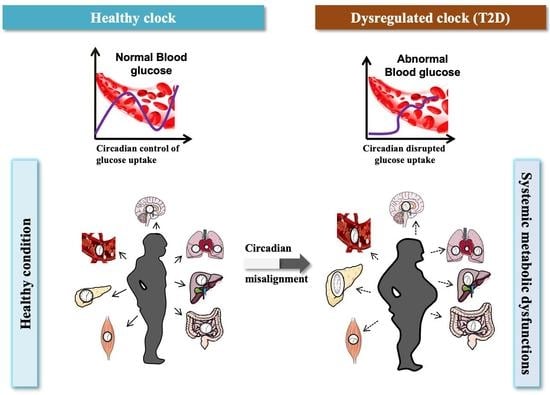
Multi-Omics Reveal Interplay between Circadian Dysfunction and Type2 Diabetes
Abstract
:Simple Summary
Abstract
1. Introduction
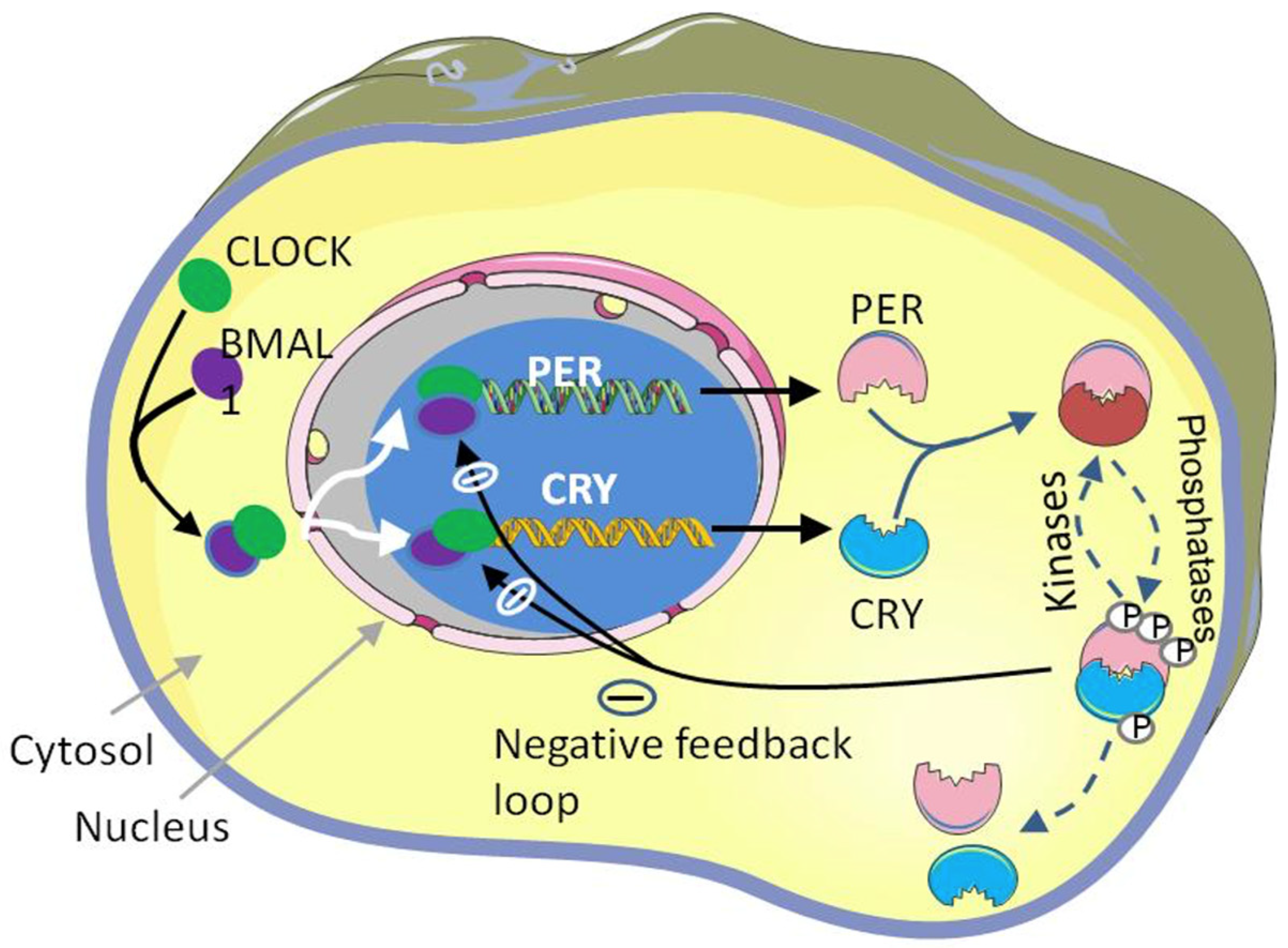
1.1. Molecular Mechanism of Circadian Clock
1.1.1. Transcriptional and Translational Feedback Loop (TTFL)
1.1.2. Non-Transcriptional Circadian Rhythm in Mammals
2. Multi-Omics Approaches to Understanding Circadian Dysfunction in Various Tissues in Type 2 Diabetes
2.1. Master Circadian Clock; Suprachiasmatic Nucleus (SCN)
2.2. Gastrointestinal Track (GI Tract) and Microbiome
2.3. Liver
2.4. Skeletal Muscles
2.5. Pancreas
2.6. Cardiovascular System
2.7. Renal System
2.8. Biological Fluids
3. Management of T2D through Circadian Intervention
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| SCN | Suprachiasmatic Nucleus |
| T2D | Type 2 Diabetes |
| ND | Non diabetic |
| HFD | High fat diet |
| ZT | Zeitgeber time |
| OW/OB | Overwight/obese |
| Q-PCR | Quantitative polymerase chain reaction |
| LC-MS | Liquid Chromatography–mass spectrometry |
| WAT | White adipose tissue |
| BAT | Brown adipose tissue |
| DALs | Differentially abundant lipids |
| HIT | High intensity interval training |
| BLT | Bright light therapy |
| TRF | Time-restricted feeding |
| CR | Calorie restriction |
| IF | Intermittent Fasting |
| AMI | Acute myocardial infarction |
| AI | Atrial fibrillation |
| GFR | Glomerular filtration rate |
| ERPF | Effective renal plasma flow |
| BP | Blood pressure |
| SGLT | sodium glucose co-transporter |
| DCT | distal convoluted tubule |
| CNT | connecting tubule |
| CCD | cortical collecting duct |
References
- Albrecht, U. Timing to Perfection: The Biology of Central and Peripheral Circadian Clocks. Neuron 2012, 74, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Fan, R.; Xie, L.; Shi, X.; Dong, K.; Zhang, S.; Tao, J.; Xu, W.; Ma, D.; Chen, J.; et al. A Growing Link between Circadian Rhythms, Type 2 Diabetes Mellitus and Alzheimer’s disease. Int. J. Mol. Sci. 2022, 23, 504. [Google Scholar] [CrossRef]
- Zhao, E.; Tait, C.; Minacapelli, C.D.; Catalano, C.; Rustgi, V.K. Circadian Rhythms, the Gut Microbiome, and Metabolic Disorders. Gastro Hep Adv. 2022, 1, 93–105. [Google Scholar] [CrossRef]
- Buijs, R.M.; Scheer, F.A.; Kreier, F.; Yi, C.; Bos, N.; Goncharuk, V.D.; Kalsbeek, A. Chapter 20: Organization of Circadian Functions: Interaction with the Body. Prog. Brain Res. 2006, 153, 341–360. [Google Scholar] [CrossRef]
- Coomans, C.P.; Van Den Berg, S.A.A.; Lucassen, E.A.; Houben, T.; Pronk, A.C.M.; Van Der Spek, R.D.; Kalsbeek, A.; Biermasz, N.R.; Van Dijk, K.W.; Romijn, J.A.; et al. The Suprachiasmatic Nucleus Controls Circadian Energy Metabolism and Hepatic Insulin Sensitivity. Diabetes 2013, 62, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Buhr, E.D.; Takahashi, J.S. Molecular Components of the Mammalian Circadian Clock. Handb. Exp. Pharmacol. 2013, 217, 3–27. [Google Scholar] [CrossRef]
- Lowrey, P.L.; Takahashi, J.S. Genetics of Circadian Rhythms in Mammalian Model Organisms. Adv. Genet. 2011, 74, 175–230. [Google Scholar] [CrossRef] [PubMed]
- Reinke, H.; Asher, G. Crosstalk between Metabolism and Circadian Clocks. Nat. Rev. Mol. Cell Biol. 2019, 20, 227–241. [Google Scholar] [CrossRef]
- Camacho, F.; Cilio, M.; Guo, Y.; Virshup, D.M.; Patel, K.; Khorkova, O.; Styren, S.; Morse, B.; Yao, Z.; Keesler, G.A. Human Casein Kinase Iδ Phosphorylation of Human Circadian Clock Proteins Period 1 and 2. FEBS Lett. 2001, 489, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Lamia, K.A.; Sachdeva, U.M.; Di Tacchio, L.; Williams, E.C.; Alvarez, J.G.; Egan, D.F.; Vasquez, D.S.; Juguilon, H.; Panda, S.; Shaw, R.J.; et al. AMPK Regulates the Circadian Clock by Cryptochrome Phosphorylation and Degradation. Science 2009, 326, 437–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, H.W.; Jiang, J.; Edery, I.; Koh, K.; Zheng, X.; Sehgal, A.; Somers, D.E.; Schultz, T.F.; Milnamow, M.; Kay, S.A.; et al. Fig. 4. Increased Cry Stability in Organotypic Tissue Slices from Afh/Afh Mice and in Mammalian Cells Expressing Fbxl3. Science 2007, 316, 900–905. [Google Scholar]
