Dissecting the Protective Effect of CD8+ T Cells in Response to SARS-CoV-2 mRNA Vaccination and the Potential Link with Lymph Node CD8+ T Cells
Abstract
:Simple Summary
Abstract
1. Introduction
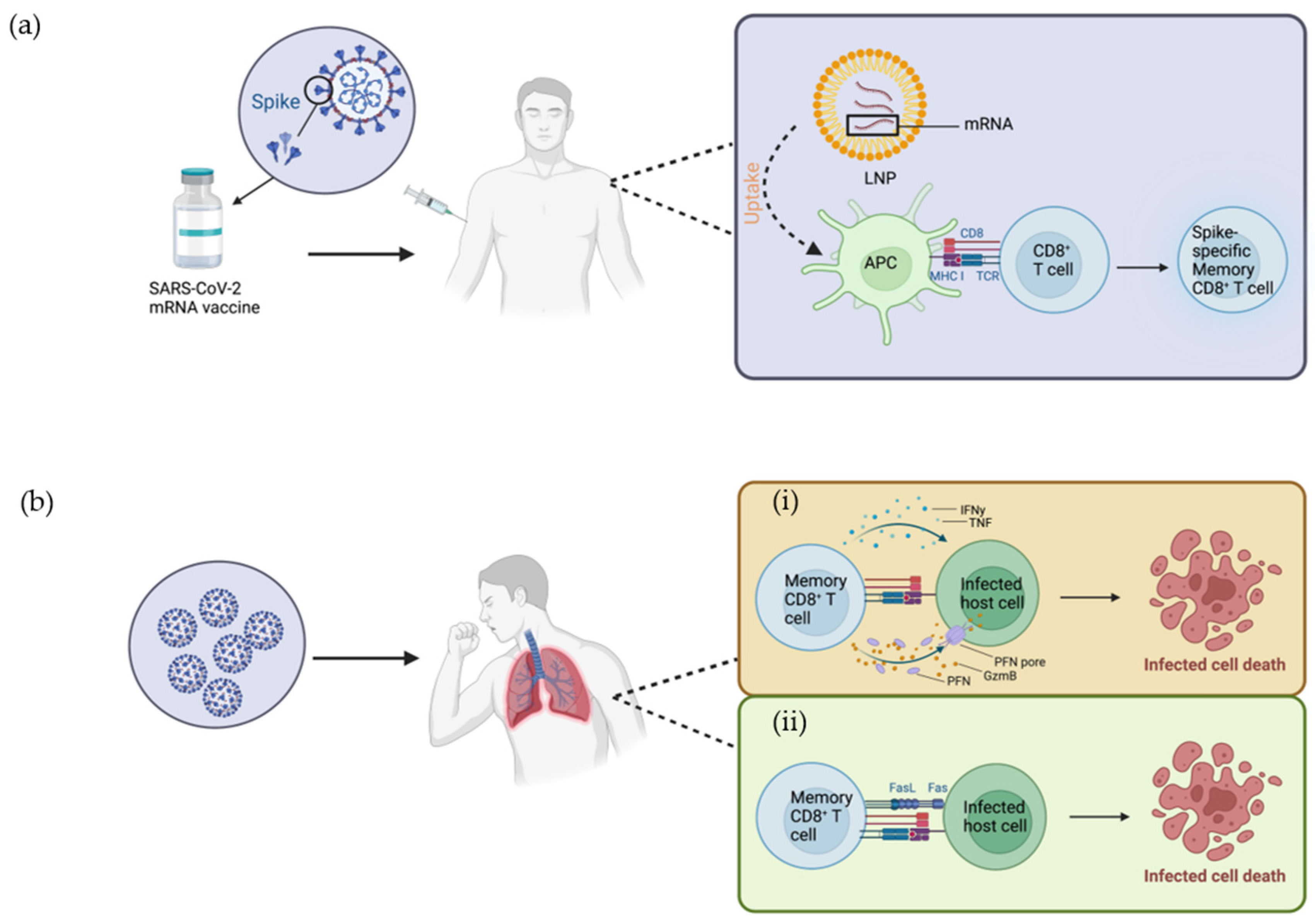
2. Mode of Action of Vaccine in Activating Immune Cells
3. CD8+ T Cell Development and Cytolytic Mechanisms in Response to Viral Infection
4. The Protective Effect of CD8+ T Cells in Response to SARS-CoV-2 mRNA Vaccine in Peripheral Blood
5. CD8+ T Cells and SARS-CoV-2 Epitopes
6. Vaccine-Elicited CD8+ Memory T Cells
7. CD8+ T Cells in Lymph Nodes
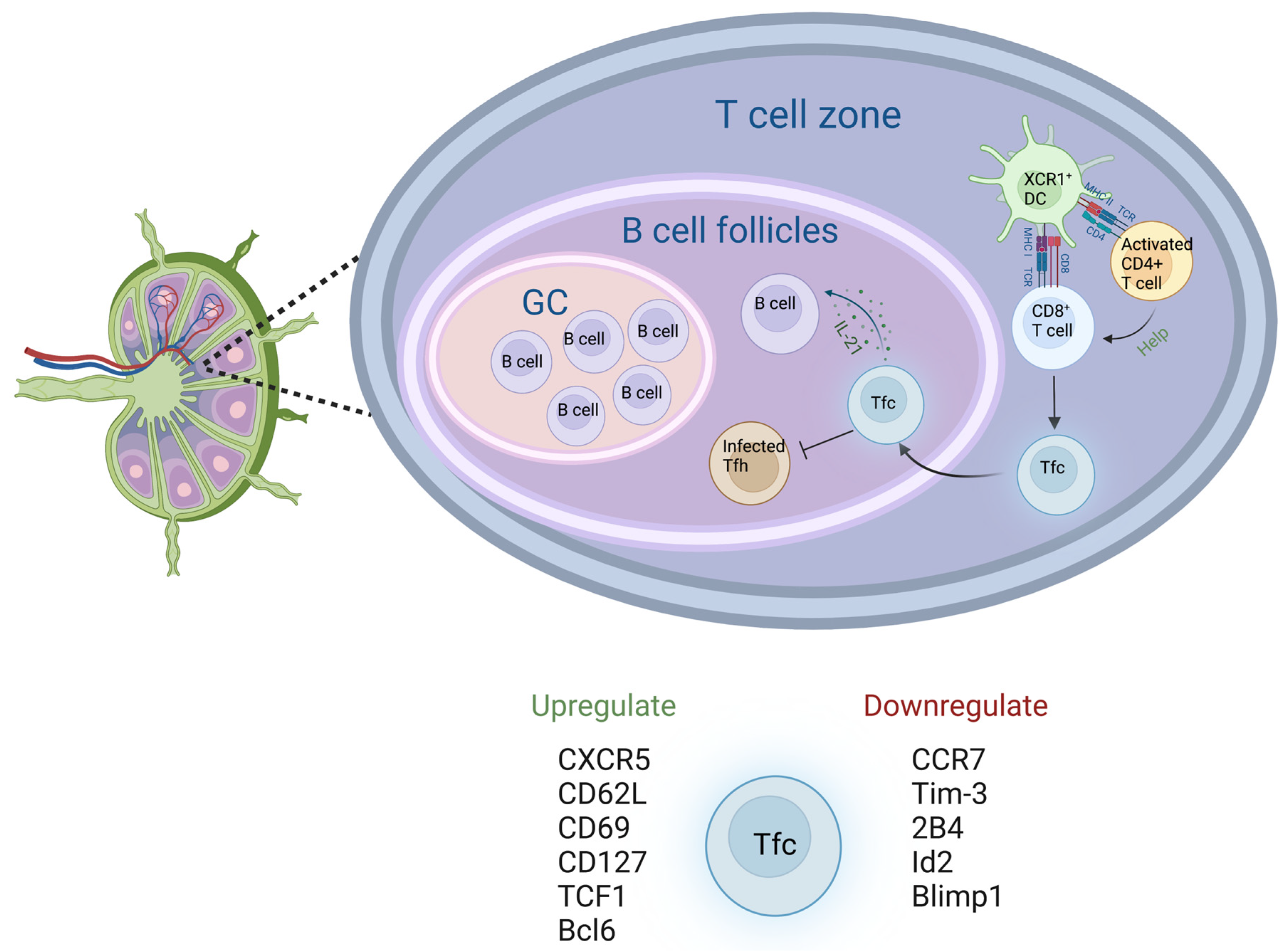
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cucinotta, D.; Vanelli, M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020, 91, 157–160. [Google Scholar] [PubMed]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Zheng, J. SARS-CoV-2: An Emerging Coronavirus that Causes a Global Threat. Int. J. Biol. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef] [Green Version]
- Laws, R.L.; Chancey, R.J.; Rabold, E.M.; Chu, V.T.; Lewis, N.M.; Fajans, M.; Reses, H.E.; Duca, L.M.; Dawson, P.; Conners, E.E.; et al. Symptoms and Transmission of SARS-CoV-2 Among Children-Utah and Wisconsin, March–May 2020. Pediatrics 2021, 147, e2020027268. [Google Scholar] [CrossRef]
- Rudberg, A.S.; Havervall, S.; Manberg, A.; Jernbom Falk, A.; Aguilera, K.; Ng, H.; Gabrielsson, L.; Salomonsson, A.C.; Hanke, L.; Murrell, B.; et al. SARS-CoV-2 exposure, symptoms and seroprevalence in healthcare workers in Sweden. Nat. Commun. 2020, 11, 5064. [Google Scholar] [CrossRef]
- Weng, L.M.; Su, X.; Wang, X.Q. Pain Symptoms in Patients with Coronavirus Disease (COVID-19): A Literature Review. J. Pain. Res. 2021, 14, 147–159. [Google Scholar] [CrossRef]
- Rahmani, K.; Shavaleh, R.; Forouhi, M.; Disfani, H.F.; Kamandi, M.; Oskooi, R.K.; Foogerdi, M.; Soltani, M.; Rahchamani, M.; Mohaddespour, M.; et al. The effectiveness of COVID-19 vaccines in reducing the incidence, hospitalization, and mortality from COVID-19: A systematic review and meta-analysis. Front. Public Health 2022, 10, 873596. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Echaide, M.; Chocarro de Erauso, L.; Bocanegra, A.; Blanco, E.; Kochan, G.; Escors, D. mRNA Vaccines against SARS-CoV-2: Advantages and Caveats. Int. J. Mol. Sci. 2023, 24, 5944. [Google Scholar] [CrossRef]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. The dawn of mRNA vaccines: The COVID-19 case. J. Control Release 2021, 333, 511–520. [Google Scholar] [CrossRef]
- Offit, P.A. Bivalent COVID-19 Vaccines-A Cautionary Tale. N. Engl. J. Med. 2023, 388, 481–483. [Google Scholar] [CrossRef]
- Bettini, E.; Locci, M. SARS-CoV-2 mRNA Vaccines: Immunological Mechanism and Beyond. Vaccines 2021, 9, 147. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Vogler, I.; Derhovanessian, E.; Kranz, L.M.; Vormehr, M.; Quandt, J.; Bidmon, N.; Ulges, A.; Baum, A.; et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 2021, 595, 572–577. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef]
- Oberhardt, V.; Luxenburger, H.; Kemming, J.; Schulien, I.; Ciminski, K.; Giese, S.; Csernalabics, B.; Lang-Meli, J.; Janowska, I.; Staniek, J.; et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature 2021, 597, 268–273. [Google Scholar] [CrossRef]
- Kent, S.J.; Khoury, D.S.; Reynaldi, A.; Juno, J.A.; Wheatley, A.K.; Stadler, E.; John Wherry, E.; Triccas, J.; Sasson, S.C.; Cromer, D.; et al. Disentangling the relative importance of T cell responses in COVID-19: Leading actors or supporting cast? Nat. Rev. Immunol. 2022, 22, 387–397. [Google Scholar] [CrossRef]
- Koutsakos, M.; Lee, W.S.; Reynaldi, A.; Tan, H.X.; Gare, G.; Kinsella, P.; Liew, K.C.; Taiaroa, G.; Williamson, D.A.; Kent, H.E.; et al. The magnitude and timing of recalled immunity after breakthrough infection is shaped by SARS-CoV-2 variants. Immunity 2022, 55, 1316–1326. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Sidney, J.; Methot, N.; Yu, E.D.; Zhang, Y.; Dan, J.M.; Goodwin, B.; Rubiro, P.; Sutherland, A.; Wang, E.; et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021, 2, 100355. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.; Liu, X.; Li, M.; Zhang, Z.; Song, L.; Zhu, B.; Wu, X.; Liu, J.; Zhao, D.; Li, Y. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 2022, 7, 94. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control Release 2015, 217, 345–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijkers, G.T.; Weterings, N.; Obregon-Henao, A.; Lepolder, M.; Dutt, T.S.; van Overveld, F.J.; Henao-Tamayo, M. Antigen Presentation of mRNA-Based and Virus-Vectored SARS-CoV-2 Vaccines. Vaccines 2021, 9, 848. [Google Scholar] [CrossRef]
- Trombetta, E.S.; Mellman, I. Cell biology of antigen processing in vitro and in vivo. Annu. Rev. Immunol. 2005, 23, 975–1028. [Google Scholar] [CrossRef]
- Guermonprez, P.; Valladeau, J.; Zitvogel, L.; Thery, C.; Amigorena, S. Antigen presentation and T cell stimulation by dendritic cells. Annu. Rev. Immunol. 2002, 20, 621–667. [Google Scholar] [CrossRef]
- Swain, S.L.; McKinstry, K.K.; Strutt, T.M. Expanding roles for CD4(+) T cells in immunity to viruses. Nat. Rev. Immunol. 2012, 12, 136–148. [Google Scholar] [CrossRef]
- Gressier, E.; Schulte-Schrepping, J.; Petrov, L.; Brumhard, S.; Stubbemann, P.; Hiller, A.; Obermayer, B.; Spitzer, J.; Kostevc, T.; Whitney, P.G.; et al. CD4(+) T cell calibration of antigen-presenting cells optimizes antiviral CD8(+) T cell immunity. Nat. Immunol. 2023, 24, 979–990. [Google Scholar] [CrossRef]
- Liao, S.; von der Weid, P.Y. Lymphatic system: An active pathway for immune protection. Semin. Cell Dev. Biol. 2015, 38, 83–89. [Google Scholar] [CrossRef]
- Goenka, R.; Barnett, L.G.; Silver, J.S.; O’Neill, P.J.; Hunter, C.A.; Cancro, M.P.; Laufer, T.M. Cutting edge: Dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J. Immunol. 2011, 187, 1091–1095. [Google Scholar] [CrossRef] [Green Version]
- Law, H.; Mach, M.; Howe, A.; Obeid, S.; Milner, B.; Carey, C.; Elfis, M.; Fsadni, B.; Ognenovska, K.; Phan, T.G.; et al. Early expansion of CD38+ICOS+ GC Tfh in draining lymph nodes during influenza vaccination immune response. iScience 2022, 25, 103656. [Google Scholar] [CrossRef]
- Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014, 41, 529–542. [Google Scholar] [CrossRef] [Green Version]
- Shulman, Z.; Gitlin, A.D.; Targ, S.; Jankovic, M.; Pasqual, G.; Nussenzweig, M.C.; Victora, G.D. T follicular helper cell dynamics in germinal centers. Science 2013, 341, 673–677. [Google Scholar] [CrossRef] [Green Version]
- Teijaro, J.R.; Farber, D.L. COVID-19 vaccines: Modes of immune activation and future challenges. Nat. Rev. Immunol. 2021, 21, 195–197. [Google Scholar] [CrossRef]
- Germain, R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002, 2, 309–322. [Google Scholar] [CrossRef]
- Zlotoff, D.A.; Bhandoola, A. Hematopoietic progenitor migration to the adult thymus. Ann. N. Y. Acad. Sci. 2011, 1217, 122–138. [Google Scholar] [CrossRef]
- Godfrey, D.I.; Zlotnik, A. Control points in early T-cell development. Immunol. Today 1993, 14, 547–553. [Google Scholar] [CrossRef]
- Goldman, K.P.; Park, C.S.; Kim, M.; Matzinger, P.; Anderson, C.C. Thymic cortical epithelium induces self tolerance. Eur. J. Immunol. 2005, 35, 709–717. [Google Scholar] [CrossRef]
- Blackman, M.; Kappler, J.; Marrack, P. The role of the T cell receptor in positive and negative selection of developing T cells. Science 1990, 248, 1335–1341. [Google Scholar] [CrossRef]
- Gaudino, S.J.; Kumar, P. Cross-Talk Between Antigen Presenting Cells and T Cells Impacts Intestinal Homeostasis, Bacterial Infections, and Tumorigenesis. Front. Immunol. 2019, 10, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, M.J.; Kelly, J.M.; Sutton, V.R.; Davis, J.E.; Browne, K.A.; Sayers, T.J.; Trapani, J.A. Unlocking the secrets of cytotoxic granule proteins. J. Leukoc. Biol. 2001, 70, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Sutton, V.R.; Brennan, A.J.; Ellis, S.; Danne, J.; Thia, K.; Jenkins, M.R.; Voskoboinik, I.; Pejler, G.; Johnstone, R.W.; Andrews, D.M.; et al. Serglycin determines secretory granule repertoire and regulates natural killer cell and cytotoxic T lymphocyte cytotoxicity. Febs J. 2016, 283, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Praper, T.; Sonnen, A.; Viero, G.; Kladnik, A.; Froelich, C.J.; Anderluh, G.; Dalla Serra, M.; Gilbert, R.J.C. Human Perforin Employs Different Avenues to Damage Membranes. J. Biol. Chem. 2011, 286, 2946–2955. [Google Scholar] [CrossRef] [Green Version]
- Lopez, J.A.; Susanto, O.; Jenkins, M.R.; Lukoyanova, N.; Sutton, V.R.; Law, R.H.P.; Johnston, A.; Bird, C.H.; Bird, P.I.; Whisstock, J.C.; et al. Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack. Blood 2013, 121, 2659–2668. [Google Scholar] [CrossRef] [Green Version]
- Metkar, S.S.; Marchioretto, M.; Antonini, V.; Lunelli, L.; Wang, B.; Gilbert, R.J.C.; Anderluh, G.; Roth, R.; Pooga, M.; Pardo, J.; et al. Perforin oligomers form arcs in cellular membranes: A locus for intracellular delivery of granzymes. Cell Death Differ. 2015, 22, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Goping, I.S.; Barry, M.; Liston, P.; Sawchuk, T.; Constantinescu, G.; Michalak, K.M.; Shostak, I.; Roberts, D.L.; Hunter, A.M.; Korneluk, R.; et al. Granzyme B-induced apoptosis requires both direct caspase activation and relief of caspase inhibition. Immunity 2003, 18, 355–365. [Google Scholar] [CrossRef] [Green Version]
- Bhat, P.; Leggatt, G.; Waterhouse, N.; Frazer, I.H. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 2017, 8, e2836. [Google Scholar] [CrossRef] [Green Version]
- Whitmire, J.K.; Tan, J.T.; Whitton, J.L. Interferon-gamma acts directly on CD8+ T cells to increase their abundance during virus infection. J. Exp. Med. 2005, 201, 1053–1059. [Google Scholar] [CrossRef]
- Schmidt, M.E.; Varga, S.M. The CD8 T Cell Response to Respiratory Virus Infections. Front. Immunol. 2018, 9, 678. [Google Scholar] [CrossRef] [Green Version]
- Moller, P.; Koretz, K.; Leithauser, F.; Bruderlein, S.; Henne, C.; Quentmeier, A.; Krammer, P.H. Expression of Apo-1 (Cd95), a Member of the Ngf/Tnf Receptor Superfamily, in Normal and Neoplastic Colon Epithelium. Int. J. Cancer 1994, 57, 371–377. [Google Scholar] [CrossRef]
- Strasser, A.; Jost, P.J.; Nagata, S. The Many Roles of FAS Receptor Signaling in the Immune System. Immunity 2009, 30, 180–192. [Google Scholar] [CrossRef] [Green Version]
- Volpe, E.; Sambucci, M.; Battistini, L.; Borsellino, G. Fas-Fas Ligand: Checkpoint of T Cell Functions in Multiple Sclerosis. Front. Immunol. 2016, 7, 382. [Google Scholar] [CrossRef] [Green Version]
- Suda, T.; Nagata, S. Purification and Characterization of the Fas-Ligand That Induces Apoptosis. J. Exp. Med. 1994, 179, 873–879. [Google Scholar] [CrossRef]
- Nagata, S. Fas-mediated apoptosis. Mol. Biol. Cell 1996, 7, 2939. [Google Scholar]
- Spoerl, S.; Kremer, A.N.; Aigner, M.; Eisenhauer, N.; Koch, P.; Meretuk, L.; Loffler, P.; Tenbusch, M.; Maier, C.; Uberla, K.; et al. Upregulation of CCR4 in activated CD8(+) T cells indicates enhanced lung homing in patients with severe acute SARS-CoV-2 infection. Eur. J. Immunol. 2021, 51, 1436–1448. [Google Scholar] [CrossRef]
- Gozzi-Silva, S.C.; Oliveira, L.M.; Alberca, R.W.; Pereira, N.Z.; Yoshikawa, F.S.; Pietrobon, A.J.; Yendo, T.M.; de Souza Andrade, M.M.; Ramos, Y.A.L.; Brito, C.A.; et al. Generation of Cytotoxic T Cells and Dysfunctional CD8 T Cells in Severe COVID-19 Patients. Cells 2022, 11, 3359. [Google Scholar] [CrossRef]
- Palgen, J.L.; Feraoun, Y.; Dzangue-Tchoupou, G.; Joly, C.; Martinon, F.; Le Grand, R.; Beignon, A.S. Optimize Prime/Boost Vaccine Strategies: Trained Immunity as a New Player in the Game. Front. Immunol. 2021, 12, 612747. [Google Scholar] [CrossRef]
- Gao, F.M.V.; Arunachalam, P.S.; van der Ploeg, K.; Manohar, M.; Röltgen, K.; Yang, F.; Wirz, O.; Hoh, R.; Haraguchi, E.; Lee, J.Y.; et al. Spheromers reveal robust T cell responses to the Pfizer/BioNTech vaccine and attenuated peripheral CD8+ T cell responses post SARS-CoV-2 infection. Immunity 2023, 56, 864–878. [Google Scholar] [CrossRef]
- Painter, M.M.; Mathew, D.; Goel, R.R.; Apostolidis, S.A.; Pattekar, A.; Kuthuru, O.; Baxter, A.E.; Herati, R.S.; Oldridge, D.A.; Gouma, S.; et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity 2021, 54, 2133–2142.e3. [Google Scholar] [CrossRef] [PubMed]
- Young, A. T cells in SARS-CoV-2 infection and vaccination. Ther. Adv. Vaccines Immunother. 2022, 10, 25151355221115011. [Google Scholar] [CrossRef] [PubMed]
- Guerrera, G.; Picozza, M.; D’Orso, S.; Placido, R.; Pirronello, M.; Verdiani, A.; Termine, A.; Fabrizio, C.; Giannessi, F.; Sambucci, M.; et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T cells with a stem cell memory phenotype. Sci. Immunol. 2021, 6, eabl5344. [Google Scholar] [CrossRef] [PubMed]
- Pardieck, I.N.; van der Sluis, T.C.; van der Gracht, E.T.I.; Veerkamp, D.M.B.; Behr, F.M.; van Duikeren, S.; Beyrend, G.; Rip, J.; Nadafi, R.; Beyranvand Nejad, E.; et al. A third vaccination with a single T cell epitope confers protection in a murine model of SARS-CoV-2 infection. Nat. Commun. 2022, 13, 3966. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Yu, J.Y.; McMahan, K.; Jacob-Dolan, C.; He, X.; Giffin, V.; Wu, C.; Sciacca, M.; Powers, O.; Nampanya, F.; et al. CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Sci. Immunol. 2022, 7, eabq7647. [Google Scholar] [CrossRef]
- Mateus, J.; Dan, J.M.; Zhang, Z.L.; Moderbacher, C.R.; Lammers, M.; Goodwin, B.; Sette, A.; Crotty, S.; Weiskopf, D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science 2021, 374, eabj9853. [Google Scholar] [CrossRef]
- Ozbay Kurt, F.G.; Lepper, A.; Gerhards, C.; Roemer, M.; Lasser, S.; Arkhypov, I.; Bitsch, R.; Bugert, P.; Altevogt, P.; Gouttefangeas, C.; et al. Booster dose of mRNA vaccine augments waning T cell and antibody responses against SARS-CoV-2. Front. Immunol. 2022, 13, 1012526. [Google Scholar] [CrossRef]
- Palatnik-de-Sousa, C.B.; Soares, I.D.; Rosa, D.S. Editorial: Epitope discovery and Synthetic Vaccine design. Front. Immunol. 2018, 9, 826. [Google Scholar] [CrossRef] [Green Version]
- Grifoni, A.; Sidney, J.; Vita, R.; Peters, B.; Crotty, S.; Weiskopf, D.; Sette, A. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe 2021, 29, 1076–1092. [Google Scholar] [CrossRef]
- Kombe, A.J.K.; Biteghe, F.A.N.; Ndoutoume, Z.N.; Jin, T.C. CD8(+) T-cell immune escape by SARS-CoV-2 variants of concern. Front. Immunol. 2022, 13, 962079. [Google Scholar] [CrossRef]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lubke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021, 22, 74–85. [Google Scholar] [CrossRef]
- Schulien, I.; Kemming, J.; Oberhardt, V.; Wild, K.; Seidel, L.M.; Killmer, S.; Sagar; Daul, F.; Lago, M.S.; Decker, A.; et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2021, 27, 78–85. [Google Scholar] [CrossRef]
- Nguyen, T.H.O.; Rowntree, L.C.; Petersen, J.; Chua, B.Y.; Hensen, L.; Kedzierski, L.; van de Sandt, C.E.; Chaurasia, P.; Tan, H.X.; Habel, J.R.; et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity 2021, 54, 1066–1082.e5. [Google Scholar] [CrossRef]
- Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernandez, J.; Prati, D.; Baselli, G.; Asselta, R.; et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar]
- Martin, M.D.; Badovinac, V.P. Defining Memory CD8 T Cell. Front. Immunol. 2018, 9, 2692. [Google Scholar] [CrossRef] [Green Version]
- Farber, D.L.; Yudanin, N.A.; Restifo, N.P. Human memory T cells: Generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 2014, 14, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Esser, M.T.; Marchese, R.D.; Kierstead, L.S.; Tussey, L.G.; Wang, F.B.; Chirmule, N.; Washabaugh, M.W. Memory T cells and vaccines. Vaccine 2003, 21, 419–430. [Google Scholar] [CrossRef]
- Flynn, J.K.; Gorry, P.R. Stem memory T cells (TSCM)-their role in cancer and HIV immunotherapies. Clin. Transl. Immunol. 2014, 3, e20. [Google Scholar] [CrossRef]
- Brasu, N.; Elia, I.; Russo, V.; Montacchiesi, G.; Stabile, S.A.; De Intinis, C.; Fesi, F.; Gizzi, K.; Macagno, M.; Montone, M.; et al. Memory CD8(+) T cell diversity and B cell responses correlate with protection against SARS-CoV-2 following mRNA vaccination. Nat. Immunol. 2022, 23, 1445–1456. [Google Scholar] [CrossRef]
- Neidleman, J.; Luo, X.; Frouard, J.; Xie, G.; Gill, G.; Stein, E.S.; McGregor, M.; Ma, T.; George, A.F.; Kosters, A.; et al. SARS-CoV-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep. Med. 2020, 1, 100081. [Google Scholar] [CrossRef]
