SLC16a6, mTORC1, and Autophagy Regulate Ketone Body Excretion in the Intestinal Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Reagents
2.3. Cell Culture
2.4. Preparation of shRNA Constitutively Expressing Cell Lines
2.5. mRNA Expression Analysis
2.6. Protein Extraction
2.7. Protein Concentration Measurement and Western Blotting
2.8. Measurement of Ketone Body Concentration
2.9. Capillary Electrophoresis (CE)–Mass Spectrometry (MS) and pH Analysis
2.10. Statistical Analysis
3. Results
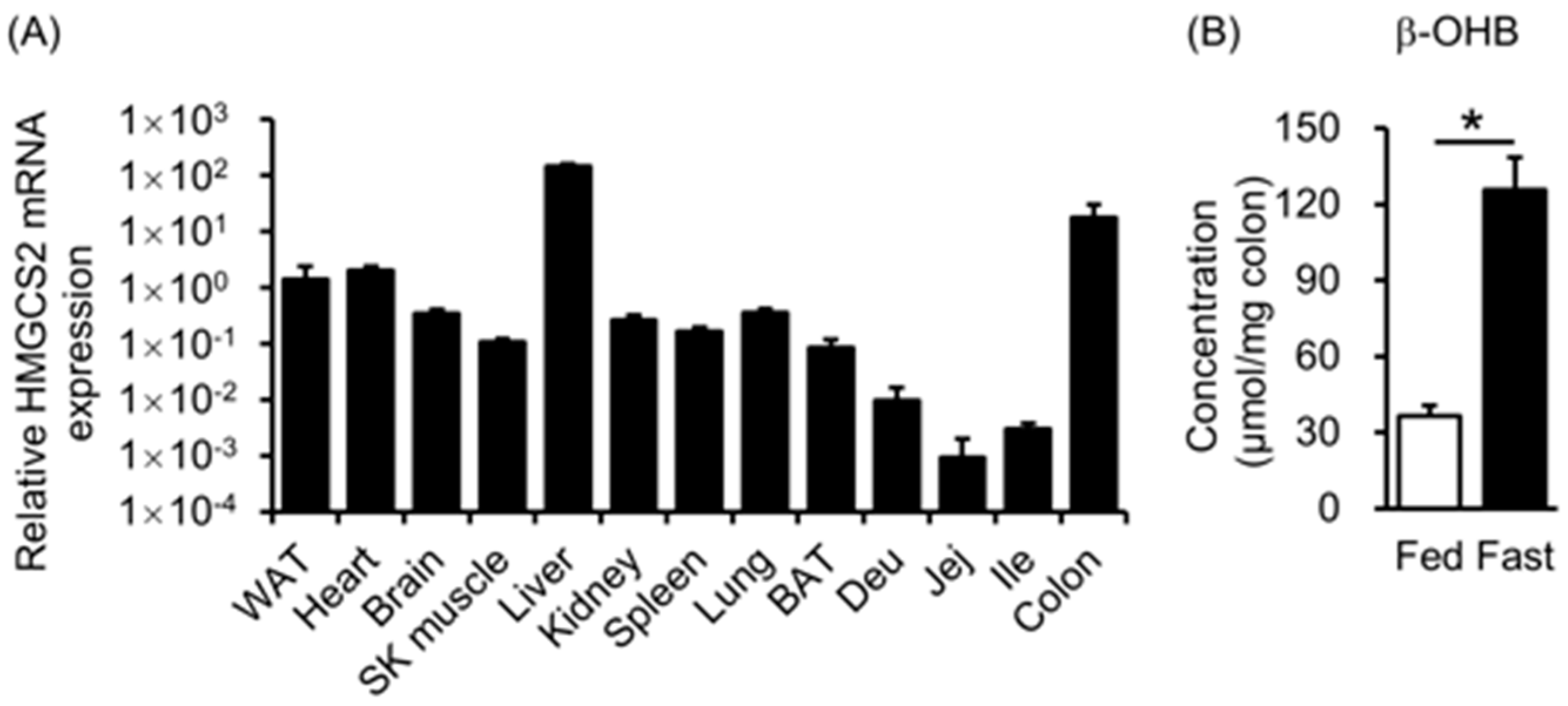
3.1. HMGCS2 Expression and Ketone Body Concentration in Mouse Colons
3.2. Ketone Body Production in Colorectal Cells
3.3. Autophagy Regulates Ketone Body Excretion in Colorectal Cells
3.4. Basigin-Associated MCT Involves Ketone Body Import in Colorectal Cells
3.5. MCT7 Involves Ketone Body Export in Colorectal Cells
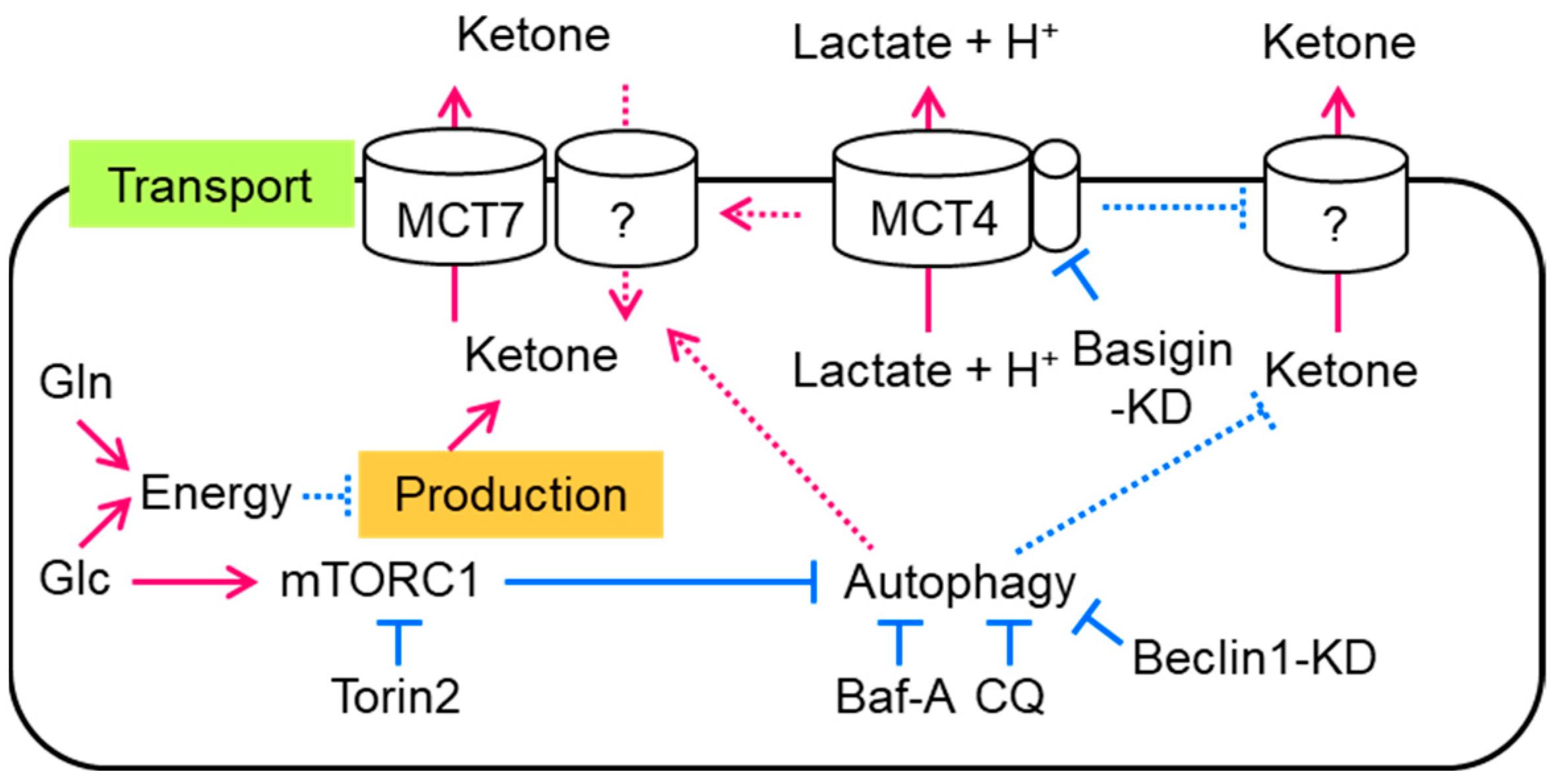
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newman, J.C.; Verdin, E. Ketone Bodies as Signaling Metabolites. Trends Endocrinol. Metab. 2014, 25, 42–52. [Google Scholar] [CrossRef]
- Hegardt, F.G. Mitochondrial 3-Hydroxy-3-Methylglutaryl-CoA Synthase: A Control Enzyme in Ketogenesis. Biochem. J. 1999, 338 Pt 3, 569–582. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-Dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Shimazu, T.; Hirschey, M.D.; Newman, J.; He, W.; Shirakawa, K.; Le Moan, N.; Grueter, C.A.; Lim, H.; Saunders, L.R.; Stevens, R.D.; et al. Suppression of Oxidative Stress by β-Hydroxybutyrate, an Endogenous Histone Deacetylase Inhibitor. Science 2013, 339, 211–214. [Google Scholar] [CrossRef]
- Kimura, I.; Inoue, D.; Maeda, T.; Hara, T.; Ichimura, A.; Miyauchi, S.; Kobayashi, M.; Hirasawa, A.; Tsujimoto, G. Short-Chain Fatty Acids and Ketones Directly Regulate Sympathetic Nervous System via G Protein-Coupled Receptor 41 (GPR41). Proc. Natl. Acad. Sci. USA 2011, 108, 8030–8035. [Google Scholar] [CrossRef]