- Guillaumond, F.; Dardente, H.; Giguère, V.; Cermakian, N. Differential Control of Bmal1 Circadian Transcription by REV-ERB and ROR Nuclear Receptors. J. Biol. Rhythm. 2005, 20, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacner, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK Protein in the Mammalian Circadian Mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. Erratum: The Orphan Nuclear Receptor REV-ERBα Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell 2002, 110, 535. [Google Scholar] [CrossRef]
- Robinson, I.; Reddy, A.B. Molecular Mechanisms of the Circadian Clockwork in Mammals. FEBS Lett. 2014, 588, 2477–2483. [Google Scholar] [CrossRef]
- Doi, M.; Hirayama, J.; Sassone-Corsi, P. Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell 2006, 125, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 Regulates Circadian Clock Gene Expression through PER2 Deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Bessho, Y. The Circadian NAD+ Metabolism: Impact on Chromatin Remodeling and Aging. Biomed. Res. Int. 2016, 2016, 3208429. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J.S.; Van Ooijen, G.; Dixon, L.E.; Troein, C.; Corellou, F.; Bouget, F.Y.; Reddy, A.B.; Millar, A.J. Circadian Rhythms Persist without Transcription in a Eukaryote. Nature 2011, 469, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Ch, R.; Rey, G.; Ray, S.; Jha, P.K.; Driscoll, P.C.; Dos Santos, M.S.; Malik, D.M.; Lach, R.; Weljie, A.M.; MacRae, J.I.; et al. Rhythmic Glucose Metabolism Regulates the Redox Circadian Clockwork in Human Red Blood Cells. Nat. Commun. 2021, 12, 377. [Google Scholar] [CrossRef] [PubMed]
- Henslee, E.A.; Crosby, P.; Kitcatt, S.J.; Parry, J.S.W.; Bernardini, A.; Abdallat, R.G.; Braun, G.; Fatoyinbo, H.O.; Harrison, E.J.; Edgar, R.S.; et al. Rhythmic Potassium Transport Regulates the Circadian Clock in Human Red Blood Cells. Nat. Commun. 2017, 8, 1978. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.B.; Morison, S.E. References and Notes. Some Early Tools Am. Sci. 2014, 320, 177–190. [Google Scholar] [CrossRef]
- Mason, I.C.; Qian, J.; Adler, G.K.; Scheer, F.A.J.L. Impact of Circadian Disruption on Glucose Metabolism: Implications for Type 2 Diabetes. Diabetologia 2020, 63, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Qian, J.; Man, C.D.; Morris, C.J.; Cobelli, C.; Program, C.; Disorders, C.; Hospital, W. Differential Effects of the Circadian System and Circadian Misalignment. Diabetes Obes. Metab. 2019, 20, 2481–2485. [Google Scholar] [CrossRef]
- Kielbasa, S.M.; Heine, M.; Dame, C.; Bozek, K.; Relo, A. Regulation of Clock-Controlled Genes in Mammals. PloS ONE 2009, 4, e4882. [Google Scholar] [CrossRef]
- Yu, L.R.; Stewart, N.A.; Veenstra, T.D. Proteomics: The Deciphering of the Functional Genome. In Essentials of Genomic and Personalized Medicine; Academic Press: Cambridge, MA, USA, 2010; pp. 89–96. [Google Scholar] [CrossRef]
- Stenvers, D.J.; Jongejan, A.; Atiqi, S.; Vreijling, J.P.; Limonard, E.J.; Endert, E.; Baas, F.; Moerland, P.D.; Fliers, E.; Kalsbeek, A.; et al. Diurnal Rhythms in the White Adipose Tissue Transcriptome Are Disturbed in Obese Individuals with Type 2 Diabetes Compared with Lean Control Individuals. Diabetologia 2019, 62, 704–716. [Google Scholar] [CrossRef]
- Baker, C.L.; Kettenbach, A.N.; Loros, J.J.; Gerber, S.A.; Dunlap, J.C. Quantitative Proteomics Reveals a Dynamic Interactome and Phase-Specific Phosphorylation in the Neurospora Circadian Clock. Mol. Cell 2009, 34, 354–363. [Google Scholar] [CrossRef]
- Ang, J.E.; Revell, V.; Mann, A.; Mäntele, S.; Otway, D.T.; Johnston, J.D.; Thumser, A.E.; Skene, D.J.; Raynaud, F. Identification of Human Plasma Metabolites Exhibiting Time-of-Day Variation Using an Untargeted Liquid Chromatographymass Spectrometry Metabolomic Approach. Chronobiol. Int. 2012, 29, 868–881. [Google Scholar] [CrossRef] [Green Version]
- Isherwood, C.M.; Van der Veen, D.R.; Johnston, J.D.; Skene, D.J. Twenty-Four-Hour Rhythmicity of Circulating Metabolites: Effect of Body Mass and Type 2 Diabetes. FASEB J. 2017, 31, 5557–5567. [Google Scholar] [CrossRef] [PubMed]
- Thorens, B. GLUT2, Glucose Sensing and Glucose Homeostasis. Diabetologia 2015, 58, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Boucher, J.; Kleinridders, A.; Kahn, C.R. Insulin Receptor Signaling in Normal. Cold Spring Harb. Perspect. Biol. 2014, 6, a009191. [Google Scholar] [CrossRef] [PubMed]
