Molecular Anatomy of the Class I Ligase Ribozyme for Elucidation of the Activity-Generating Unit
Abstract
:Simple Summary
Abstract
1. Introduction

2. Materials and Methods
2.1. Preparation of Class I Ligase Ribozyme, Its Mutants, and RNA Substrate
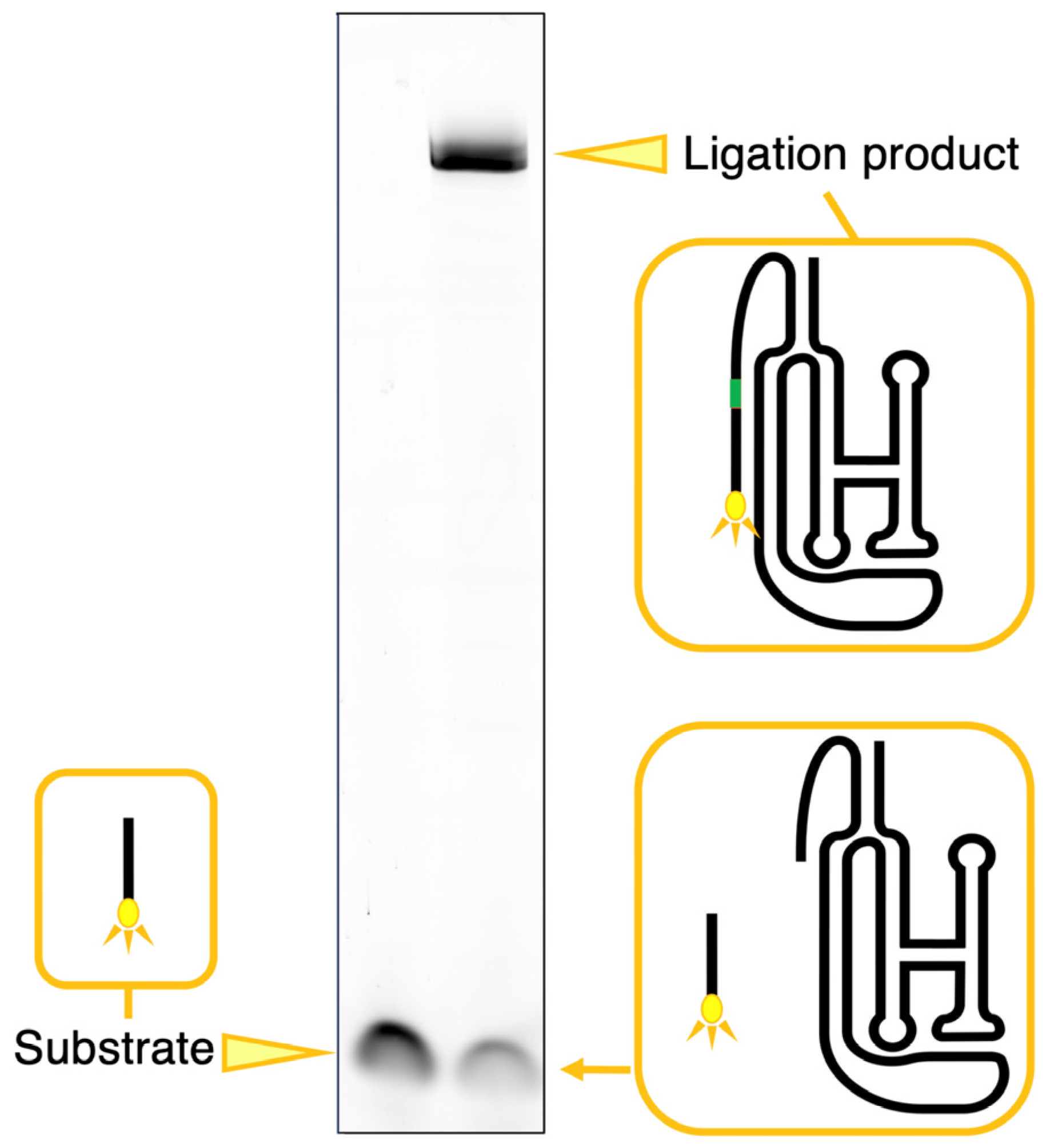
2.2. Analysis of Ligation
3. Results
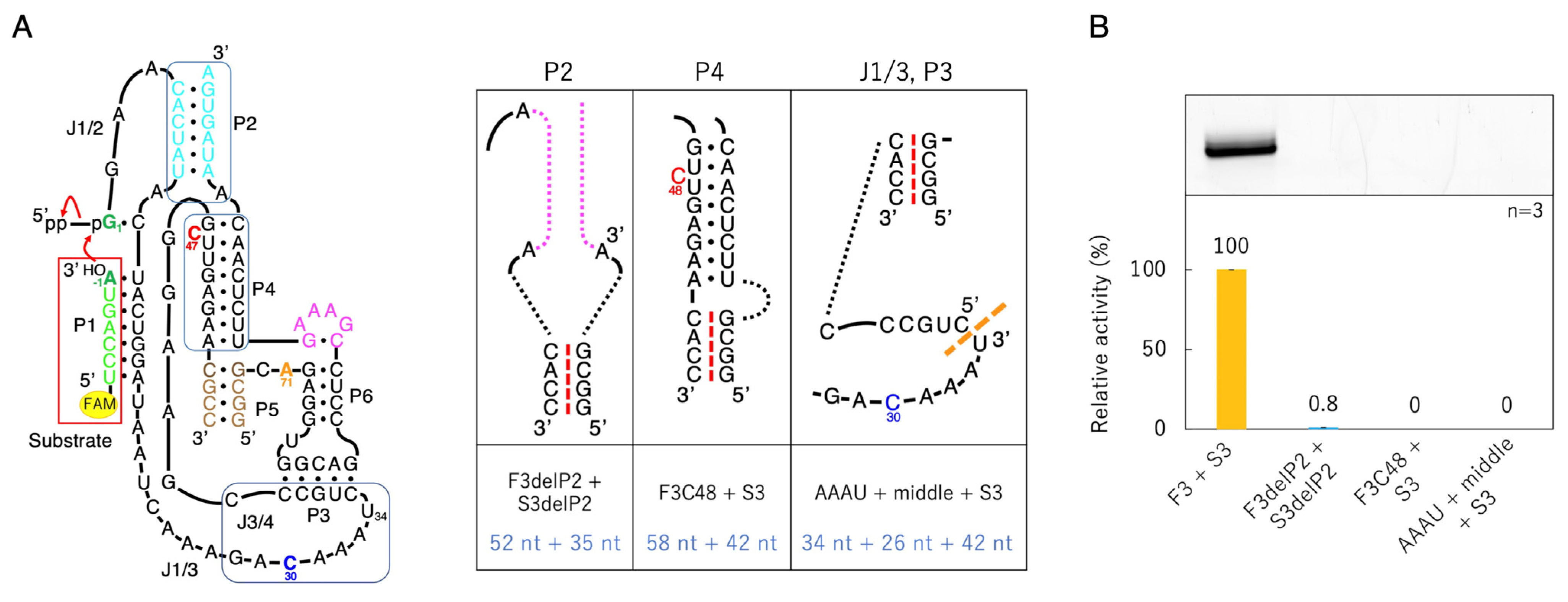
3.1. Ligation Activities of Class I Ribozyme and Deletion Mutants in Regions P7 and P5
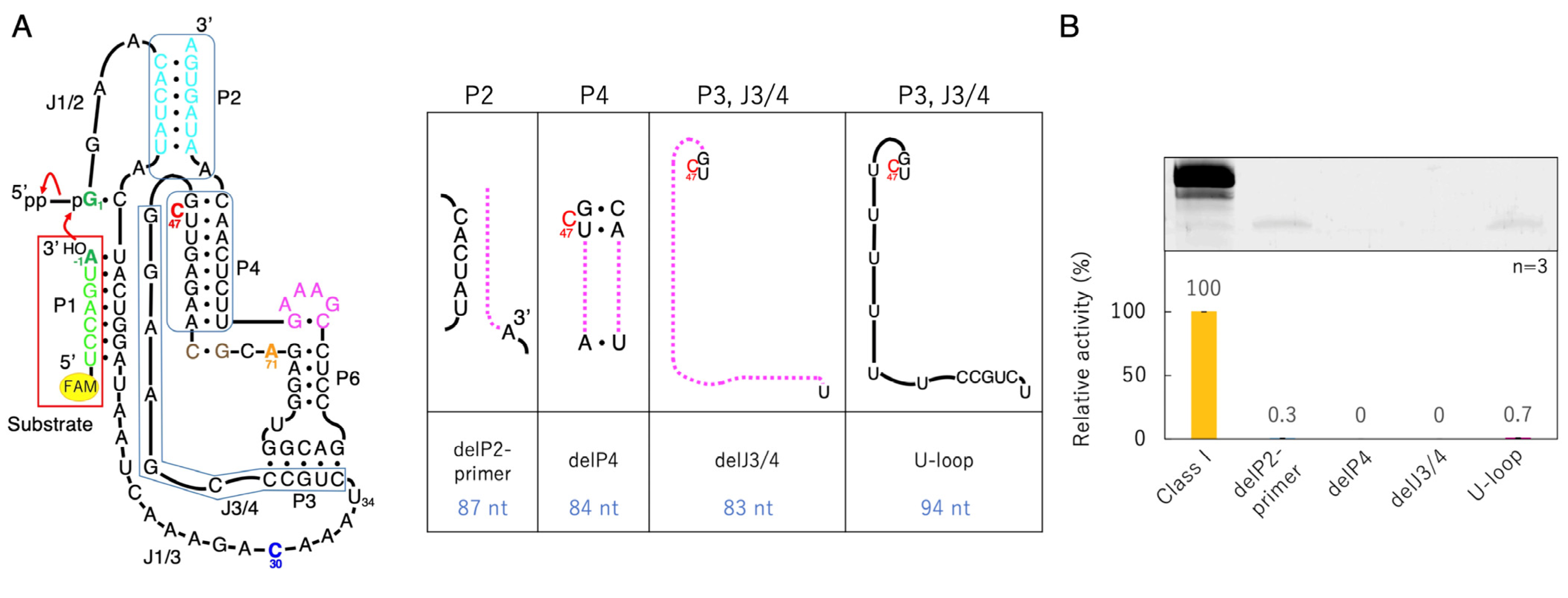
3.2. Ligation Activities of Class I Ribozyme and Deletion Mutants in Regions P2, P4, and J3/4
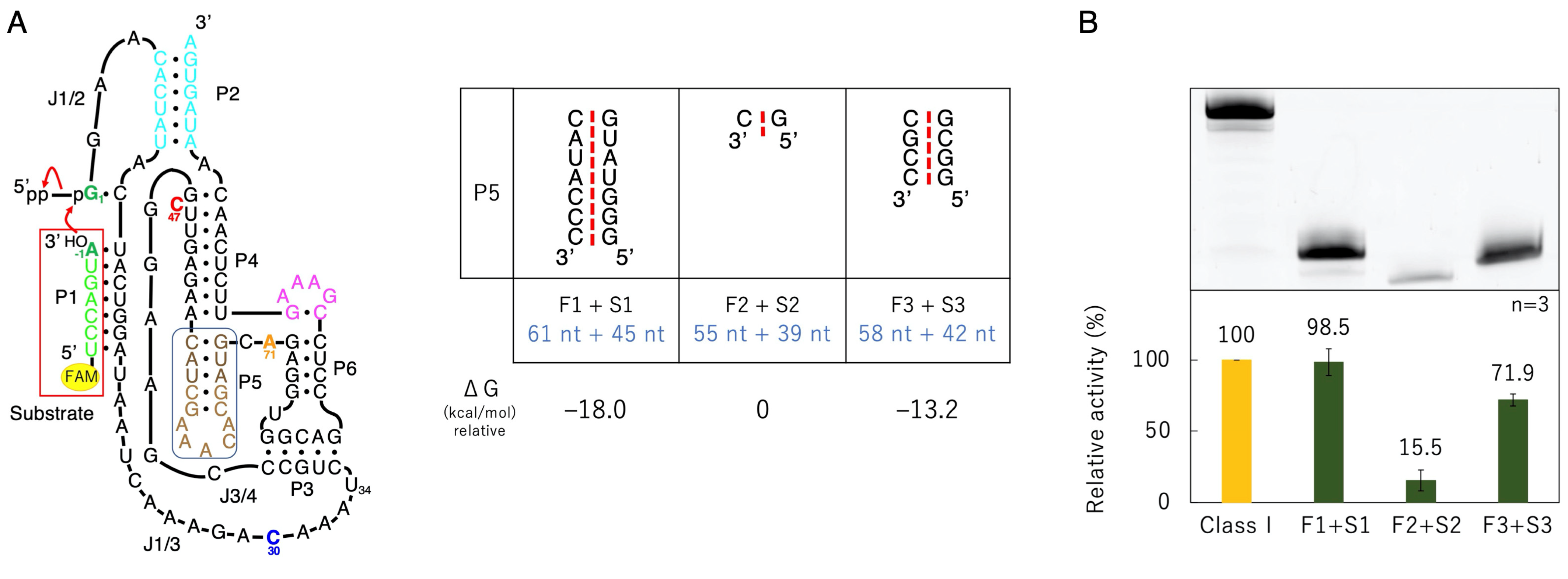
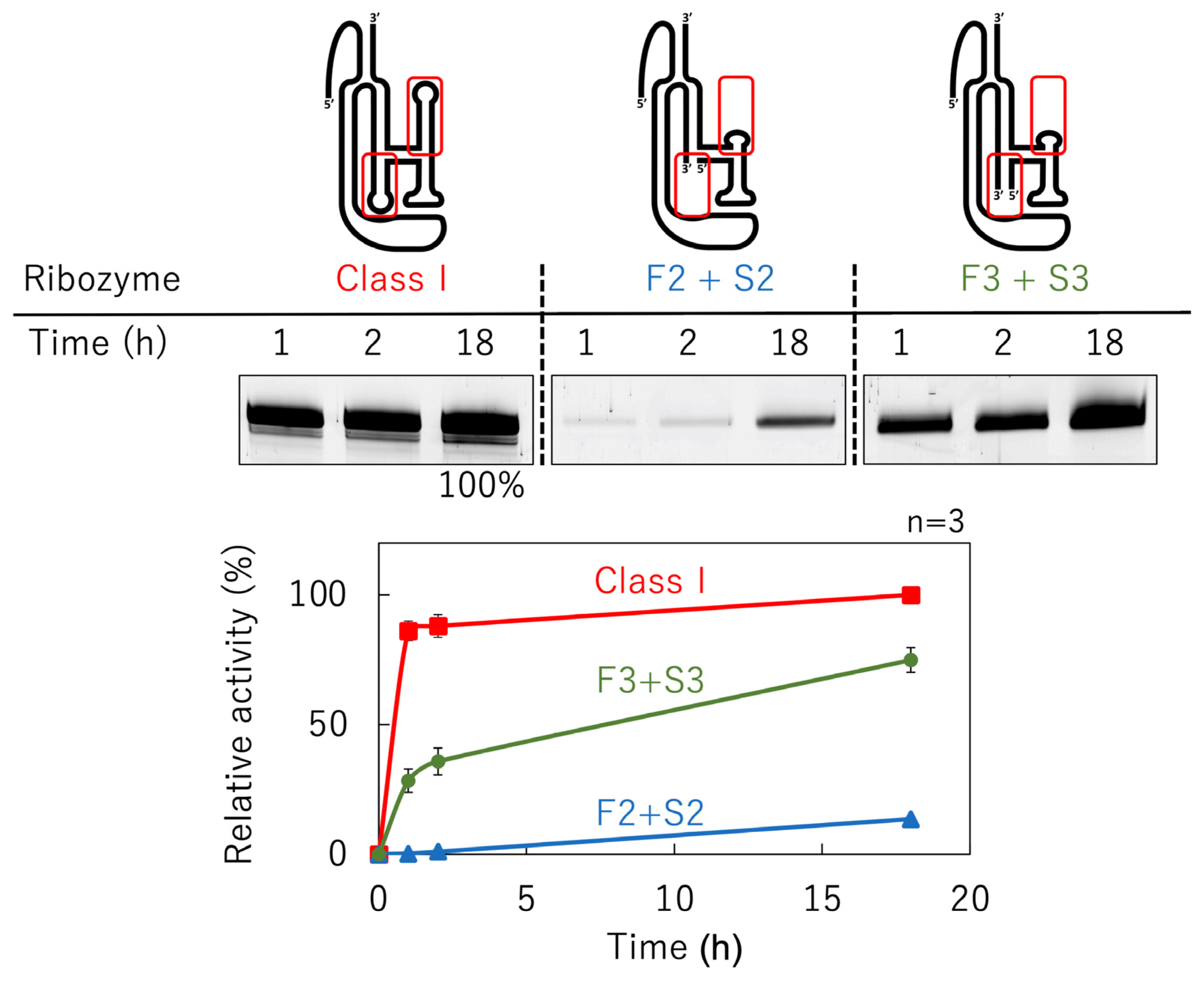
3.3. Ligation Activities of Mutants Split into Two Molecules
3.4. Ligation Activities of Deletion Mutants Based on F3 + S3
3.5. Ligation Activities of Mutants with Active Site Substitution Based on F3 + S3
3.6. Ligation Activities of Mutants with the 47th Base Fixed to A
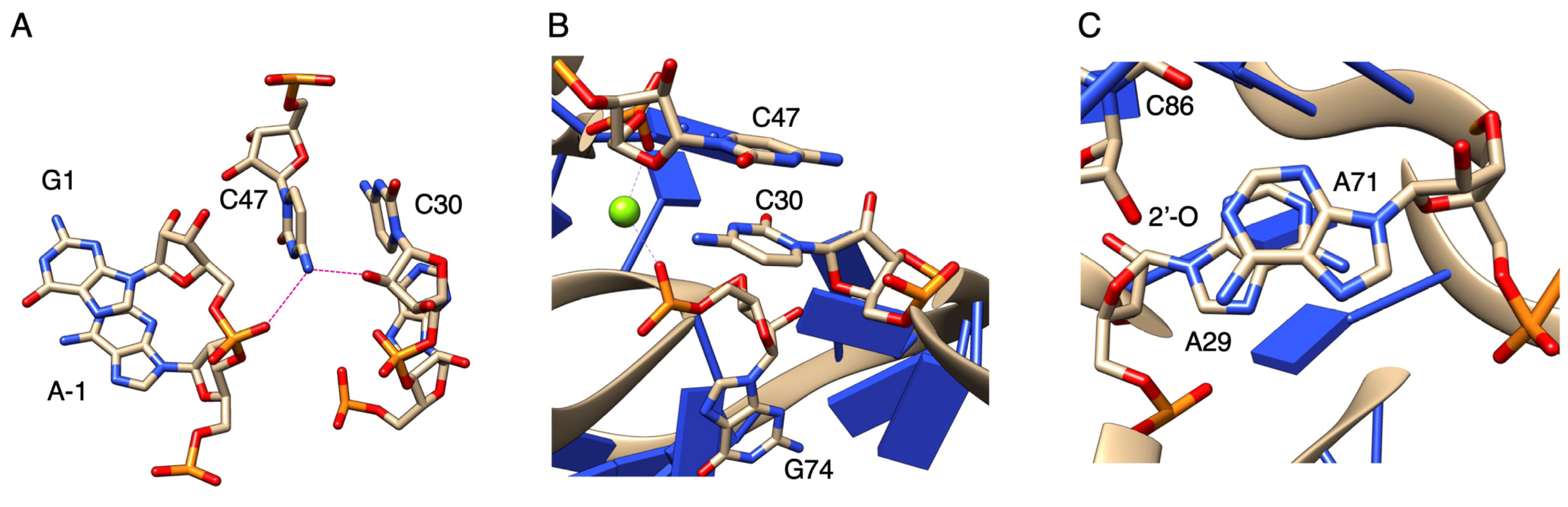
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crick, F.H.C. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 139–163. [Google Scholar]
