Sample Treatment with Trypsin for RT-LAMP COVID-19 Diagnosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Standard and Clinical Samples
2.2. RNA Extraction
2.3. Sample Treatment
2.4. RT-qPCR
2.5. RT-LAMP
2.6. Sequencing
2.7. Conservation
2.8. Statistical Analysis
3. Results
3.1. Sensitivity and Specificity RT-LAMP Assay using Synthetic SARS-CoV-2 RNA
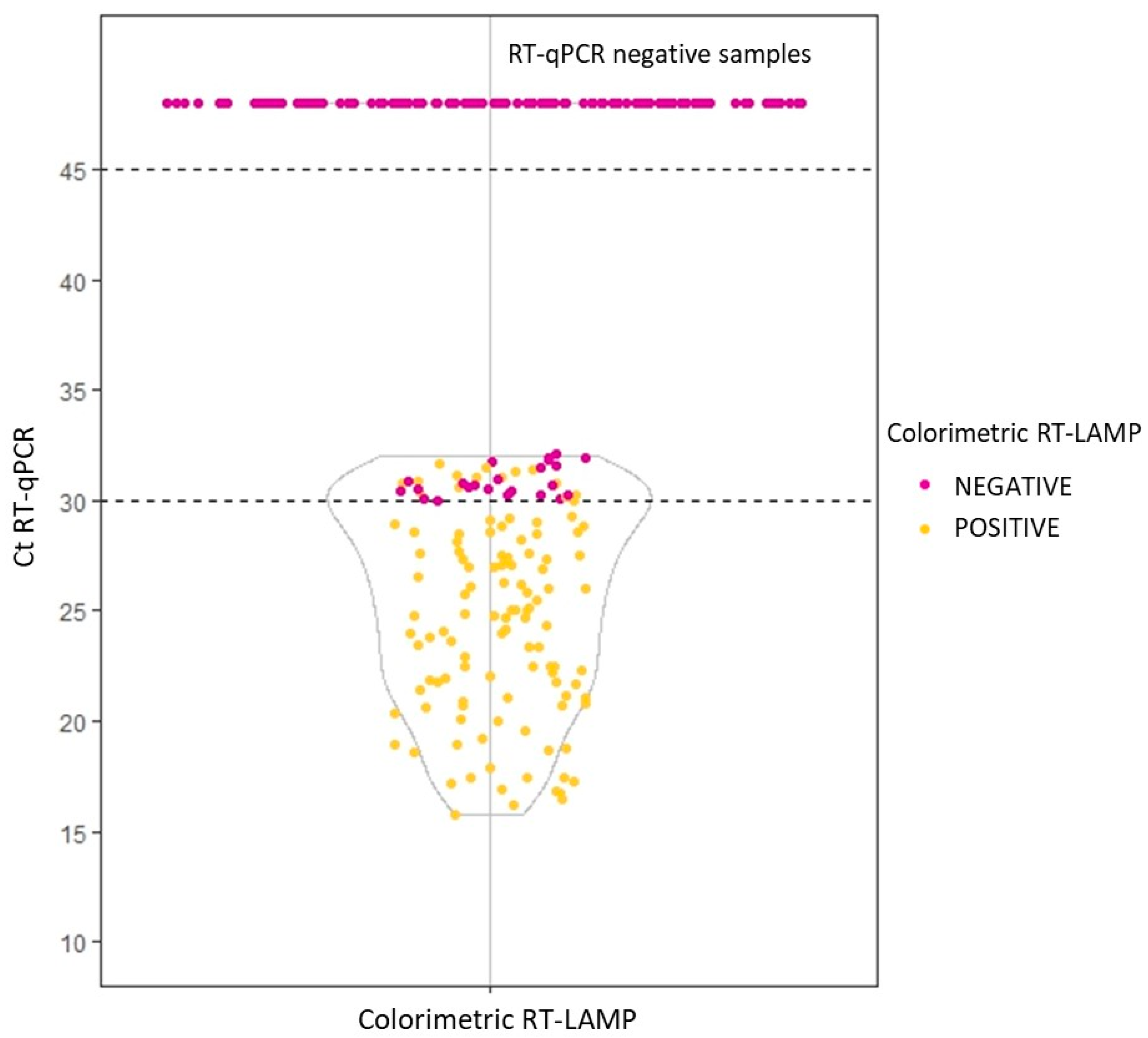
3.2. Sample Treatment and Validation on Clinical Samples with RT-LAMP Assay
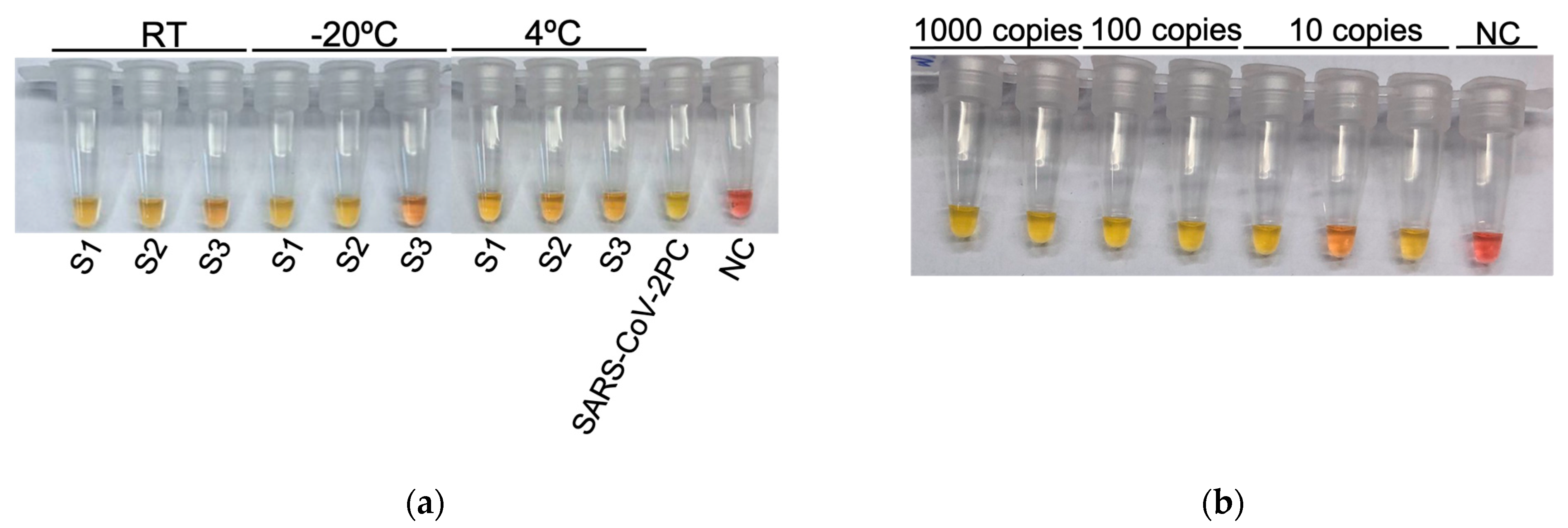
3.3. Evaluation of Sample Conservation in Trypsin Solution and Maintenance of Colorimetric RT-LAMP Reaction Mix
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghinai, I.; McPherson, T.D.; Hunter, J.C.; Kirking, H.L.; Christiansen, D.; Joshi, K.; Rubin, R.; Morales-Estrada, S.; Black, S.R.; Pacilli, M.; et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet 2020, 395, 1137–1144. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: https://covid19.who.int/table (accessed on 20 February 2023).