- Kalsbeek, A.; Yi, C.X.; La Fleur, S.E.; Fliers, E. The Hypothalamic Clock and Its Control of Glucose Homeostasis. Trends Endocrinol. Metab. 2010, 21, 402–410. [Google Scholar] [CrossRef]
- Mühlbauer, E.; Wolgast, S.; Finckh, U.; Peschke, D.; Peschke, E. Indication of Circadian Oscillations in the Rat Pancreas. FEBS Lett. 2004, 564, 91–96. [Google Scholar] [CrossRef]
- Silver, R.; LeSauter, J.; Tresco, P.A.; Lehman, M.N. A Diffusible Coupling Signal from the Transplanted Suprachiasmatic Nucleus Controlling Circadian Locomotor Rhythms. Nature 1996, 382, 810–813. [Google Scholar] [CrossRef]
- De la Iglesia, H.O.; Meyer, J.; Schwartz, W.J. Lateralization of Circadian Pacemaker Output: Activation of Left- and Right-Sided Luteinizing Hormone-Releasing Hormone Neurons Involves a Neural Rather than a Humoral Pathway. J. Neurosci. 2003, 23, 7412–7414. [Google Scholar] [CrossRef]
- Welsh, D.K.; Takahashi, J.S.; Kay, S.A. Suprachiasmatic Nucleus: Cell Autonomy and Network Properties. Annu. Rev. Physiol. 2009, 72, 551–577. [Google Scholar] [CrossRef]
- Herzog, E.D. Neurons and Networks in Daily Rhythms. Nat. Rev. Neurosci. 2007, 8, 790–802. [Google Scholar] [CrossRef]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and Peripheral Clocks. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [Green Version]
- Astiz, M.; Heyde, I.; Oster, H. Mechanisms of Communication in the Mammalian Circadian Timing System. Int. J. Mol. Sci. 2019, 20, 343. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Honma, S.; Shinohara, K.; Honma, K.I. Substance P Receptor Regulates the Photic Induction of Fos-like Protein in the Suprachiasmatic Nucleus of Syrian Hamsters. Brain Res. 1996, 708, 135–142. [Google Scholar] [CrossRef] [PubMed]
- La Fleur, S.E.; Kalsbeek, A.; Wortel, J.; Fekkes, M.L.; Buijs, R.M. A Daily Rhythm in Glucose Tolerance: A Role for the Suprachiasmatic Nucleus. Diabetes 2001, 50, 1237–1243. [Google Scholar] [CrossRef]
- Voigt, R.M.; Forsyth, C.B.; Keshavarzian, A. Circadian Rhythms: A Regulator of Gastrointestinal Health and Dysfunction. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 411–424. [Google Scholar] [CrossRef]
- Vasey, C.; McBride, J.; Penta, K. Circadian Rhythm Dysregulation and Restoration: The Role of Melatonin. Nutrients 2021, 13, 3480. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Terada, T.; Shimakura, J.; Katsura, T.; Inui, K.I. Regulatory Mechanism Governing the Diurnal Rhythm of Intestinal H +/Peptide Cotransporter 1 (PEPT1). Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the Gut Microbiota in Nutrition and Health. BMJ 2018, 361, 36–44. [Google Scholar] [CrossRef]
- Liang, X.; Bushman, F.D.; FitzGerald, G.A. Rhythmicity of the Intestinal Microbiota Is Regulated by Gender and the Host Circadian Clock. Proc. Natl. Acad. Sci. USA 2015, 112, 10479–10484. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Sailani, M.R.; Contrepois, K.; Zhou, Y.; Ahadi, S.; Leopold, S.R.; Zhang, M.J.; Rao, V.; Avina, M.; Mishra, T.; et al. Longitudinal Multi-Omics of Host–Microbe Dynamics in Prediabetes. Nature 2019, 569, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.; Wang, Y.; Li, Y.; Ye, C.; Ruhn, K.A.; Behrendt, C.L.; Olson, E.N.; Hooper, L.V. The Intestinal Microbiota Programs Diurnal Rhythms in Host Metabolism through Histone Deacetylase 3. Science 2019, 365, 1428–1434. [Google Scholar] [CrossRef]
- Reitmeier, S.; Kiessling, S.; Clavel, T.; List, M.; Almeida, E.L.; Ghosh, T.S.; Neuhaus, K.; Grallert, H.; Linseisen, J.; Skurk, T.; et al. Arrhythmic Gut Microbiome Signatures Predict Risk of Type 2 Diabetes. Cell Host Microbe 2020, 28, 258–272.e6. [Google Scholar] [CrossRef] [PubMed]
- Beli, E.; Prabakaran, S.; Krishnan, P.; Evans-Molina, C.; Grant, M.B. Loss of Diurnal Oscillatory Rhythms in Gut Microbiota Correlates with Changes in Circulating Metabolites in Type 2 Diabetic Db/Db Mice. Nutrients 2019, 11, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.; Shibata, S. Circadian Rhythms of Liver Physiology and Disease: Experimental and Clinical Evidence. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Ishida, A.; Mutoh, T.; Ueyama, T.; Bando, H.; Masubuchi, S.; Nakahara, D.; Tsujimoto, G.; Okamura, H. Light Activates the Adrenal Gland: Timing of Gene Expression and Glucocorticoid Release. Cell Metab. 2005, 2, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Koronowski, K.B.; Kinouchi, K.; Welz, P.S.; Smith, J.G.; Zinna, V.M.; Shi, J.; Samad, M.; Chen, S.; Magnan, C.N.; Kinchen, J.M.; et al. Defining the Independence of the Liver Circadian Clock. Cell 2019, 177, 1448–1462.e14. [Google Scholar] [CrossRef]