- Temin, H.M.; Mizutani, S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Tamura, K. Origins and early evolution of the tRNA molecule. Life 2015, 5, 1687–1699. [Google Scholar] [CrossRef] [Green Version]
- Zielinski, W.; Orgel, L. Autocatalytic synthesis of a tetranucleotide analogue. Nature 1987, 327, 346–347. [Google Scholar] [CrossRef]
- Sievers, D.; von Kiedrowski, G. Self-replication of complementary nucleotide-based oligomers. Nature 1994, 369, 221–224. [Google Scholar] [CrossRef]
- Rogers, J.; Joyce, G.F. The effect of cytidine on the structure and function of an RNA ligase ribozyme. RNA 2001, 7, 395–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, M.P.; Ellington, A.D. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 1999, 17, 62–66. [Google Scholar] [CrossRef]
- Ikawa, Y.; Tsuda, K.; Matsumura, S.; Inoue, T. De novo synthesis and development of an RNA enzyme. Proc. Natl. Acad. Sci. USA 2004, 101, 13750–13755. [Google Scholar] [CrossRef]
- Ekland, E.H.; Szostak, J.W.; Bartel, D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 1995, 269, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Johnston, W.K.; Unrau, P.J.; Lawrence, M.S.; Glasner, M.E.; Bartel, D.P. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 2001, 292, 1319–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, M.S.; Bartel, D.P. New ligase-derived RNA polymerase ribozymes. RNA 2005, 11, 1173–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaher, H.S.; Unrau, P.J. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 2007, 13, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Robertson, M.P.; Scott, W.G. The structural basis of ribozyme-catalyzed RNA assembly. Science 2007, 315, 1549–1553. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, E.; Uchida, S.; Umehara, T.; Tamura, K. Development of a functionally minimized mutant of the R3C ligase ribozyme offers insight into the plausibility of the RNA world hypothesis. Biology 2014, 3, 452–465. [Google Scholar] [CrossRef] [Green Version]
- Nomura, Y.; Yokobayashi, Y. Systematic minimization of RNA ligase ribozyme through large-scale design-synthesis-sequence cycles. Nucleic Acids Res. 2019, 47, 8950–8960. [Google Scholar] [CrossRef] [Green Version]
- Tanizawa, K.; Uchida, S.; Kurihara, E.; Umehara, T.; Tamura, K. The kiss switch brings inactive R3C ligase ribozyme back to life. Biology 2018, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamachi, K.; Mutsuro-Aoki, H.; Tanizawa, K.; Hirasawa, I.; Umehara, T.; Tamura, K. Effects of complementary loop composition in truncated R3C ligase ribozymes on kiss switch activation. Biosystems 2019, 177, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Mutsuro-Aoki, H.; Hamachi, K.; Kurihara, R.; Tamura, K. Aminoacylation of short hairpin RNAs through kissing-loop interactions indicates evolutionary trend of RNA molecules. Biosystems 2020, 197, 104206. [Google Scholar] [CrossRef] [PubMed]
- Mutsuro-Aoki, H.; Tamura, K. Acquisition of dual ribozyme-functions in nonfunctional short hairpin RNAs through kissing-loop interactions. Life 2022, 12, 1561. [Google Scholar] [CrossRef] [PubMed]
- Shechner, D.M.; Grant, R.A.; Bagby, S.C.; Koldobskaya, Y.; Piccirilli, J.A.; Bartel, D.P. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 2009, 326, 1271–1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shechner, D.M.; Bartel, D.P. The structural basis of RNA-catalyzed RNA polymerization. Nat. Struct. Mol. Biol. 2011, 18, 1036–1042. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. Hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften 1977, 64, 541–565. [Google Scholar] [CrossRef]