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel coronavirus disease (COVID-19): Paving the road for rapid detection and point-of-care diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert. Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef]
- Afzal, A. Molecular diagnostic technologies for COVID-19: Limitations and challenges. J. Adv. Res. 2020, 26, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Machado, B.A.S.; Hodel, K.V.S.; Barbosa-Júnior, V.G.; Soares, M.B.P.; Badaró, R. The main molecular and serological methods for diagnosing covid-19: An overview based on the literature. Viruses 2021, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Majumder, J.; Minko, T. Recent Developments on Therapeutic and Diagnostic Approaches for COVID-19. AAPS J. 2021, 23, 14. [Google Scholar] [CrossRef]
- Syal, K. Guidelines on newly identified limitations of diagnostic tools for COVID-19 and consequences. J. Med. Virol. 2021, 93, 1837–1842. [Google Scholar] [CrossRef]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef]
- Ji, T.; Liu, Z.; Wang, G.Q.; Guo, X.; Akbar Khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Nyan, D.C.; Swinson, K.L. A novel multiplex isothermal amplification method for rapid detection and identification of viruses. Sci. Rep. 2015, 5, 17925. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Daskou, M.; Dimitriou, T.G.; Alexopoulou, D.S.; Tsakogiannis, D.; Amoutzias, G.D.; Mossialos, D.; Kyriakopoulou, Z.; Markoulatos, P. WarmStart colorimetric RT-LAMP for the rapid, sensitive and specific detection of Enteroviruses A–D targeting the 5′UTR region. J. Appl. Microbiol. 2021, 130, 292–301. [Google Scholar] [CrossRef]
- Peltzer, D.; Tobler, K.; Fraefel, C.; Maley, M.; Bachofen, C. Rapid and simple colorimetric loop-mediated isothermal amplification (LAMP) assay for the detection of Bovine alphaherpesvirus 1. J. Virol. Methods 2021, 289, 114041. [Google Scholar] [CrossRef]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-mediated isothermal amplification (Lamp): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of covid-19 pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.M.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- World Health Organization. Available online: https://cdn.who.int/media/docs/default-source/blue-print/who-rd-blueprint-diagnostics-tpp-final-v1-0-28-09-jc-ppc-final-cmp92616a80172344e4be0edf315b582021.pdf?sfvrsn=e3747f20_1&download=true (accessed on 24 January 2023).
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; Von Stetten, F. Loop-mediated isothermal amplification (LAMP)-review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef]
- Lee, Y.H.; Zhou, H.; Reiss, J.K.; Yan, X.; Zhang, L.; Chia, D.; Wong, D.T.W. Direct saliva transcriptome analysis. Clin. Chem. 2011, 57, 1295–1302. [Google Scholar] [CrossRef]
- Aoki, M.N.; de Oliveira Coelho, B.; Góes, L.G.B.; Minoprio, P.; Durigon, E.L.; Morello, L.G.; Marchini, F.K.; Riediger, I.N.; do Carmo Debur, M.; Nakaya, H.I.; et al. Colorimetric RT-LAMP SARS-CoV-2 diagnostic sensitivity relies on color interpretation and viral load. Sci. Rep. 2021, 11, 9026. [Google Scholar] [CrossRef] [PubMed]
- Wölfl-Duchek, M.; Bergmann, F.; Jorda, A.; Weber, M.; Müller, M.; Seitz, T.; Zoufaly, A.; Strassl, R.; Zeitlinger, M.; Herkner, H.; et al. Sensitivity and Specificity of SARS-CoV-2 Rapid Antigen Detection Tests Using Oral, Anterior Nasal, and Nasopharyngeal Swabs: A Diagnostic Accuracy Study. Microbiol. Spectr. 2022, 10, e02029-21. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, K.K.; Jensen, J.S.; Todsen, T.; Tolsgaard, M.G.; Kirkby, N.; Lippert, F.; Vangsted, A.M.; Martel, C.J.M.; Klokker, M.; von Buchwald, C. Accuracy and cost description of rapid antigen test compared with reverse transcriptase-polymerase chain reaction for SARS-CoV-2 detection. Dan. Med. J. 2021, 68, A03210217. [Google Scholar] [PubMed]
- Jefferson, T.; Spencer, E.; Brassey, J.; Heneghan, C. Viral cultures for COVID-19 infectious potential assessment—A systematic review. Clin. Infect. Dis. 2020, e3884–e3899. [Google Scholar] [CrossRef]
- Abe, H.; Ushijima, Y.; Bikangui, R.; Ondo, G.N. