The “Infernaccio” Gorges: Microbial Diversity of Black Deposits and Isolation of Manganese-Solubilizing Bacteria
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Geological Description of the Sampling Area
2.2. Sampling and Sample Handling
2.3. DNA Extraction and 16S rRNA Gene Metabarcoding
2.4. Mineralogical Investigation by XRPD
2.5. Sample Characterization by SEM-EDS
2.6. Isolation and Characterization of Culturable Solubilizing Bacteria
2.7. 16s Barcoding and Phylogenetic Analysis
2.8. Statistical Analysis
3. Results
3.1. DNA Extraction and 16S rRNA Gene Metabarcoding
3.2. Isolation and Characterization of Culturable Solubilizing Bacteria
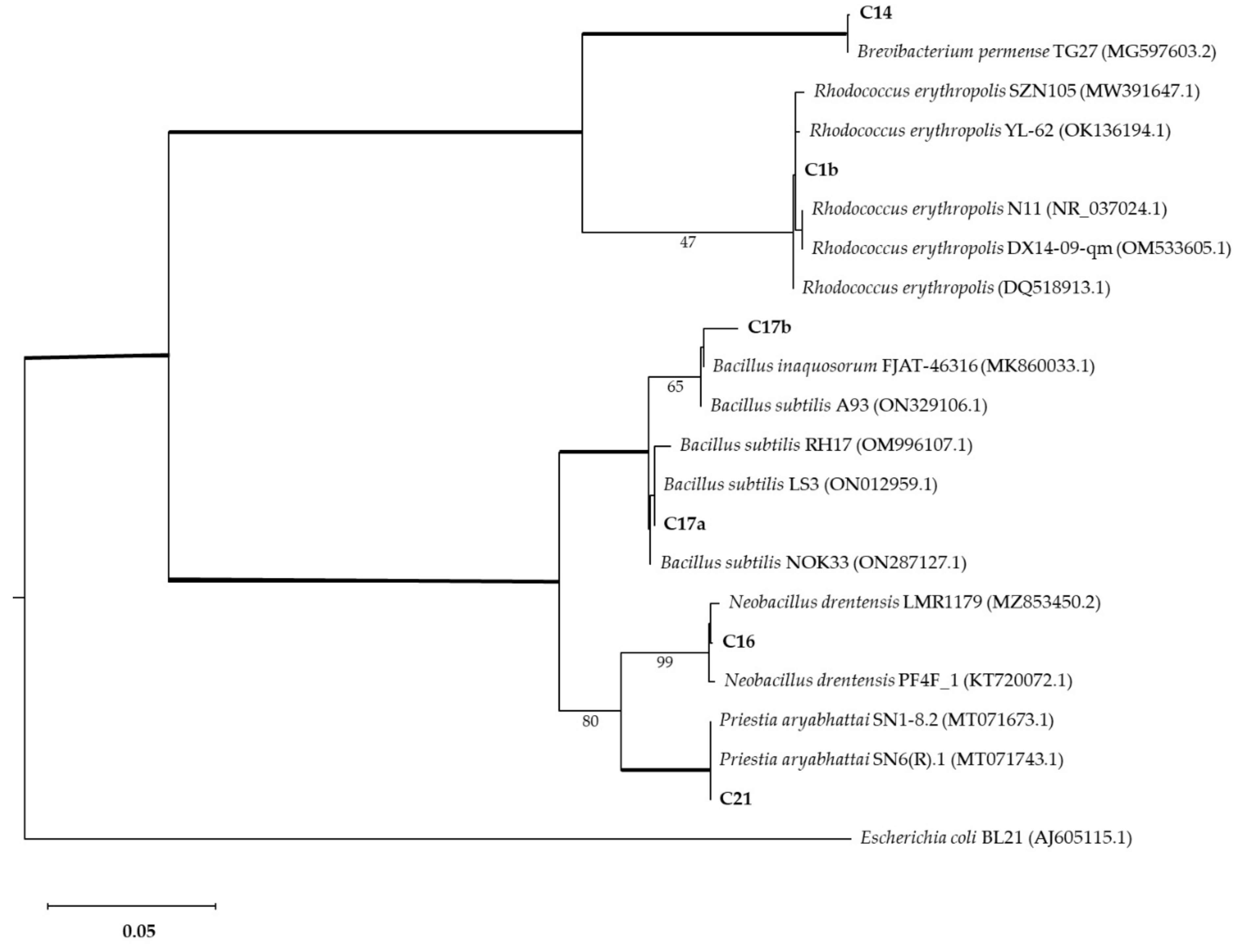
3.3. 16s Barcoding and Phylogenetic Analysis
3.4. Mineralogical Investigation by XRPD
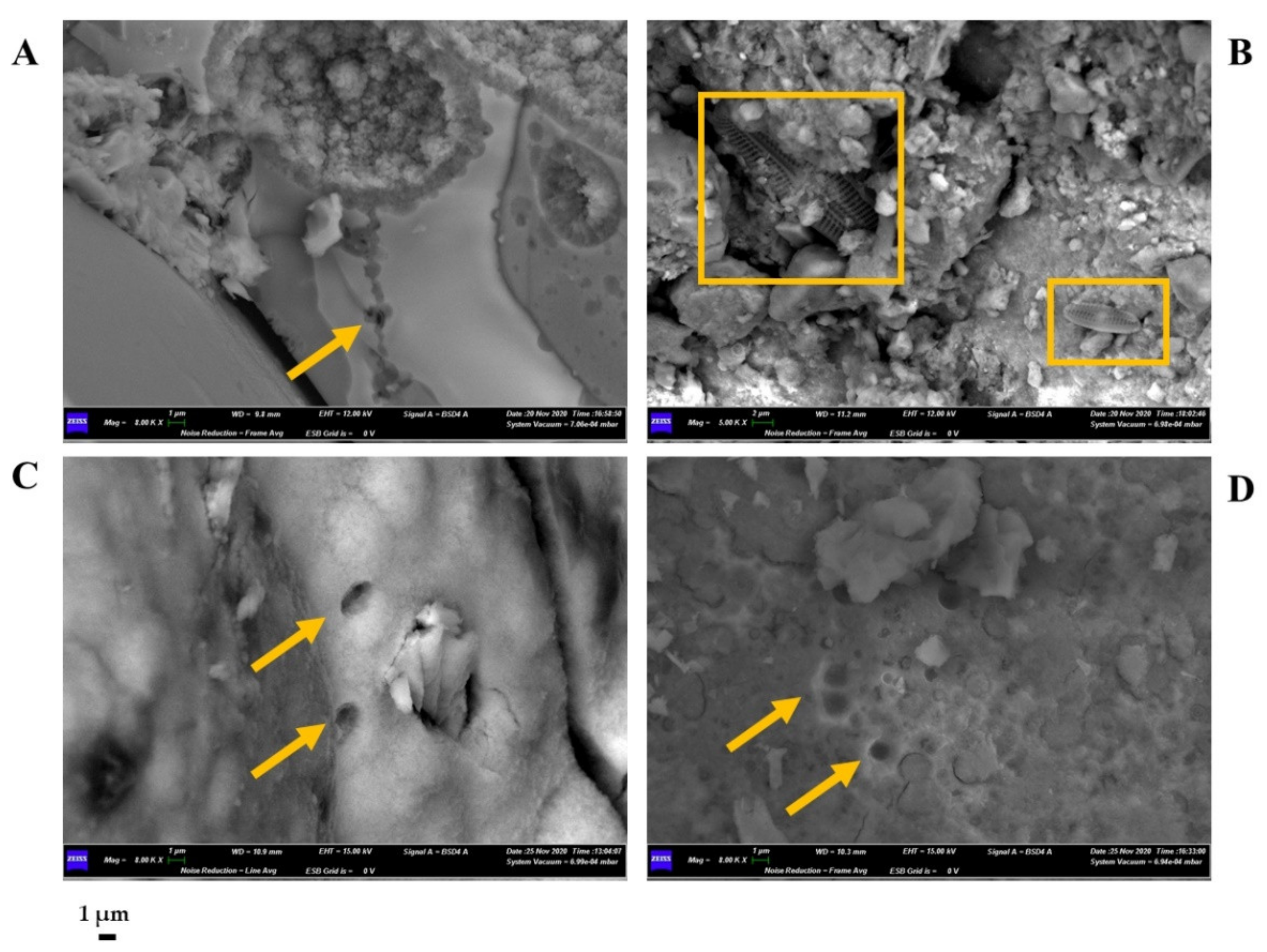
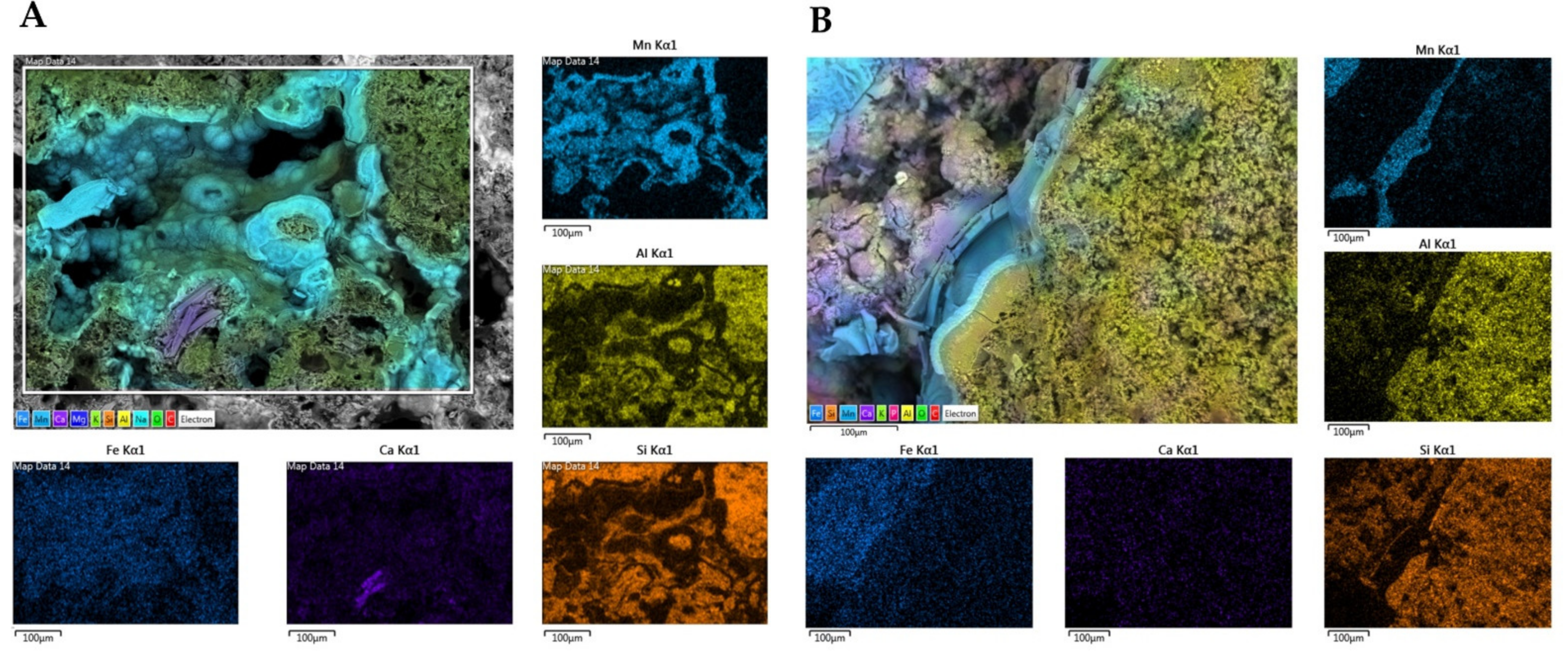
3.5. Sample Characterization by SEM-EDS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Geszvain, K.; Butterfield, C.; Davis, R.E.; Madison, A.S.; Lee, S.-W.; Parker, D.L.; Soldatova, A.; Spiro, T.G.; Luther, G.W.; Tebo, B.M. The Molecular Biogeochemistry of Manganese(II) Oxidation. Biochem. Soc. Trans. 2012, 40, 1244–1248. [Google Scholar] [CrossRef]
- Marshall, K.C. Chapter 5 Biogeochemistry of Manganese Minerals. In Studies in Environmental Science; Elsevier: Amsterdam, The Netherlands, 1979; Volume 3, pp. 253–292. ISBN 978-0-444-41745-9. [Google Scholar]
- Gad, S.C. Manganese. In Encyclopedia of Toxicology; Elsevier: Amsterdam, The Netherlands, 2005; pp. 11–13. [Google Scholar]
- Wang, X.; Xie, G.-J.; Tian, N.; Dang, C.-C.; Cai, C.; Ding, J.; Liu, B.-F.; Xing, D.-F.; Ren, N.-Q. Anaerobic Microbial Manganese Oxidation and Reduction: A Critical Review. Sci. Total Environ. 2022, 822, 153513. [Google Scholar] [CrossRef]
- Polgari, M.; Hein, J.R.; Toth, A.L.; Pal-Molnár, E.; Vigh, T.; Biró, L.; Fintor, K. Microbial Action Formed Jurassic Mn-Carbonate Ore Deposit in Only a Few Hundred Years (Úrkút, Hungary). Geology 2012, 40, 903–906. [Google Scholar] [CrossRef]