- Reinscheid, M.; Luxenburger, H.; Karl, V.; Graeser, A.; Giese, S.; Ciminski, K.; Reeg, D.B.; Oberhardt, V.; Roehlen, N.; Lang-Meli, J.; et al. COVID-19 mRNA booster vaccine induces transient CD8+T effector cell responses while conserving the memory pool for subsequent reactivation. Nat. Commun. 2022, 13, 4631. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, A.; Stavridou, F.; Giannaki, M.; Paschoudi, K.; Chatzopoulou, F.; Gavriilaki, E.; Georgolopoulos, G.; Anagnostopoulos, A.; Yannaki, E. Robust SARS-COV-2-specific T-cell immune memory persists long-term in immunocompetent individuals post BNT162b2 double shot. Heliyon 2022, 8, e09863. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Kageyama, T.; Tanaka, S.; Otsuka, K.; Tsukumo, S.I.; Mashimo, Y.; Onouchi, Y.; Nakajima, H.; Yasutomo, K. Markers of Memory CD8 T Cells Depicting the Effect of the BNT162b2 mRNA COVID-19 Vaccine in Japan. Front. Immunol. 2022, 13, 836923. [Google Scholar] [CrossRef] [PubMed]
- Dunne, P.J.; Faint, J.M.; Gudgeon, N.H.; Fletcher, J.M.; Plunkett, F.J.; Soares, M.V.; Hislop, A.D.; Annels, N.E.; Rickinson, A.B.; Salmon, M.; et al. Epstein-Barr virus-specific CD8(+) T cells that re-express CD45RA are apoptosis-resistant memory cells that retain replicative potential. Blood 2002, 100, 933–940. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.H.; Rha, M.S.; Sa, M.; Choi, H.K.; Jeon, J.H.; Seok, H.; Park, D.W.; Park, S.H.; Jeong, H.W.; Choi, W.S.; et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat. Commun. 2021, 12, 4043. [Google Scholar] [CrossRef]
- Fuertes Marraco, S.A.; Soneson, C.; Cagnon, L.; Gannon, P.O.; Allard, M.; Abed Maillard, S.; Montandon, N.; Rufer, N.; Waldvogel, S.; Delorenzi, M.; et al. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci. Transl. Med. 2015, 7, 282ra48. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Wang, G.; Wang, Y.; Zhang, Q.; Ren, L.; Gu, X.; Huang, T.; Zhong, J.; Wang, Y.; Wang, X.; et al. SARS-CoV-2-specific antibody and T-cell responses 1 year after infection in people recovered from COVID-19: A longitudinal cohort study. Lancet Microbe 2022, 3, e348–e356. [Google Scholar] [CrossRef]
- Rius, C.; Attaf, M.; Tungatt, K.; Bianchi, V.; Legut, M.; Bovay, A.; Donia, M.; Thor Straten, P.; Peakman, M.; Svane, I.M.; et al. Peptide-MHC Class I Tetramers Can Fail To Detect Relevant Functional T Cell Clonotypes and Underestimate Antigen-Reactive T Cell Populations. J. Immunol. 2018, 200, 2263–2279. [Google Scholar] [CrossRef] [Green Version]
- Ferragut, F.; Cruz, K.M.; Gallardo, J.P.; Fernandez, M.; Hernandez Vasquez, Y.; Gomez, K.A. Activation-induced marker assays for identification of Trypanosoma cruzi-specific CD4 or CD8 T cells in chronic Chagas disease patients. Immunology 2022, 169, 185–203. [Google Scholar] [CrossRef]
- Reiss, S.; Baxter, A.E.; Cirelli, K.M.; Dan, J.M.; Morou, A.; Daigneault, A.; Brassard, N.; Silvestri, G.; Routy, J.P.; Havenar-Daughton, C.; et al. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE 2017, 12, e0186998. [Google Scholar] [CrossRef] [Green Version]
- Mudd, P.A.; Martins, M.A.; Ericsen, A.J.; Tully, D.C.; Power, K.A.; Bean, A.T.; Piaskowski, S.M.; Duan, L.; Seese, A.; Gladden, A.D.; et al. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature 2012, 491, 129–133. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.C.; Dayball, K.; Wan, Y.H.; Bramson, J. Detailed analysis of the CD8+ T-cell response following adenovirus vaccination. J. Virol. 2003, 77, 13407–13411. [Google Scholar] [CrossRef] [Green Version]
