in six winter wheat genotypes. PLoS ONE 2019, 14, e0212411. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.H.; Kim, H.J.; Pack, I.S.; Kim, H.J.; Chung, Y.S.; Kim, S.Y.; Kim, C.G. Global metabolite profiling based on GC–MS and LC–MS/MS analyses in ABF3-overexpressing soybean with enhanced drought tolerance. Appl. Biol. Chem. 2019, 62, 15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Chen, J.; Huang, W.; Song, X.; Niu, J. Transcriptomics and metabolomics reveal purine and phenylpropanoid metabolism response to drought stress in Dendrobium sinense, an endemic orchid species in Hainan Island. Front. Genet. 2021, 12, 692702. [Google Scholar] [CrossRef]
- Guo, X.; Xin, Z.; Yang, T.; Ma, X.; Zhang, Y.; Wang, Z.; Ren, Y.; Lin, T. Metabolomics response for drought stress tolerance in Chinese wheat genotypes (Triticum aestivum). Plants 2020, 9, 520. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Cai, S.; Chen, M.; Ye, L.; Chen, Z.; Zhang, H.; Dai, F.; Wu, F.; Zhang, G. Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS ONE 2013, 8, e55431. [Google Scholar] [CrossRef]
- Sarri, E.; Termentzi, A.; Abraham, E.M.; Papadopoulos, G.K.; Baira, E.; Machera, K.; Loukas, V.; Komaitis, F.; Tani, E. Salinity stress alters the secondary metabolic profile of M. sativa, M. arborea and Their Hybrid (Alborea). Int. J. Mol. Sci. 2021, 22, 4882. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Chen, H.; Chen, J.; Yang, R.; Zou, L.; Wang, C.; Chen, J.; Tan, M.; Mei, Y.; Wei, L.; et al. Metabolomics characterizes the metabolic changes of Lonicerae Japonicae Flos under different salt stresses. PLoS ONE 2020, 15, e0243111. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, B.; Liu, D.; Zou, C.; Wu, P.; Wang, Z.; Wang, Y.; Li, C. Transcriptomic and metabolomic analyses reveal mechanisms of adaptation to salinity in which carbon and nitrogen metabolism is altered in sugar beet roots. BMC Plant Biol. 2020, 20, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, M.K.; Kumar, M.; Li, W.; Luo, Y.; Burritt, D.J.; Alkan, N.; Tran, L.S.P. Enhancing salt tolerance of plants: From metabolic reprogramming to exogenous chemical treatments and molecular approaches. Cells 2020, 9, 2492. [Google Scholar] [CrossRef]
- Jamaloddin, M.; Maliha, A.; Gokulan, C.G.; Gaur, N.; Patel, H.K. Metabolomics-assisted breeding for crop improvement: An emerging approach. In Omics Technologies for Sustainable Agriculture and Global food Security; Kumar, A., Kumar, R., Shukla, P., Pandey, M.K., Eds.; Springer: Singapore, 2021; pp. 241–279. [Google Scholar]
- Agarrwal, R.; Nair, S. Metabolomics-assisted crop improvement. In Advancement in Crop Improvement Techniques; Tuteja, N., Tuteja, R., Passricha, N., Saifi, S.K., Eds.; Woodhead: Duxford, UK, 2020; Chapter 16; pp. 263–274. [Google Scholar]
- Moose, S.P.; Dudley, J.W.; Rocheford, T.R. Maize selection passes the century mark: A unique resource for 21st century genomics. Trends Plant Sci. 2004, 9, 358–364. [Google Scholar] [CrossRef]
- Fernie, A.R.; Tadmor, Y.; Zamir, D. Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 2006, 9, 196–202. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Piater, L.A.; Steenkamp, P.A.; Dubery, I.A.; Tugizimana, F.; Mhlongo, M.I. Comparative metabolite profiling of wheat cultivars (Triticum aestivum) reveals signatory markers for resistance and susceptibility to stripe rust and aluminium (Al3+) toxicity. Metabolites 2022, 12, 98. [Google Scholar] [CrossRef]
- Hamany Djande, C.Y.; Pater, L.A.; Steenkamp, P.A.; Tugizimana, F.; Dubery, I.A. A metabolomics approach and chemometric tools for differentiation of barley cultivars and biomarker discovery. Metabolites 2021, 11, 578. [Google Scholar] [CrossRef]
- Toubiana, D.; Fait, A. Metabolomics-assisted crop breeding towards improvement in seed quality and yield. In Seed Development: Omics Technologies toward Improvement of Seed Quality and Crop Yield; Agrawal, G.K., Rakwal, R., Eds.; Springer Science: Heidelberg, Germany; London, UK; New York, NY, USA, 2012; Chapter 22; pp. 453–475. [Google Scholar]
- Miles, C.; Wayne, M. Quantitative trait locus (QTL) analysis. Nat. Educ. 2008, 1, 208. Available online: https://www.nature.com/scitable/topicpage/quantitative-trait-locus-qtl-analysis-53904/ (accessed on 1 May 2022).
- Shi, T.; Zhu, A.; Jia, J.; Hu, X.; Chen, J.; Liu, W.; Ren, X.; Sun, D.; Fernie, A.R.; Cui, F.; et al. Metabolomics analysis and metabolite-agronomic trait associations using kernels of wheat (Triticum aestivum) recombinant inbred lines. Plant J. 2020, 103, 279–292. [Google Scholar] [CrossRef] [Green Version]
- Piasecka, A.; Sawikowska, A.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Krystkowiak, K.; Gudyś, K.; Guzy-Wróbelska, J.; Krajewski, P.; Kachlicki, P. Drought-related secondary metabolites of barley (Hordeum vulgare L.) leaves and their metabolomic quantitative trait loci. Plant J. 2017, 89, 898–913. [Google Scholar] [CrossRef] [Green Version]
- Alseekh, S.; Tong, H.; Scossa, F.; Brotman, Y.; Vigroux, F.; Tohge, T.; Ofner, I.; Zamir, D.; Nikoloski, Z.; Fernie, A.R. Canalization of tomato fruit metabolism. Plant Cell 2017, 29, 2753–2765. [Google Scholar] [CrossRef]
- Templer, S.E.; Ammon, A.; Pscheidt, D.; Ciobotea, O.; Schuy, C.; McCollum, C.; Sonnewald, U.; Hanemann, A.; Förster, J.; Ordon, F.; et al. Metabolite profiling of barley flag leaves under drought and combined heat and drought stress reveals metabolic QTLs for metabolites associated with antioxidant defense. J. Exp. Bot. 2017, 68, 1697–1713. [Google Scholar] [CrossRef] [Green Version]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 1–21. [Google Scholar] [CrossRef]
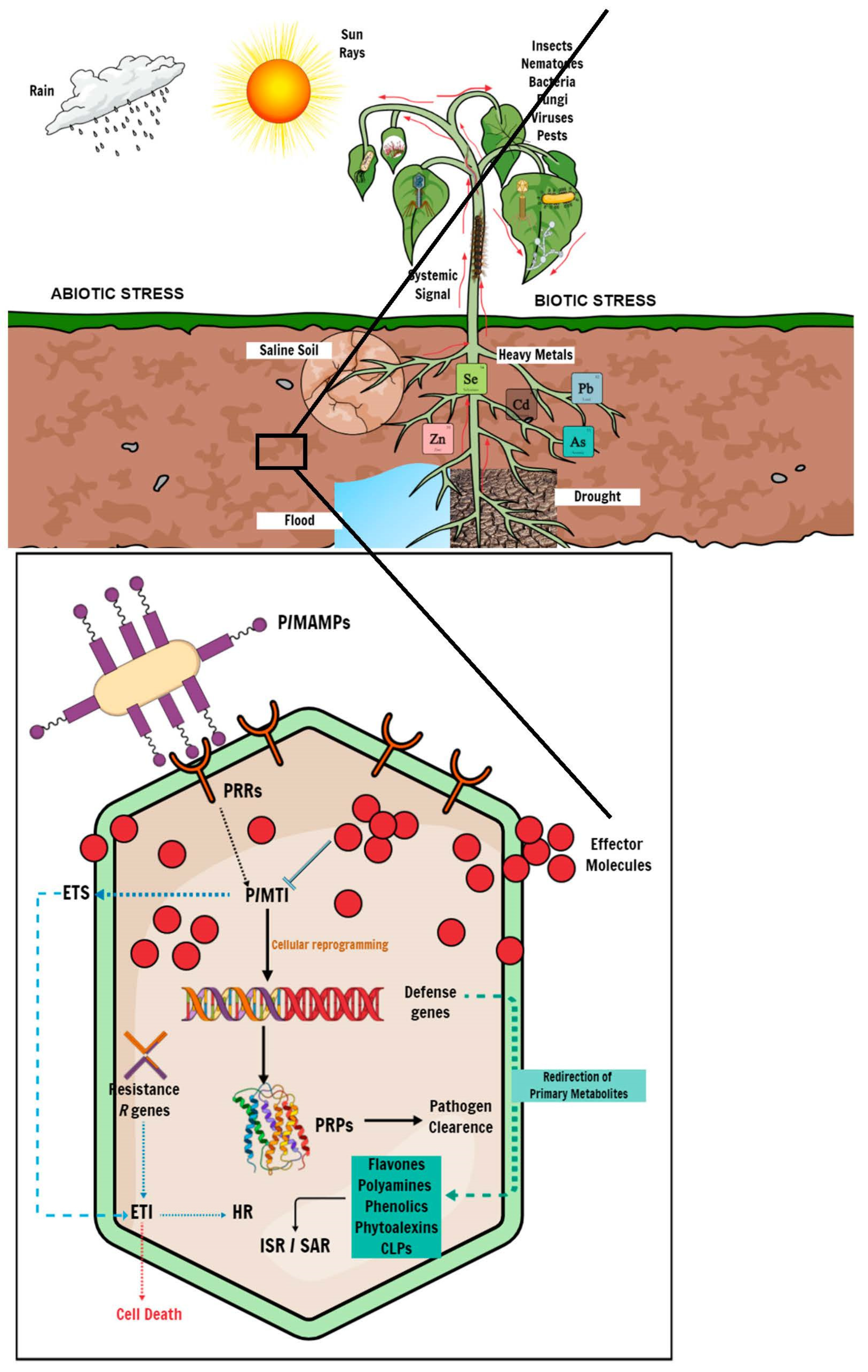
| Biotic Stress | |||
|---|---|---|---|
| Method | Plant | Summary of Study | Ref. |
| UHPLC-ESI-MS; q-TOF-MS | Solanum lycopersicum | Elevated and concentrated levels of potential biomarkers or stress-signaling molecules were seen during R infection, providing insight into the underlying association of metabolites and defense. | [35] |
| UHPLC–MS; UHPLC–QqQ-MS | Solanum lycopersicum | Time-dependent metabolic changes and tissue-specific reprogramming were observed in response to Phytophthora capsica infection. | [36] |
| UHPLC-ESI-qTOF-MS; UHPLC-QqQ-MS | Solanum lycopersicum | Differential reprogramming of amino acids and phytohormones were observed in primary metabolism in response to Phytophthora capsica infection. | [72] |
| 1H NMR; 2D TOCSY; HSQC | Triticum aestivum | Showed that the elevated changes taking place in the host metabolic profile were dependent on wheat inoculated with Fusarium graminearum both at ambient and increased CO2 levels. | [33] |
| NMR; GC/LC-MS/MS | Oryza sativa L. cv. Hwacheong | Demonstrated metabolic changes in Magnaporthe grisea-induced rice cultivars. | [74] |
| UHPLC-MS; GC-MS | Oryza sativa L. | Primary, carbohydrate, and secondary metabolism form a significant part of rice defense mechanisms against Chilo suppressalis. | [75] |
| LC/TOF/MS; LC/QE/MS | Triticum turgidum ssp. durum | Activation of defense-related phytohormone, and terpenoid-related and shikimate-mediated secondary metabolism in rice responding to C. suppressalis feeding and a significant induction of benzoxazoles in wheat genotypes subjected to aphid feeding. | [76] |