- Vaccarelli, I.; Matteucci, F.; Pellegrini, M.; Bellatreccia, F.; del Gallo, M. Exploring Microbial Biosignatures in Mn-Deposits of Deep Biosphere: A Preliminary Cross-Disciplinary Approach to Investigate Geomicrobiological Interactions in a Cave in Central Italy. Front. Earth Sci. 2021, 9. [Google Scholar] [CrossRef]
- Biondi, J.C.; Polgári, M.; Gyollai, I.; Fintor, K.; Kovács, I.; Fekete, J.; Mojzsis, S.J. Biogenesis of the Neoproterozoic Kremydilite Manganese Ores from Urucum (Brazil)—A New Manganese Ore Type. Precambrian Res. 2020, 340, 105624. [Google Scholar] [CrossRef]
- Tebo, B.M.; Bargar, J.R.; Clement, B.G.; Dick, G.J.; Murray, K.J.; Parker, D.; Verity, R.; Webb, S.M. Biogenic Manganese Oxides: Properties and Mechanisms of Formation. Annu. Rev. Earth Planet. Sci. 2004, 32, 287–328. [Google Scholar] [CrossRef]
- Barboza, N.R.; Guerra-Sá, R.; Leão, V.A. Mechanisms of Manganese Bioremediation by Microbes: An Overview. J. Chem. Technol. Biotechnol. 2016, 91, 2733–2739. [Google Scholar] [CrossRef]
- Caspi, R.; Haygood, M.G.; Tebo, B.M. Unusual Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Genes from a Marine Manganese-Oxidizing Bacterium. Microbiology 1996, 142, 2549–2559. [Google Scholar] [CrossRef] [PubMed]
- Ercole, C.; Altieri, F.; Piccone, C. Influence of Manganese Dioxide and Manganic Ions on the Production of Two Proteins in Arthrobacter Sp. Geomicrobiol. J. 1999, 16, 95–103. [Google Scholar] [CrossRef]
- Lovley, D.R.; Chapelle, F.H. Deep Subsurface Microbial Processes. Rev. Geophys. 1995, 33, 365. [Google Scholar] [CrossRef]
- Trimble, R.B.; Ehrlich, H.L. Bacteriology of Manganese Nodules. Appl. Microbiol. 1968, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Sedlakova-Kadukova, J. Microorganisms in Metal Recovery—Tools or Teachers. In Microbial Syntrophy-Mediated Eco-Enterprising; Elsevier: Amsterdam, The Netherlands, 2022; pp. 71–86. [Google Scholar]
- Johnson, D.B. Biomining—Biotechnologies for Extracting and Recovering Metals from Ores and Waste Materials. Curr. Opin. Biotechnol. 2014, 30, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Acharya, C.; Kar, R.N.; Sukla, L.B. Studies on Reaction Mechanism of Bioleaching of Manganese Ore. Miner. Eng. 2003, 16, 1027–1030. [Google Scholar] [CrossRef]
- Nkuna, R.; Ijoma, G.N.; Matambo, T.S.; Chimwani, N. Accessing Metals from Low-Grade Ores and the Environmental Impact Considerations: A Review of the Perspectives of Conventional versus Bioleaching Strategies. Minerals 2022, 12, 506. [Google Scholar] [CrossRef]
- Mizrahi-Man, O.; Davenport, E.R.; Gilad, Y. Taxonomic Classification of Bacterial 16S RRNA Genes Using Short Sequencing Reads: Evaluation of Effective Study Designs. PLoS ONE 2013, 8, e53608. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, S.; Bellatreccia, F.; Columbu, A.; Vaccarelli, I.; Pellegrini, M.; Jurado, V.; del Gallo, M.; Saiz-Jimenez, C.; Sodo, A.; Millo, C.; et al. Morpho-Mineralogical and Bio-Geochemical Description of Cave Manganese Stromatolite-Like Patinas (Grotta Del Cervo, Central Italy) and Hints on Their Paleohydrological-Driven Genesis. Front. Earth Sci. 2021, 9. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Welsh, C.T. Microbiology: A Laboratory Manual, Global Edition, 11th ed.; Cappuccino, J.G., Welsh, C.T., Eds.; Pearson Education Limited: Essex, UK, 2017. [Google Scholar]
- Stamatakis, A.; Ludwig, T.; Meier, H. RAxML-III: A Fast Program for Maximum Likelihood-Based Inference of Large Phylogenetic Trees. Bioinformatics 2005, 21, 456–463. [Google Scholar] [CrossRef]
- Bauer, M.A.; Kainz, K.; Carmona-Gutierrez, D.; Madeo, F. Microbial Wars: Competition in Ecological Niches and within the Microbiome. Microb. Cell 2018, 5, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Akob, D.M.; Bohu, T.; Beyer, A.; Schäffner, F.; Händel, M.; Johnson, C.A.; Merten, D.; Büchel, G.; Totsche, K.U.; Küsel, K. Identification of Mn(II)-Oxidizing Bacteria from a Low-PH Contaminated Former Uranium Mine. Appl. Environ. Microbiol. 2014, 80, 5086–5097. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Zhang, P.; Wei, D.; Zhang, J.; Yan, B.; Cao, L.; Zhou, Y.; Luo, L. Simultaneous Removal of Iron and Manganese from Acid Mine Drainage by Acclimated Bacteria. J. Hazard. Mater. 2020, 396, 122631. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Tang, X.; Luo, X.; Li, G.; Liang, H.; Snyder, S. Oxidants-Assisted Sand Filter to Enhance the Simultaneous Removals of Manganese, Iron and Ammonia from Groundwater: Formation of Active MnOx and Involved Mechanisms. J. Hazard. Mater. 2021, 415, 125707. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tsang, Y.F.; Wang, H.; Sun, Y.; Song, Y.; Pan, X.; Luo, S. Effective Stabilization of Arsenic in Contaminated Soils with Biogenic Manganese Oxide (BMO) Materials. Environ. Pollut. 2020, 258, 113481. [Google Scholar] [CrossRef] [PubMed]
- Volokhin, Y.G.; Mikhailik, P.E.; Mikhailik, E.V. Minerals in Manganese Deposits of Belyaevsky Volcano, the Sea of Japan. Russ. J. Pac. Geol. 2020, 14, 340–362. [Google Scholar] [CrossRef]
- Carmichael, I.S.E. The Mineralogy and Petrology of the Volcanic Rocks from the Leucite Hills, Wyoming. Contrib. Mineral. Petrol. 1967, 15, 24–66. [Google Scholar] [CrossRef]
- Singer, A.; Zarei, M.; Lange, F.M.; Stahr, K. Halloysite Characteristics and Formation in the Northern Golan Heights. Geoderma 2004, 123, 279–295. [Google Scholar] [CrossRef]
- Barile, M. Caratterizzazione Funzionale Della Comunita’ Macrobentonica Dei Fiumi Della Tuscia e Analisi Delle Relazioni Con Le Variabili Abiotiche a Diversa Scala Spaziale. Ph.D. Thesis, Università degli Studi della Tuscia di Viterbo, Viterbo, Italy, 2010. Available online: https://dspace.unitus.it/handle/2067/1128 (accessed on 31 March 2021).