- Jacobi, D.; Liu, S.; Burkewitz, K.; Kory, N.; Knudsen, N.H.; Alexander, R.K.; Unluturk, U.; Li, X.; Kong, X.; Hyde, A.L.; et al. Hepatic Bmal1 Regulates Rhythmic Mitochondrial Dynamics and Promotes Metabolic Fitness. Cell Metab. 2015, 22, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Vollmers, C.; Gill, S.; DiTacchio, L.; Pulivarthy, S.R.; Le, H.D.; Panda, S. Time of Feeding and the Intrinsic Circadian Clock Drive Rhythms in Hepatic Gene Expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21453–21458. [Google Scholar] [CrossRef] [PubMed]
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017, 25, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Mauvoisin, D.; Atger, F.; Dayon, L.; Núñez Galindo, A.; Wang, J.; Martin, E.; Da Silva, L.; Montoliu, I.; Collino, S.; Martin, F.P.; et al. Circadian and Feeding Rhythms Orchestrate the Diurnal Liver Acetylome. Cell Rep. 2017, 20, 1729–1743. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Han, Z.; Yang, P.; Zhu, L.; Hua, Z.; Zhang, J. Loss of Clock Gene MPer2 Promotes Liver Fibrosis Induced by Carbon Tetrachloride. Hepatol. Res. 2010, 40, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Grimshaw, J.M. What Do We Really Know about the Afromontane Archipelago? Syst. Geogr. Plants 2001, 71, 949–957. [Google Scholar] [CrossRef]
- van Moorsel, D.; Hansen, J.; Havekes, B.; Scheer, F.A.J.L.; Jörgensen, J.A.; Hoeks, J.; Schrauwen-Hinderling, V.B.; Duez, H.; Lefebvre, P.; Schaper, N.C.; et al. Demonstration of a Day-Night Rhythm in Human Skeletal Muscle Oxidative Capacity. Mol. Metab. 2016, 5, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Shavlakadze, T.; Anwari, T.; Soffe, Z.; Cozens, G.; Mark, P.J.; Gondro, C.; Grounds, M.D. Impact of Fasting on the Rhythmic Expression of Myogenic and Metabolic Factors in Skeletal Muscle of Adult Mice. Am. J. Physiol. Cell Physiol. 2013, 305, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Harfmann, B.D.; Schroder, E.A.; Kachman, M.T.; Hodge, B.A.; Zhang, X.; Esser, K.A. Muscle-Specific Loss of Bmal1 Leads to Disrupted Tissue Glucose Metabolism and Systemic Glucose Homeostasis. Skelet. Muscle 2016, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Wefers, J.; Van Moorsel, D.; Hansen, J.; Connell, N.J.; Havekes, B.; Hoeks, J.; Van MarkenLichtenbelt, W.D.; Duez, H.; Phielix, E.; Kalsbeek, A.; et al. Circadian Misalignment Induces Fatty Acid Metabolism Gene Profiles and Compromises Insulin Sensitivity in Human Skeletal Muscle. Proc. Natl. Acad. Sci. USA 2018, 115, 7789–7794. [Google Scholar] [CrossRef]
- Gabriel, B.M.; Altintaş, A.; Smith, J.A.B.; Sardon-Puig, L.; Zhang, X.; Basse, A.L.; Laker, R.C.; Gao, H.; Liu, Z.; Dollet, L.; et al. Disrupted Circadian Oscillations in Type 2 Diabetes Are Linked to Altered Rhythmic Mitochondrial Metabolism in Skeletal Muscle. Sci. Adv. 2021, 7, eabi9654. [Google Scholar] [CrossRef]
- Elia, M.; Carter, A.; Bacon, S.; Winearls, C.G.; Smith, R. Clinical Usefulness of Urinary 3-Methylhistidine Excretion in Indicating Muscle Protein Breakdown. Br. Med. J. 1981, 282, 351–354. [Google Scholar] [CrossRef]
- Dyar, K.A.; Lutter, D.; Artati, A.; Ceglia, N.J.; Liu, Y.; Armenta, D.; Jastroch, M.; Schneider, S.; de Mateo, S.; Cervantes, M.; et al. Atlas of Circadian Metabolism Reveals System-Wide Coordination and Communication between Clocks. Cell 2018, 174, 1571–1585.e11. [Google Scholar] [CrossRef]
- Mårtensson, C.U.; Doan, K.N.; Becker, T. Effects of Lipids on Mitochondrial Functions. Biochim. Biophys. ActaMol. Cell Biol. Lipids 2017, 1862, 102–113. [Google Scholar] [CrossRef]
- Harfmann, B.D.; Schroder, E.A.; Esser, K.A. Circadian Rhythms, the Molecular Clock, and Skeletal Muscle. J. Biol. Rhythm. 2015, 30, 84–94. [Google Scholar] [CrossRef]
- Savikj, M.; Stocks, B.; Sato, S.; Caidahl, K.; Krook, A.; Deshmukh, A.S.; Zierath, J.R.; Wallberg-Henriksson, H. Exercise Timing Influences Multi-Tissue Metabolome and Skeletal Muscle Proteome Profiles in Type 2 Diabetic Patients—A Randomized Crossover Trial. Metabolism 2022, 135, 155268. [Google Scholar] [CrossRef] [PubMed]
- Hoehn, K.L.; Salmon, A.B.; Hohnen-Behrens, C.; Turner, N.; Hoy, A.J.; Maghzal, G.J.; Stocker, R.; Van Remmen, H.; Kraegen, E.W.; Cooney, G.J.; et al. Insulin Resistance Is a Cellular Antioxidant Defense Mechanism. Proc. Natl. Acad. Sci. USA 2009, 106, 17787–17792. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Wong, F.S.; Pearson, J.A. Circadian Rhythms and Pancreas Physiology: A Review. Front. Endocrinol. 2022, 13, 1865. [Google Scholar] [CrossRef] [PubMed]
- Czech, M.P. Insulin Action and Resistance in Obesity and Type 2 Diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Leproult, R.; Holmbäck, U.; Van Cauter, E. Circadian Misalignment Augments Markers of Insulin Resistance and Inflammation, Independently of Sleep Loss. Diabetes 2014, 63, 1860–1869. [Google Scholar] [CrossRef]
- Petrenko, V.; Gandasi, N.R.; Sage, D.; Tengholm, A.; Barg, S.; Dibner, C. In Pancreatic Islets from Type 2 Diabetes Patients, the Dampened Circadian Oscillators Lead to Reduced Insulin and Glucagon Exocytosis. Proc. Natl. Acad. Sci. USA 2020, 117, 2484–2495. [Google Scholar] [CrossRef]
- Stamenkovic, J.A.; Olsson, A.H.; Nagorny, C.L.; Malmgren, S.; Dekker-Nitert, M.; Ling, C.; Mulder, H. Regulation of Core Clock Genes in Human Islets. Metabolism 2012, 61, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Takamura, T.; Matsuzawa-Nagata, N.; Shima, K.R.; Eto, T.; Misu, H.; Shiramoto, M.; Tsuru, T.; Irie, S.; Fujimura, A.; et al. Clock Gene Expression in Peripheral Leucocytes of Patients with Type 2 Diabetes. Diabetologia 2009, 52, 329–335. [Google Scholar] [CrossRef]
- Hannich, J.T.; Loizides-Mangold, U.; Sinturel, F.; Harayama, T.; Vandereycken, B.; Saini, C.; Gosselin, P.; Brulhart-Meynet, M.C.; Robert, M.; Chanon, S.; et al. Ether Lipids, Sphingolipids and Toxic 1-Deoxyceramides as Hallmarks for Lean and Obese Type 2 Diabetic Patients. Acta Physiol. 2021, 232, e13610. [Google Scholar] [CrossRef]
- Petrenko, V.; Sinturel, F.; Loizides-Mangold, U.; Montoya, J.P.; Chera, S.; Riezman, H.; Dibner, C. Type 2 Diabetes Disrupts Circadian Orchestration of Lipid Metabolism and Membrane Fluidity in Human Pancreatic Islets. PLoS Biol. 2022, 20, e3001725. [Google Scholar] [CrossRef]
- Crnko, S.; Du Pré, B.C.; Sluijter, J.P.G.; Van Laake, L.W. Circadian Rhythms and the Molecular Clock in Cardiovascular Biology and Disease. Nat. Rev. Cardiol. 2019, 16, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Cheng, Y.; Kapoor, S.; Reilly, D.; Price, T.S.; FitzGerald, G.A. Circadian Variation of Blood Pressure and the Vascular Response to Asynchronous Stress. Proc. Natl. Acad. Sci. USA 2007, 104, 3450–3455. [Google Scholar] [CrossRef] [PubMed]
- Martín-Timón, I. Type 2 Diabetes and Cardiovascular Disease: Have All Risk Factors the Same Strength? World J. Diabetes 2014, 5, 444. [Google Scholar] [CrossRef] [PubMed]
- Paneni, F.; Beckman, J.A.; Creager, M.A.; Cosentino, F. Diabetes and Vascular Disease: Pathophysiology, Clinical Consequences, and Medical Therapy: Part I. Eur. Heart J. 2013, 34, 2436–2446. [Google Scholar] [CrossRef]
- Škrlec, I.; Milić, J.; Cilenšek, I.; Petrovič, D.; Wagner, J.; Peterlin, B. Circadian Clock Genes and Myocardial Infarction in Patients with Type 2 Diabetes Mellitus. Gene 2019, 701, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.E. Circadian Variation and Triggering of Acute Coronary Events. Am. Heart J. 1999, 137, S1–S8. [Google Scholar] [CrossRef]
- Moschos, N.; Christoforaki, M.; Antonatos, P. Seasonal Distribution of Acute Myocardial Infarction and Its Relation to Acute Infections in a Mild Climate. Int. J. Cardiol. 2004, 93, 39–44. [Google Scholar] [CrossRef]
- Guo, Y.F.; Stein, P.K. Circadian Rhythm in the Cardiovascular System: Chronocardiology. Am. Heart J. 2003, 145, 779–786. [Google Scholar] [CrossRef]
- Daios, S.; Savopoulos, C.; Kanellos, I.; Goudis, C.A.; Nakou, I.; Petalloti, S.; Hadjidimitriou, N.; Pilalas, D.; Ziakas, A.; Kaiafa, G. Article Circadian Pattern of Acute Myocardial Infarction and Atrial Fibrillation in a Mediterranean Country: A Study in Diabetic Patients. Medicina 2021, 57, 41. [Google Scholar] [CrossRef]
- Lautt, W.W. Postprandial Insulin Resistance as an Early Predictor of Cardiovascular Risk. Ther. Clin. Risk Manag. 2007, 3, 761–770. [Google Scholar] [CrossRef]