- Hirai, T.; Yoshioka, Y. Considerations of CD8(+) T Cells for Optimized Vaccine Strategies Against Respiratory Viruses. Front. Immunol. 2022, 13, 918611. [Google Scholar] [CrossRef]
- Li, C.; Lee, A.; Grigoryan, L.; Arunachalam, P.S.; Scott, M.K.D.; Trisal, M.; Wimmers, F.; Sanyal, M.; Weidenbacher, P.A.; Feng, Y.; et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat. Immunol. 2022, 23, 543–555. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Charles, T.P.; Joag, V.; Bollimpelli, V.S.; Scott, M.K.D.; Wimmers, F.; Burton, S.L.; Labranche, C.C.; Petitdemange, C.; Gangadhara, S.; et al. T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers. Nat. Med. 2020, 26, 932–940. [Google Scholar] [CrossRef]
- Perdomo-Celis, F.; Taborda, N.A.; Rugeles, M.T. Follicular CD8(+) T Cells: Origin, Function and Importance during HIV Infection. Front. Immunol. 2017, 8, 1241. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Ye, L.L. A Portrait of CXCR5(+) Follicular Cytotoxic CD8(+) T cells. Trends Immunol. 2018, 39, 965–979. [Google Scholar] [CrossRef]
- Xiao, M.; Chen, X.; He, R.; Ye, L. Differentiation and Function of Follicular CD8 T Cells During Human Immunodeficiency Virus Infection. Front. Immunol. 2018, 9, 1095. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Masters, A.R.; Fortner, K.A.; Champagne, D.P.; Yanguas-Casas, N.; Silberger, D.J.; Weaver, C.T.; Haynes, L.; Rincon, M. IL-6 promotes the differentiation of a subset of naive CD8+ T cells into IL-21-producing B helper CD8+ T cells. J. Exp. Med. 2016, 213, 2281–2291. [Google Scholar] [CrossRef]
- Lv, Y.; Ricard, L.; Gaugler, B.; Huang, H.; Ye, Y. Biology and clinical relevance of follicular cytotoxic T cells. Front. Immunol. 2022, 13, 1036616. [Google Scholar] [CrossRef]
- Collins, D.R.; Hitschfel, J.; Urbach, J.M.; Mylvaganam, G.H.; Ly, N.L.; Arshad, U.; Racenet, Z.J.; Yanez, A.G.; Diefenbach, T.J.; Walker, B.D. Cytolytic CD8(+) T cells infiltrate germinal centers to limit ongoing HIV replication in spontaneous controller lymph nodes. Sci. Immunol. 2023, 8, eade5872. [Google Scholar] [CrossRef] [PubMed]
- Perdomo-Celis, F.; Taborda, N.A.; Rugeles, M.T. Circulating CXCR5-Expressing CD8+ T-Cells Are Major Producers of IL-21 and Associate With Limited HIV Replication. J. Acquir. Immune Defic. Syndr. 2018, 78, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Quigley, M.F.; Gonzalez, V.D.; Granath, A.; Andersson, J.; Sandberg, J.K. CXCR5(+) CCR7(-) CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur. J. Immunol. 2007, 37, 3352–3362. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Luo, X.; Wu, Q.; Huang, J.; Xiao, G.; Wang, L.; Yang, B.; Li, H.; Wu, C. A Subset of CXCR5(+)CD8(+) T Cells in the Germinal Centers from Human Tonsils and Lymph Nodes Help B Cells Produce Immunoglobulins. Front. Immunol. 2018, 9, 2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elzein, S.M.; Zimmerer, J.M.; Han, J.L.; Ringwald, B.A.; Bumgardner, G.L. CXCR5(+)CD8(+) T cells: A Review of their Antibody Regulatory Functions and Clinical Correlations. J. Immunol. 2021, 206, 2775–2783. [Google Scholar] [CrossRef] [PubMed]
- Valentine, K.M.; Hoyer, K.K. CXCR5+ CD8 T Cells: Protective or Pathogenic? Front. Immunol. 2019, 10, 1322. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Zhou, J.; Ye, L. Role of CXCR5(+) CD8(+) T cells in human immunodeficiency virus-1 infection. Front. Microbiol. 2022, 13, 998058. [Google Scholar] [CrossRef]
- Leong, Y.A.; Chen, Y.P.; Ong, H.S.; Wu, D.; Man, K.V.; Deleage, C.; Minnich, M.; Meckiff, B.J.; Wei, Y.B.; Hou, Z.H.; et al. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 2016, 17, 1187–1196. [Google Scholar] [CrossRef]
- He, R.; Hou, S.; Liu, C.; Zhang, A.; Bai, Q.; Han, M.; Yang, Y.; Wei, G.; Shen, T.; Yang, X.; et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 2016, 537, 412–428. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, H.; Yu, Q.; Liu, J.; Chen, S.; Zhao, Z. Protective role of follicular CXCR5(+)CD8(+) T cells against dengue virus 2 infection. Int. J. Infect. Dis. 2019, 83, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Reuter, M.A.; Del Rio Estrada, P.M.; Buggert, M.; Petrovas, C.; Ferrando-Martinez, S.; Nguyen, S.; Sada Japp, A.; Ablanedo-Terrazas, Y.; Rivero-Arrieta, A.; Kuri-Cervantes, L.; et al. HIV-Specific CD8(+) T Cells Exhibit Reduced and Differentially Regulated Cytolytic Activity in Lymphoid Tissue. Cell Rep. 2017, 21, 3458–3470. [Google Scholar] [CrossRef] [Green Version]
- Chu, F.; Li, H.S.; Liu, X.; Cao, J.; Ma, W.; Ma, Y.; Weng, J.; Zhu, Z.; Cheng, X.; Wang, Z.; et al. CXCR5(+)CD8(+) T cells are a distinct functional subset with an antitumor activity. Leukemia 2019, 33, 2640–2653. [Google Scholar] [CrossRef]
- Hey-Nguyen, W.J.; Xu, Y.; Pearson, C.F.; Bailey, M.; Suzuki, K.; Tantau, R.; Obeid, S.; Milner, B.; Field, A.; Carr, A.; et al. Quantification of Residual Germinal Center Activity and HIV-1 DNA and RNA Levels Using Fine Needle Biopsies of Lymph Nodes During Antiretroviral Therapy. AIDS Res. Hum. Retroviruses 2017, 33, 648–657. [Google Scholar] [CrossRef]
- Mudd, P.A.; Minervina, A.A.; Pogorelyy, M.V.; Turner, J.S.; Kim, W.; Kalaidina, E.; Petersen, J.; Schmitz, A.J.; Lei, T.; Haile, A.; et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell 2022, 185, 603–613.e15. [Google Scholar] [CrossRef]
- Laidlaw, B.J.; Craft, J.E.; Kaech, S.M. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat. Rev. Immunol. 2016, 16, 102–111. [Google Scholar] [CrossRef] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Venturi, V.; Munier, C.M.L. Dissecting the Protective Effect of CD8+ T Cells in Response to SARS-CoV-2 mRNA Vaccination and the Potential Link with Lymph Node CD8+ T Cells. Biology 2023, 12, 1035. https://doi.org/10.3390/biology12071035
Chen M, Venturi V, Munier CML. Dissecting the Protective Effect of CD8+ T Cells in Response to SARS-CoV-2 mRNA Vaccination and the Potential Link with Lymph Node CD8+ T Cells. Biology. 2023; 12(7):1035. https://doi.org/10.3390/biology12071035
Chicago/Turabian StyleChen, Mengfei, Vanessa Venturi, and C. Mee Ling Munier. 2023. "Dissecting the Protective Effect of CD8+ T Cells in Response to SARS-CoV-2 mRNA Vaccination and the Potential Link with Lymph Node CD8+ T Cells" Biology 12, no. 7: 1035. https://doi.org/10.3390/biology12071035
APA StyleChen, M., Venturi, V., & Munier, C. M. L. (2023). Dissecting the Protective Effect of CD8+ T Cells in Response to SARS-CoV-2 mRNA Vaccination and the Potential Link with Lymph Node CD8+ T Cells. Biology, 12(7), 1035. https://doi.org/10.3390/biology12071035