- Beal, E.J.; House, C.H.; Orphan, V.J. Manganese- and Iron-Dependent Marine Methane Oxidation. Science 2009, 325, 184–187. [Google Scholar] [CrossRef]
- Rajabzadeh, M.A.; Haddad, F.; Polgári, M.; Fintor, K.; Walter, H.; Molnár, Z.; Gyollai, I. Investigation on the Role of Microorganisms in Manganese Mineralization from Abadeh-Tashk Area, Fars Province, Southwestern Iran by Using Petrographic and Geochemical Data. Ore Geol. Rev. 2017, 80, 229–249. [Google Scholar] [CrossRef]
- Yu, W.; Polgári, M.; Gyollai, I.; Fintor, K.; Szabó, M.; Kovács, I.; Fekete, J.; Du, Y.; Zhou, Q. Microbial Metallogenesis of Cryogenian Manganese Ore Deposits in South China. Precambrian Res. 2019, 322, 122–135. [Google Scholar] [CrossRef]
- Ijaz, A.; Mumtaz, M.Z.; Wang, X.; Ahmad, M.; Saqib, M.; Maqbool, H.; Zaheer, A.; Wang, W.; Mustafa, A. Insights into Manganese Solubilizing Bacillus Spp. for Improving Plant Growth and Manganese Uptake in Maize. Front. Plant Sci. 2021, 12, 719504. [Google Scholar] [CrossRef]
- Sanket, A.S.; Ghosh, S.; Sahoo, R.; Nayak, S.; Das, A.P. Molecular Identification of Acidophilic Manganese (Mn)-oxide solubilizing Bacteria from Mining Effluents and Their Application in Mineral Beneficiation. Geomicrobiol. J. 2017, 34, 71–80. [Google Scholar] [CrossRef]
- Kanso, S.; Greene, A.C.; Patel, B.K.C. Bacillus Subterraneus Sp. Nov., an Iron- and Manganese-Reducing Bacterium from a Deep Subsurface Australian Thermal Aquifer. Int. J. Syst. Evol. Microbiol. 2002, 52, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Sujith, P.P.; Mourya, B.S.; Krishnamurthi, S.; Meena, R.M.; Loka Bharathi, P.A. Mobilization of Manganese by Basalt Associated Mn(II)-Oxidizing Bacteria from the Indian Ridge System. Chemosphere 2014, 95, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust Demarcation of 17 Distinct Bacillus Species Clades, Proposed as Novel Bacillaceae Genera, by Phylogenomics and Comparative Genomic Analyses: Description of Robertmurraya Kyonggiensis Sp. Nov. and Proposal for an Emended Genus Bacillus Limiting It only to the members of the Subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar] [CrossRef]
- Schmid, J.; Auling, G. Manganese Transport in Brevibacterium Ammoniagenes ATCC J. Bacteriol. 1987, 169, 3385–3387. [Google Scholar] [CrossRef]
- Willing, A.; Follmann, H.; Auling, G. Ribonucleotide Reductase of Brevibacterium Ammoniagenes Is a Manganese Enzyme. Eur. J. Biochem. 1988, 170, 603–611. [Google Scholar] [CrossRef]
- Gillard, B.; Chatzievangelou, D.; Thomsen, L.; Ullrich, M.S. Heavy-Metal-Resistant Microorganisms in Deep-Sea Sediments Disturbed by Mining Activity: An Application Toward the Development of Experimental in Vitro Systems. Front. Mar. Sci. 2019, 6, 462. [Google Scholar] [CrossRef]
- Shulse, C.N.; Maillot, B.; Smith, C.R.; Church, M.J. Polymetallic Nodules, Sediments, and Deep Waters in the Equatorial North Pacific Exhibit Highly Diverse and Distinct Bacterial, Archaeal, and Microeukaryotic Communities. Microbiologyopen 2017, 6, e00428. [Google Scholar] [CrossRef]
- Lindh, M.v.; Maillot, B.M.; Shulse, C.N.; Gooday, A.J.; Amon, D.J.; Smith, C.R.; Church, M.J. From the Surface to the Deep-Sea: Bacterial Distributions across Polymetallic Nodule Fields in the Clarion-Clipperton Zone of the Pacific Ocean. Front. Microbiol. 2017, 8, 1696. [Google Scholar] [CrossRef]
- Gillan, D.C.; Baeyens, W.; Bechara, R.; Billon, G.; Denis, K.; Grosjean, P.; Leermakers, M.; Lesven, L.; Pede, A.; Sabbe, K.; et al. Links between Bacterial Communities in Marine Sediments and Trace Metal Geochemistry as Measured by in Situ DET/DGT Approaches. Mar. Pollut. Bull. 2012, 64, 353–362. [Google Scholar] [CrossRef]