- Ferris, J.P. Montmorillonite-catalysed formation of RNA oligomers: The possible role of catalysis in the origins of life. Phil. Trans. R. Soc. B 2006, 361, 1777–1786. [Google Scholar] [CrossRef] [Green Version]
- Robertson, M.P.; Hesselberth, J.R.; Ellington, A.D. Optimization and optimality of a short ribozyme ligase that joins non-Watson-Crick base pairings. RNA 2001, 7, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Hayden, E.J.; Lehman, N. Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem. Biol. 2006, 13, 909–918. [Google Scholar] [CrossRef] [Green Version]
- Wachowius, F.; Holliger, P. Non-enzymatic assembly of a minimized RNA polymerase ribozyme. ChemSystemsChem 2019, 1, 1–4. [Google Scholar] [CrossRef]
- Akoopie, A.; Müller, U.F. Lower temperature optimum of a smaller, fragmented triphosphorylation ribozyme. Phys. Chem. Chem. Phys. 2016, 18, 20118–20125. [Google Scholar] [CrossRef] [Green Version]
- Bergman, N.H.; Lau, N.C.; Lehnert, V.; Westhof, E.; Bartel, D.P. The three-dimensional architecture of the class I ligase ribozyme. RNA 2004, 10, 176–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamachi, K.; Hayashi, H.; Shimamura, M.; Yamaji, Y.; Kaneko, A.; Fujisawa, A.; Umehara, T.; Tamura, K. Glycols modulate terminator stem stability and ligand-dependency of a glycine riboswitch. BioSystems 2013, 113, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, L.; Michel, F.; Westhof, E. Involvement of a GNRA tetraloop in long-range RNA tertiary interaction. J. Mol. Biol. 1994, 236, 1271–1276. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.; Bartel, D.P.; Szostak, J.W. Kinetic and mechanistic analysis of nonenzymatic, template-directed oligoribonucleotide ligation. J. Am. Chem. Soc. 1996, 118, 3332–3339. [Google Scholar] [CrossRef] [PubMed]
- Tinoco, I., Jr.; Borer, P.N.; Dengler, B.; Levin, M.D.; Uhlenbeck, O.C.; Crothers, D.M.; Bralla, J. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 1973, 246, 40–41. [Google Scholar] [CrossRef]
- Joyce, G.F. A glimpse of biology’s first enzyme. Science 2007, 315, 1507–1508. [Google Scholar] [CrossRef] [Green Version]
- Saenger, W. Principles of Nucleic Acid Structure; Springer: New York, NY, USA, 1984. [Google Scholar]