- Storch, K.; Lipan, O.; Leykin, I.; Viswanathan, N. Extensive and Divergent Circadian Gene Expression in Liver and Heart. Nature 2002, 417, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Viswambharan, H.; Carvas, J.M.; Antic, V.; Marecic, A.; Jud, C.; Zaugg, C.E.; Ming, X.F.; Montani, J.P.; Albrecht, U.; Yang, Z. Mutation of the Circadian Clock Gene Per2 Alters Vascular Endothelial Function. Circulation 2007, 115, 2188–2195. [Google Scholar] [CrossRef] [PubMed]
- Nakao, T.; Kohsaka, A.; Otsuka, T.; Thein, Z.L.; Le, H.T.; Waki, H.; Gouraud, S.S.; Ihara, H.; Nakanishi, M.; Sato, F.; et al. Impact of Heart-Specific Disruption of the Circadian Clock on Systemic Glucose Metabolism in Mice. Chronobiol. Int. 2018, 35, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.M.; Carter, A.M.; Grant, P.J. Association between Polymorphisms in the Clock Gene, Obesity and the Metabolic Syndrome in Man. Int. J. Obes. 2008, 32, 658–662. [Google Scholar] [CrossRef]
- Hsieh, P.N.; Zhang, L.; Jain, M.K. Coordination of Cardiac Rhythmic Output and Circadian Metabolic Regulation in the Heart. Cell. Mol. Life Sci. 2018, 75, 403–416. [Google Scholar] [CrossRef]
- Podobed, P.; Glen Pyle, W.; Ackloo, S.; Alibhai, F.J.; Tsimakouridze, E.V.; Ratcliffe, W.F.; Mackay, A.; Simpson, J.; Wright, D.C.; Kirby, G.M.; et al. The Day/Night Proteome in the Murine Heart. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, 121–137. [Google Scholar] [CrossRef]
- Mészáros, K.; Pruess, L.; Szabó, A.J.; Gondan, M.; Ritz, E.; Schaefer, F. Development of the Circadian Clockwork in the Kidney. Kidney Int. 2014, 86, 915–922. [Google Scholar] [CrossRef]
- Stow, L.R.; Gumz, M.L. The Circadian Clock in the Kidney. J. Am. Soc. Nephrol. 2011, 22, 598–604. [Google Scholar] [CrossRef]
- Johnston, J.G.; Pollock, D.M. Circadian Regulation of Renal Function. Free Radic. Biol. Med. 2018, 119, 93–107. [Google Scholar] [CrossRef]
- Scott, R.P.; Quaggin, S.E. The Cell Biology of Renal Filtration. J. Cell Biol. 2015, 209, 199–210. [Google Scholar] [CrossRef]
- Koopman, M.G.; Koomen, G.C.M.; Krediet, R.T.; De Moor, E.A.-M.; Hoek, F.J.; Arisz, L. Circadian Rhythm of Glomerular Filtration Rate in Normal Individuals. Clin. Sci. 1989, 77, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Koopman, M.G.; Koomen, G.C.M.; Van Acker, B.A.C.; Arisz, L. Circadian Rhythm in Glomerular Transport of Macromolecules through Large Pores and Shunt Pathway. Kidney Int. 1996, 49, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.M.; Chen, W.L.; Yuan, J.P.; Yang, Y.H.; Mei, Q.H.; Huang, L.X. Altered Diurnal Variation and Localization of Clock Proteins in the Remnant Kidney of 5/6 Nephrectomy Rats. Nephrology 2013, 18, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Ledoussal, C.; Lorenz, J.N.; Nieman, M.L.; Soleimani, M.; Schultheis, P.J.; Shull, G.E. Renal Salt Wasting in Mice Lacking NHE3 Na+/H+ Exchanger but Not in Mice Lacking NHE2. Am. J. Physiol. Ren. Physiol. 2001, 281, 718–727. [Google Scholar] [CrossRef]
- Kanai, Y.; Lee, W.; You, G.; Brown, D.; Hediger, M.A. The Human Kidney Low Affinity Na+/Glucose Cotransporter SGL 2. J. Clin. Investig. 1994, 93, 397–404. [Google Scholar] [CrossRef]
- Lee, W.S.; Kanai, Y.; Wells, R.G.; Hediger, M.A. The High Affinity Na+_glucose Cotransporter. Re-Evaluation of Function and Distribution of Expression. J. Biol. Chem. 1994, 269, 12032–12039. [Google Scholar] [CrossRef]
- Sturrock, N.D.C.; Georget, E.; Pound, N.; Stevenson, J.; Peck, G.M.; Sowter, H. Non-Dipping Circadian Blood Pressure and Renal Impairment Are Associated with Increased Mortality in Diabetes Mellitus. Diabet. Med. 2000, 17, 360–364. [Google Scholar] [CrossRef]
- Vörös, P.; Lengyel, Z.; Nagy, V.; Németh, C.; Rosivall, L.; Kammerer, L. Diurnal Blood Pressure Variation and Albuminuria in Normotensive Patients with Insulin-Dependent Diabetes Mellitus. Nephrol. Dial. Transplant. 1998, 13, 2257–2260. [Google Scholar] [CrossRef]
- Felício, J.S.; de Souza, A.C.C.B.; Kohlmann, N.; Kohlmann, O.; Ribeiro, A.B.; Zanella, M.T. Nocturnal Blood Pressure Fall as Predictor of Diabetic Nephropathy in Hypertensive Patients with Type 2 Diabetes. Cardiovasc. Diabetol. 2010, 9, 36. [Google Scholar] [CrossRef]
- Gumz, M.L.; Stow, L.R.; Lynch, I.J.; Greenlee, M.M.; Rudin, A.; Cain, B.D.; Weaver, D.R.; Wingo, C.S. The Circadian Clock Protein Period 1 Regulates Expression of the Renal Epithelial Sodium Channel in Mice. J. Clin. Investig. 2009, 119, 2423–2434. [Google Scholar] [CrossRef] [Green Version]