- Tunaru, S.; Kero, J.; Schaub, A.; Wufka, C.; Blaukat, A.; Pfeffer, K.; Offermanns, S. PUMA-G and HM74 Are Receptors for Nicotinic Acid and Mediate Its Anti-Lipolytic Effect. Nat. Med. 2003, 9, 352–355. [Google Scholar] [CrossRef]
- Ahmed, K.; Tunaru, S.; Offermanns, S. GPR109A, GPR109B and GPR81, a Family of Hydroxy-Carboxylic Acid Receptors. Trends Pharmacol. Sci. 2009, 30, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Youm, Y.-H.; Nguyen, K.Y.; Grant, R.W.; Goldberg, E.L.; Bodogai, M.; Kim, D.; D’Agostino, D.; Planavsky, N.; Lupfer, C.; Kanneganti, T.D.; et al. The Ketone Metabolite β-Hydroxybutyrate Blocks NLRP3 Inflammasome-Mediated Inflammatory Disease. Nat. Med. 2015, 21, 263–269. [Google Scholar] [CrossRef]
- Cherbuy, C.; Darcy-Vrillon, B.; Morel, M.T.; Pégorier, J.P.; Duée, P.H. Effect of Germfree State on the Capacities of Isolated Rat Colonocytes to Metabolize N-Butyrate, Glucose, and Glutamine. Gastroenterology 1995, 109, 1890–1899. [Google Scholar] [CrossRef] [PubMed]
- Mascaró, C.; Buesa, C.; Ortiz, J.A.; Haro, D.; Hegardt, F.G. Molecular Cloning and Tissue Expression of Human Mitochondrial 3-Hydroxy-3-Methylglutaryl-CoA Synthase. Arch. Biochem. Biophys. 1995, 317, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhou, Y.; Rychahou, P.; Fan, T.W.-M.; Lane, A.N.; Weiss, H.L.; Evers, B.M. Ketogenesis Contributes to Intestinal Cell Differentiation. Cell Death Differ. 2017, 24, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.-W.; Biton, M.; Haber, A.L.; Gunduz, N.; Eng, G.; Gaynor, L.T.; Tripathi, S.; Calibasi-Kocal, G.; Rickelt, S.; Butty, V.L.; et al. Ketone Body Signaling Mediates Intestinal Stem Cell Homeostasis and Adaptation to Diet. Cell 2019, 178, 1115–1131.e15. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P. The SLC16 Gene Family—Structure, Role and Regulation in Health and Disease. Mol. Aspects Med. 2013, 34, 337–349. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Meredith, D. The SLC16 Gene Family-from Monocarboxylate Transporters (MCTs) to Aromatic Amino Acid Transporters and Beyond. Pflugers Arch. 2004, 447, 619–628. [Google Scholar] [CrossRef]
- Carneiro, L.; Pellerin, L. Monocarboxylate Transporters: New Players in Body Weight Regulation. Obes. Rev. 2015, 16 (Suppl. S1), 55–66. [Google Scholar] [CrossRef]
- Vijay, N.; Morris, M.E. Role of Monocarboxylate Transporters in Drug Delivery to the Brain. Curr. Pharm. Des. 2014, 20, 1487–1498. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.M.; Gopal, E.; Ananth, S.; Zhuang, L.; Itagaki, S.; Prasad, B.M.; Smith, S.B.; Prasad, P.D.; Ganapathy, V. Identity of SMCT1 (SLC5A8) as a Neuron-Specific Na+-Coupled Transporter for Active Uptake of L-Lactate and Ketone Bodies in the Brain. J. Neurochem. 2006, 98, 279–288. [Google Scholar] [CrossRef]
- Bröer, S.; Schneider, H.P.; Bröer, A.; Rahman, B.; Hamprecht, B.; Deitmer, J.W. Characterization of the Monocarboxylate Transporter 1 Expressed in Xenopus Laevis Oocytes by Changes in Cytosolic pH. Biochem. J. 1998, 333 Pt 1, 167–174. [Google Scholar] [CrossRef]
- Hugo, S.E.; Cruz-Garcia, L.; Karanth, S.; Anderson, R.M.; Stainier, D.Y.R.; Schlegel, A. A Monocarboxylate Transporter Required for Hepatocyte Secretion of Ketone Bodies during Fasting. Genes. Dev. 2012, 26, 282–293. [Google Scholar] [CrossRef]
- Uebanso, T.; Ohnishi, A.; Kitayama, R.; Yoshimoto, A.; Nakahashi, M.; Shimohata, T.; Mawatari, K.; Takahashi, A. Effects of Low-Dose Non-Caloric Sweetener Consumption on Gut Microbiota in Mice. Nutrients 2017, 9, 560. [Google Scholar] [CrossRef]