- Castro, C.; Smidansky, E.D.; Arnold, J.J.; Maksimchuk, K.R.; Moustafa, I.; Uchida, A.; Götte, M.; Konigsberg, W.; Cameron, C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009, 16, 212–218. [Google Scholar] [CrossRef] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasuga, M.; Mutsuro-Aoki, H.; Ando, T.; Tamura, K. Molecular Anatomy of the Class I Ligase Ribozyme for Elucidation of the Activity-Generating Unit. Biology 2023, 12, 1012. https://doi.org/10.3390/biology12071012
Kasuga M, Mutsuro-Aoki H, Ando T, Tamura K. Molecular Anatomy of the Class I Ligase Ribozyme for Elucidation of the Activity-Generating Unit. Biology. 2023; 12(7):1012. https://doi.org/10.3390/biology12071012
Chicago/Turabian StyleKasuga, Miho, Hiromi Mutsuro-Aoki, Tadashi Ando, and Koji Tamura. 2023. "Molecular Anatomy of the Class I Ligase Ribozyme for Elucidation of the Activity-Generating Unit" Biology 12, no. 7: 1012. https://doi.org/10.3390/biology12071012
APA StyleKasuga, M., Mutsuro-Aoki, H., Ando, T., & Tamura, K. (2023). Molecular Anatomy of the Class I Ligase Ribozyme for Elucidation of the Activity-Generating Unit. Biology, 12(7), 1012. https://doi.org/10.3390/biology12071012