- Depner, C.M.; Melanson, E.L.; McHill, A.W.; Wright, K.P. Mistimed Food Intake and Sleep Alters 24-Hour Time-of-Day Patterns of the Human Plasma Proteome. Proc. Natl. Acad. Sci. USA 2018, 115, E5390–E5399. [Google Scholar] [CrossRef] [PubMed]
- Barnea, M.; Haviv, L.; Gutman, R.; Chapnik, N.; Madar, Z.; Froy, O. Metformin Affects the Circadian Clock and Metabolic Rhythms in a Tissue-Specific Manner. Biochim. Biophys. ActaMol. Basis Dis. 2012, 1822, 1796–1806. [Google Scholar] [CrossRef] [PubMed]
- Gooley, J.J.; Chamberlain, K.; Smith, K.A.; Khalsa, S.B.S.; Rajaratnam, S.M.W.; Van Reen, E.; Zeitzer, J.M.; Czeisler, C.A.; Lockley, S.W. Exposure to Room Light before Bedtime Suppresses Melatonin Onset and Shortens Melatonin Duration in Humans. J. Clin. Endocrinol. Metab. 2011, 96, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Forrestel, A.C.; Miedlich, S.U.; Yurcheshen, M.; Wittlin, S.D.; Sellix, M.T. Chronomedicine and Type 2 Diabetes: Shining Some Light on Melatonin. Diabetologia 2017, 60, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Garfinkel, D.; Zorin, M.; Wainstein, J.; Matas, Z.; Laudon, M.; Zisapel, N. Efficacy and Safety of Prolonged-Release Melatonin in Insomnia Patients with Diabetes: A Randomized, Double-Blind, Crossover Study. Diabetes Metab. Syndr. Obes. Targets Ther. 2011, 4, 307–313. [Google Scholar] [CrossRef]
- Duez, H.; Staels, B. The Nuclear Receptors Rev-Erbs and RORs Integrate Circadian Rhythms and Metabolism. Diabetes Vasc. Dis. Res. 2008, 5, 82–88. [Google Scholar] [CrossRef]
- Mulvihill, E.E.; Assini, J.M.; Lee, J.K.; Allister, E.M.; Sutherland, B.G.; Koppes, J.B.; Sawyez, C.G.; Edwards, J.Y.; Telford, D.E.; Charbonneau, A.; et al. Nobiletin Attenuates VLDL Overproduction, Dyslipidemia, and Atherosclerosis in Mice with Diet-Induced Insulin Resistance. Diabetes 2011, 60, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.P.; McHill, A.W.; Birks, B.R.; Griffin, B.R.; Rusterholz, T.; Chinoy, E.D. Entrainment of the Human Circadian Clock to the Natural Light-Dark Cycle. Curr. Biol. 2013, 23, 1554–1558. [Google Scholar] [CrossRef]
- Comtet, H.; Geoffroy, P.A.; Kobayashi Frisk, M.; Hubbard, J.; Robin-Choteau, L.; Calvel, L.; Hugueny, L.; Viola, A.U.; Bourgin, P. Light Therapy with Boxes or Glasses to Counteract Effects of Acute Sleep Deprivation. Sci. Rep. 2019, 9, 18073. [Google Scholar] [CrossRef] [PubMed]
- Eymard, L.; Reverdin, G. Ocean-Atmosphere Interactions. Ocean Earth Syst. 2014, 9781848217, 105–144. [Google Scholar] [CrossRef]
- Manoogian, E.N.C.; Panda, S. Circadian Rhythms, Time-Restricted Feeding, and Healthy Aging. Ageing Res. Rev. 2017, 39, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Hawley, J.A.; Sassone-Corsi, P.; Zierath, J.R. Chrono-Nutrition for the Prevention and Treatment of Obesity and Type 2 Diabetes: From Mice to Men. Diabetologia 2020, 63, 2253–2259. [Google Scholar] [CrossRef]
- Makwana, K.; Gosai, N.; Poe, A.; Kondratov, R.V. Calorie Restriction Reprograms Diurnal Rhythms in Protein Translation to Regulate Metabolism. FASEB J. 2019, 33, 4473–4489. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Xie, Z.; Ye, Y.; Bahijri, S.; Chen, M. The Beneficial Effects of Intermittent Fasting: An Update on Mechanism, and the Role of Circadian Rhythm and Gut Microbiota. Hepatobiliary Surg. Nutr. 2020, 9, 597–602. [Google Scholar] [CrossRef]
- La Fleur, S.E.; Kalsbeek, A.; Wortel, J.; Buijs, R.M. A Suprachiasmatic Nucleus Generated Rhythm in Basal Glucose Concentrations. J. Neuroendocrinol. 1999, 11, 643–652. [Google Scholar] [CrossRef]


| Dysregulated Clock | Effects |
|---|---|
| Cardiomyocyte-specific Clock-mutant mice | 8% of the proteome exhibits a physiological variation. |
| Cardiomyocyte-specific Bmal1-KO mice | Heart metabolic abnormalities, dilated cardiomyopathy, and early mortality are all caused by the disruption of physiological variation in 10% of the transcriptome. |
| Specific vascular-smooth-muscle cell specific Bmal1-KO mice | 24-h rhythm in blood pressure is distorted, and the moment of its peak has changed. |
| Klf15-KO mice | Greater vulnerability to ventricular arrhythmias and loss of physiological rhythm in ventricular repolarization length. |
| Per2-KO mice | Endothelial dysfunction |
| Bmal1-KO mice | Loss of physiological rhythms in heart rate and blood pressure |
| Dbp−/−Hlf−/−Tef−/− mice | Low levels of aldosterone, cardiomyopathy, cardiac hypertrophy, and left ventricular dysfunction. |
| Gene | Functions | Models |
|---|---|---|
| Aqp2 | Involved in water transport | CCD cell line |
| Aqp4 | Involved in water transport | CCD cell line |
| Slc9a3 (NHE3) | Involved in Sodium/hydrogen exchange | Kidney |
| Gilz | Leucine zipper protein/regulation of sodium transport | DCT, CNT, CCD. Whole kidney |
| V1aR | Vasopressin receptor/regulation of water balance | DCT, CNT, CCD. Whole kidney |
| Slc6a9 | Involved in glycine transport | DCT/CNT |
| Scnn1a (αENaC) | Alpha subunit of epithelial sodium channel | Cortex, outer medulla and inner medulla |
| Model System | Altered Circadian Rhythm Markers | Omics | References |
|---|---|---|---|
| Human (skeletal muscles) | PER and CRY blunted and PPAR gene enhanced in circadian-misaligned men. | Genomics | [66] |
| Human (skeletal muscles) | BMAL1, CLOCK, and PER3 | Genomics | [67] |
| Male mice (BAT) | Bmal1 and Clock showed altered expression and GmprImpdh1and Ucp1 also showed drastic alterations on HFD. | Genomics | [69] |
| Human | Study-I. BMAL1, PER1, PER2, PER3 level reduced by more in T2D than ND Study-II. BMAL1, PER1 and PER3 reduced by more in T2D than ND. | Genomics | [69] |
| Human | Per1-3, Cry2, Rev-erba, Clock, and Dbp levels were lower in T2D compared to ND islet cells. | Genomics | [77] |
| Human | CLOCK, BMAL1, PER1, CRY1, and CRY2 levels were reduced in T2D compared to ND in peripheral blood leucocytes. | Genomics | [28] |
| Human | Per2, Per3, and Cry2 mRNA levels were reduced in T2D | Genomics | [78] |
| Mice | mRNA expression of Per2 and Bmal-1 significantly elevated | Genomics | [53] |
| Human (Plasma) | 62 proteins of 1129 proteins altered, associated with multiple biological functions | Proteomics | [112] |
| KO mice (Cry1/2 & Bmal1) | Deregulated acetylation level of protein involved in TCA and urea cycle. | Proteomics | [60] |
| Human (skeletal muscles) | Lipoproteins were higher and mitochondrial complex III abundance was lower after morning HIT compared to after afternoon HIT. | Proteomics | [72] |
| Male mice | PUFA, diacylglycerols, phospholipids, sphingolipids, glycerolipids, and lysolipids and arginine, proline lysine, BCAA leucine, isoleucine & valine, and dipeptides altered on HFD | Metabolomics | [69] |
| Mice | Plasma glucose levels and hepatic glycogen disturbed by shift work | Metabolomics | [63] |
| Human pancreatic islets | Phosphatidylinositol (PI) and phosphatidylethanolamine [PE]. | Metabolomics | [81] |
| Human (Plasma) | Amino acids, biogenic amines, acylcarnitine, sphingolipids significantly higher and metabolites, including glutamine, histidine, ornithine, serine, (octadecanoyl carnitine), octadecadienylcarnitine, lysoPC, PC, and 2Sphingolipids [SM C16:0 and SM (OH) C16:1], were significantly lower. | Metabolomics | [32] |
| Mice | Histidine and beta alanine showed a significant reduction and histamine and its derivatives increased in the diabetic mice. | Metabolomics | [53] |
| Human | Polyunsaturated triradylglycerol was abundant and phospholipid cardiolipin (CL) was low | Metabolomics | [80] |
| Human | Increased plasma diacylglycerols, skeletal muscle acyl-carnitines, and subcutaneous adipose tissue, and sphingomyelins and lysophospholipids. | Metabolomics | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, A.; Rathor, P.; Trivedi, P.K.; Ch, R. Multi-Omics Reveal Interplay between Circadian Dysfunction and Type2 Diabetes. Biology 2023, 12, 301. https://doi.org/10.3390/biology12020301
Tiwari A, Rathor P, Trivedi PK, Ch R. Multi-Omics Reveal Interplay between Circadian Dysfunction and Type2 Diabetes. Biology. 2023; 12(2):301. https://doi.org/10.3390/biology12020301
Chicago/Turabian StyleTiwari, Ashutosh, Priya Rathor, Prabodh Kumar Trivedi, and Ratnasekhar Ch. 2023. "Multi-Omics Reveal Interplay between Circadian Dysfunction and Type2 Diabetes" Biology 12, no. 2: 301. https://doi.org/10.3390/biology12020301
APA StyleTiwari, A., Rathor, P., Trivedi, P. K., & Ch, R. (2023). Multi-Omics Reveal Interplay between Circadian Dysfunction and Type2 Diabetes. Biology, 12(2), 301. https://doi.org/10.3390/biology12020301