- Uebanso, T.; Kano, S.; Yoshimoto, A.; Naito, C.; Shimohata, T.; Mawatari, K.; Takahashi, A. Effects of Consuming Xylitol on Gut Microbiota and Lipid Metabolism in Mice. Nutrients 2017, 9, 756. [Google Scholar] [CrossRef]
- Sengupta, S.; Peterson, T.R.; Laplante, M.; Oh, S.; Sabatini, D.M. mTORC1 Controls Fasting-Induced Ketogenesis and Its Modulation by Ageing. Nature 2010, 468, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Jun, C.B.; Ro, S.-H.; Kim, Y.-M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.-H. ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Meredith, D.; Fox, J.E.M.; Manoharan, C.; Davies, A.J.; Halestrap, A.P. Basigin (CD147) Is the Target for Organomercurial Inhibition of Monocarboxylate Transporter Isoforms 1 and 4: The Ancillary Protein for the Insensitive MCT2 Is EMBIGIN (Gp70). J. Biol. Chem. 2005, 280, 27213–27221. [Google Scholar] [CrossRef] [PubMed]
- Kirk, P.; Wilson, M.C.; Heddle, C.; Brown, M.H.; Barclay, A.N.; Halestrap, A.P. CD147 Is Tightly Associated with Lactate Transporters MCT1 and MCT4 and Facilitates Their Cell Surface Expression. EMBO J. 2000, 19, 3896–3904. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, T.; Hirschey, M.D.; Hua, L.; Dittenhafer-Reed, K.E.; Schwer, B.; Lombard, D.B.; Li, Y.; Bunkenborg, J.; Alt, F.W.; Denu, J.M.; et al. SIRT3 Deacetylates Mitochondrial 3-Hydroxy-3-Methylglutaryl CoA Synthase 2 and Regulates Ketone Body Production. Cell Metab. 2010, 12, 654–661. [Google Scholar] [CrossRef]
- Grimsrud, P.A.; Carson, J.J.; Hebert, A.S.; Hubler, S.L.; Niemi, N.M.; Bailey, D.J.; Jochem, A.; Stapleton, D.S.; Keller, M.P.; Westphall, M.S.; et al. A Quantitative Map of the Liver Mitochondrial Phosphoproteome Reveals Posttranslational Control of Ketogenesis. Cell Metab. 2012, 16, 672–683. [Google Scholar] [CrossRef]