| Abiotic Stress | |||
| UHPLC- qTOF-HDMS | Zea mays | Differential accumulations of HCAs, HCA derivatives, and flavonoids in maize plants under drought stress. | [37] |
| UHPLC-qTOF-MS | Zea mays | Significant amino acid reduction was observed in nutrient-starved maize plants in comparison to the control. | [38] |
| LC–MS and GC–MS | Solanum lycopersicum L. | Decreased amino acid levels in the leaves of tomato plants due to nutrient deficiency. | [78] |
| GC-MS | Hordeum vulgare L. | Barley plants experienced increased levels of amino acids, sugars, and organic acids when exposed to drought conditions. | [80] |
| GC-MS | Triticum ssp. | Water and nutrient uptake were metabolically activated in the roots and shoots due to a significant increase in amino acids and sugars caused by exposure to drought stress. | [30] |
| (QTRAP)-MS | Dendrobium sinense | An increase in flavonoids, alkaloids, and phenylpropanoids was recorded under drought stress. | [86] |
| UHPLC-MS/MS | Triticum aestivum | Accumulation of phenolics, alkaloids, and flavonoids in wheat genotypes exposed to drought conditions | [87] |
| FTMS | Medicago sativa and Medicago arborea | Secondary metabolites from saponins and hydroxycinnamic acids increased salinity-stress tolerance in Medicago sativa and Medicago arborea species. | [89] |
| HPLC-triple TOF-MS/MS | Lonicerae Japonicae Flos | Differential accumulation of secondary metabolites (phenolic acids, flavonoids, and iridoids) in salt-stressed plants compared to controls. | [90] |
| UPLC-MS | Beta vulgaris | Significant increases in flavonoids (Apigenin-7-glucoside and luteolin) in plants under salt stress. | [91] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mashabela, M.D.; Masamba, P.; Kappo, A.P. Metabolomics and Chemoinformatics in Agricultural Biotechnology Research: Complementary Probes in Unravelling New Metabolites for Crop Improvement. Biology 2022, 11, 1156. https://doi.org/10.3390/biology11081156
Mashabela MD, Masamba P, Kappo AP. Metabolomics and Chemoinformatics in Agricultural Biotechnology Research: Complementary Probes in Unravelling New Metabolites for Crop Improvement. Biology. 2022; 11(8):1156. https://doi.org/10.3390/biology11081156
Chicago/Turabian StyleMashabela, Manamele Dannies, Priscilla Masamba, and Abidemi Paul Kappo. 2022. "Metabolomics and Chemoinformatics in Agricultural Biotechnology Research: Complementary Probes in Unravelling New Metabolites for Crop Improvement" Biology 11, no. 8: 1156. https://doi.org/10.3390/biology11081156
APA StyleMashabela, M. D., Masamba, P., & Kappo, A. P. (2022). Metabolomics and Chemoinformatics in Agricultural Biotechnology Research: Complementary Probes in Unravelling New Metabolites for Crop Improvement. Biology, 11(8), 1156. https://doi.org/10.3390/biology11081156