- Walsh, E.A.; Kirkpatrick, J.B.; Pockalny, R.; Sauvage, J.; Spivack, A.J.; Murray, R.W.; Sogin, M.L.; D’Hondt, S. Relationship of Bacterial Richness to Organic Degradation Rate and Sediment Age in Subseafloor Sediment. Appl. Environ. Microbiol. 2016, 82, 4994–4999. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.L.; Singh, P.K. Global Environmental Problems. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Singapore, 2017; pp. 13–41. [Google Scholar]
- Das, A.P.; Ghosh, S.; Mohanty, S.; Sukla, L.B. Advances in Manganese Pollution and Its Bioremediation. In Environmental Microbial Biotechnology; Sukla, L.B., Pradhan, N., Panda, S., Mishra, K.B., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 45, pp. 313–328. [Google Scholar]
- Mourinha, C.; Palma, P.; Alexandre, C.; Cruz, N.; Rodrigues, S.M.; Alvarenga, P. Potentially Toxic Elements’ Contamination of Soils Affected by Mining Activities in the Portuguese Sector of the Iberian Pyrite Belt and Optional Remediation Actions: A Review. Environments 2022, 9, 11. [Google Scholar] [CrossRef]

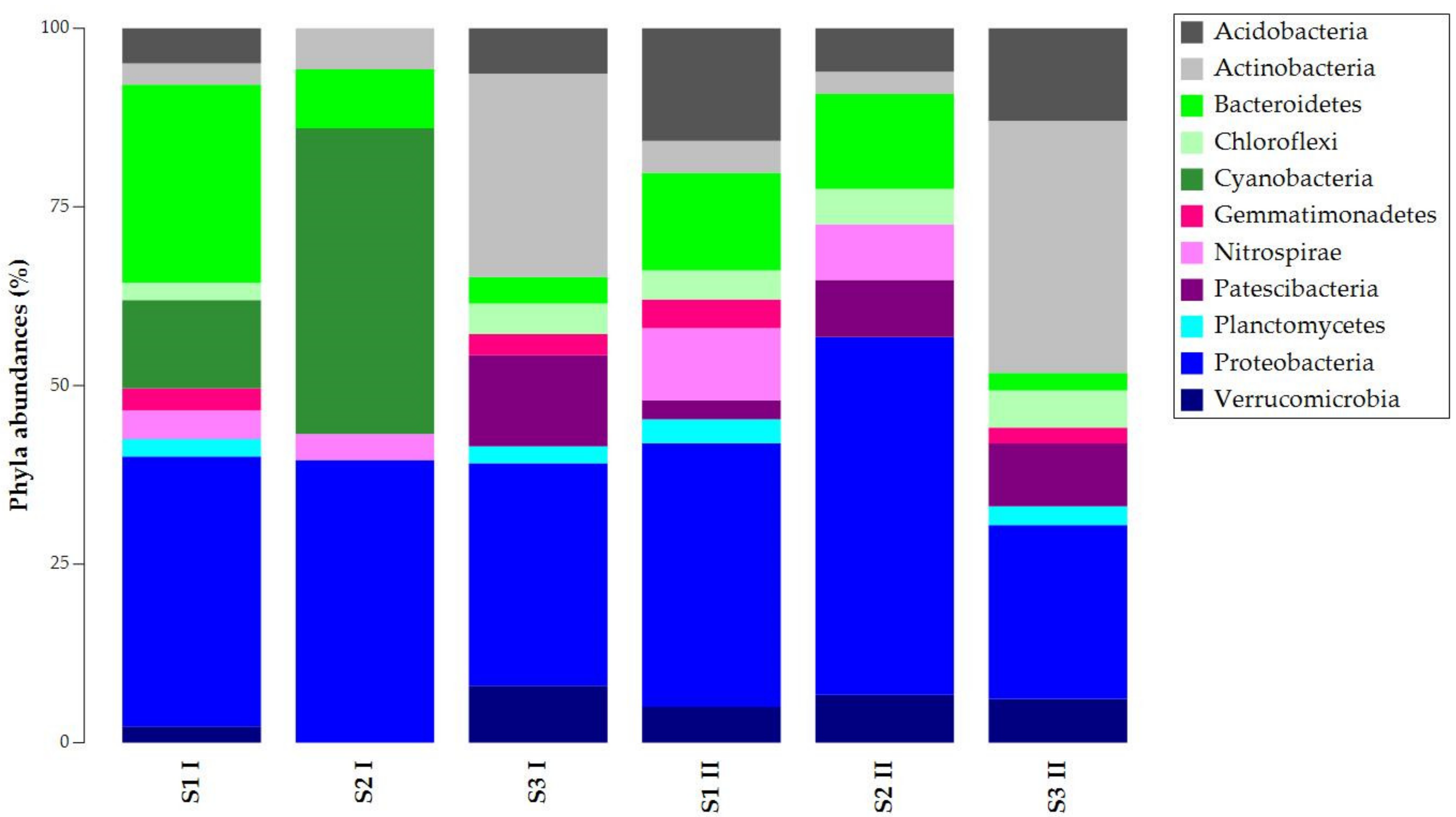
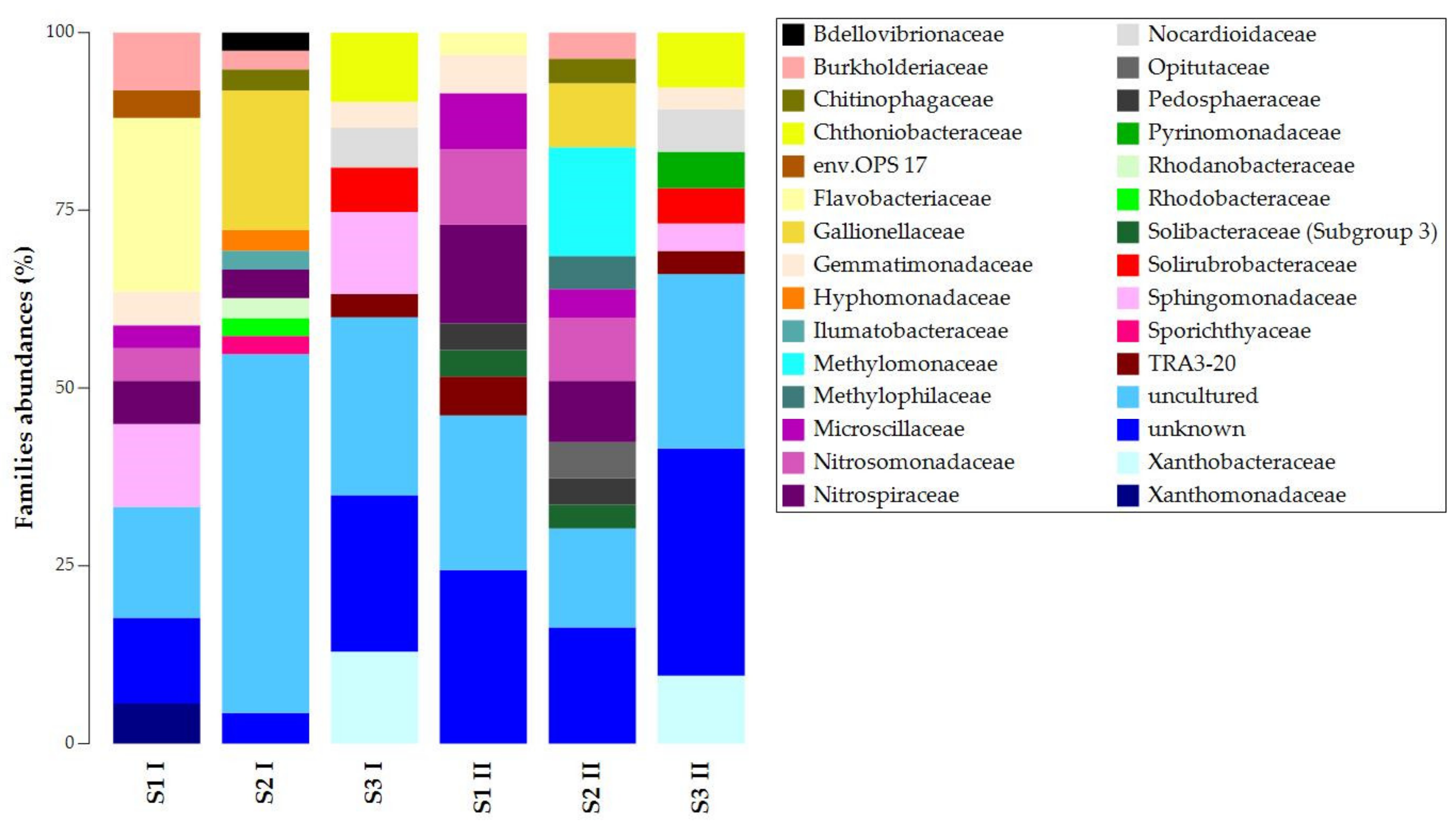
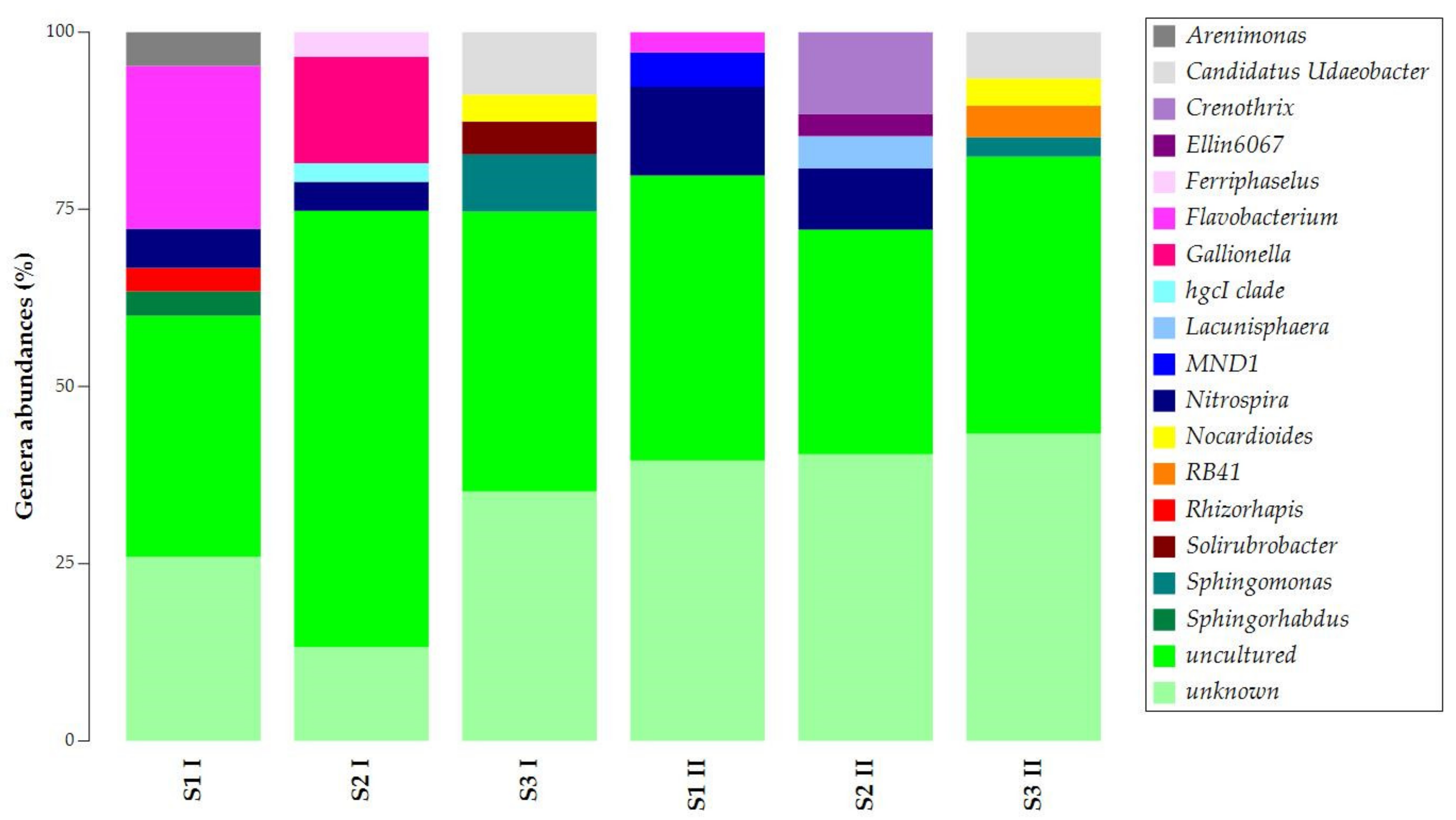

| S1 I | S2 I | S3 I | S1 II | S2 II | S3 II | |
|---|---|---|---|---|---|---|
| Taxa S | 374 | 358 | 858 | 872 | 633 | 827 |
| Individuals | 4860 | 12439 | 16651 | 11483 | 9702 | 15253 |
| Simpson 1-D | 0.992 | 0.861 | 0.995 | 0.997 | 0.995 | 0.997 |
| Shannon H’ | 5.50 | 3.76 | 6.07 | 6.39 | 5.92 | 6.25 |
| Chao-1 | 374 | 358 | 858 | 872.4 | 633.1 | 827 |
| Domain-level abundances | ||||||
| Archaea | 0.0 | 0.1 | 0.2 | 0.3 | 0.2 | 0.5 |
| Bacteria | 100.0 | 99.9 | 99.8 | 99.7 | 99.8 | 99.5 |
| C1b | C14 | C16 | C17a | C17b | C19 | C20a | C20b | C20c | C21 | C22 | C23 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMB Discoloration Rate | + | + | +++ | ++ | ++ | − | − | − | − | ++ | − | − |
| C1b | C14 | C16 | C17a | C17b | C21 | |
|---|---|---|---|---|---|---|
| Size | Large | Large | Small | Medium | Medium | Medium |
| Color | Pink | Clear pink | White | Clear brown | Clear brown | Clear brown |
| Form | Circular | Circular | Circular | Irregular | Irregular | Irregular |
| Margin | Regular | Regular | Regular | Undulating | Undulating | Undulating |
| Surface | Smooth | Smooth | Smooth | Rough | Rough | Rough |
| Transparence | Opaque | Opaque | Opaque | Opaque | Opaque | Opaque |
| Texture | Creamy | Mucosal | Creamy | Creamy | Creamy | Creamy |
| C1b | C14 | C16 | C17a | C17b | C21 | |
|---|---|---|---|---|---|---|
| Spores | no | no | yes | yes | yes | yes |
| Gram reaction | + | + | + | + | + | + |
| Morphology | coccus | rod | rod | rod | rod | rod |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farda, B.; Djebaili, R.; Del Gallo, M.; Ercole, C.; Bellatreccia, F.; Pellegrini, M. The “Infernaccio” Gorges: Microbial Diversity of Black Deposits and Isolation of Manganese-Solubilizing Bacteria. Biology 2022, 11, 1204. https://doi.org/10.3390/biology11081204
Farda B, Djebaili R, Del Gallo M, Ercole C, Bellatreccia F, Pellegrini M. The “Infernaccio” Gorges: Microbial Diversity of Black Deposits and Isolation of Manganese-Solubilizing Bacteria. Biology. 2022; 11(8):1204. https://doi.org/10.3390/biology11081204
Chicago/Turabian StyleFarda, Beatrice, Rihab Djebaili, Maddalena Del Gallo, Claudia Ercole, Fabio Bellatreccia, and Marika Pellegrini. 2022. "The “Infernaccio” Gorges: Microbial Diversity of Black Deposits and Isolation of Manganese-Solubilizing Bacteria" Biology 11, no. 8: 1204. https://doi.org/10.3390/biology11081204
APA StyleFarda, B., Djebaili, R., Del Gallo, M., Ercole, C., Bellatreccia, F., & Pellegrini, M. (2022). The “Infernaccio” Gorges: Microbial Diversity of Black Deposits and Isolation of Manganese-Solubilizing Bacteria. Biology, 11(8), 1204. https://doi.org/10.3390/biology11081204