- Kostiuk, M.A.; Keller, B.O.; Berthiaume, L.G. Palmitoylation of Ketogenic Enzyme HMGCS2 Enhances Its Interaction with PPARalpha and Transcription at the Hmgcs2 PPRE. FASEB J. 2010, 24, 1914–1924. [Google Scholar] [CrossRef]
- Thangaraju, M.; Cresci, G.; Itagaki, S.; Mellinger, J.; Browning, D.D.; Berger, F.G.; Prasad, P.D.; Ganapathy, V. Sodium-Coupled Transport of the Short Chain Fatty Acid Butyrate by SLC5A8 and Its Relevance to Colon Cancer. J. Gastrointest. Surg. 2008, 12, 1773–1781, discussion 1781–1782. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Myeroff, L.; Smiraglia, D.; Romero, M.F.; Pretlow, T.P.; Kasturi, L.; Lutterbaugh, J.; Rerko, R.M.; Casey, G.; Issa, J.-P.; et al. SLC5A8, a Sodium Transporter, Is a Tumor Suppressor Gene Silenced by Methylation in Human Colon Aberrant Crypt Foci and Cancers. Proc. Natl. Acad. Sci. USA 2003, 100, 8412–8417. [Google Scholar] [CrossRef] [PubMed]
- Dedkova, E.N.; Blatter, L.A. Role of β-Hydroxybutyrate, Its Polymer Poly-β-Hydroxybutyrate and Inorganic Polyphosphate in Mammalian Health and Disease. Front. Physiol. 2014, 5, 260. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uebanso, T.; Fukui, M.; Naito, C.; Shimohata, T.; Mawatari, K.; Takahashi, A. SLC16a6, mTORC1, and Autophagy Regulate Ketone Body Excretion in the Intestinal Cells. Biology 2023, 12, 1467. https://doi.org/10.3390/biology12121467
Uebanso T, Fukui M, Naito C, Shimohata T, Mawatari K, Takahashi A. SLC16a6, mTORC1, and Autophagy Regulate Ketone Body Excretion in the Intestinal Cells. Biology. 2023; 12(12):1467. https://doi.org/10.3390/biology12121467
Chicago/Turabian StyleUebanso, Takashi, Moeka Fukui, Chisato Naito, Takaaki Shimohata, Kazuaki Mawatari, and Akira Takahashi. 2023. "SLC16a6, mTORC1, and Autophagy Regulate Ketone Body Excretion in the Intestinal Cells" Biology 12, no. 12: 1467. https://doi.org/10.3390/biology12121467





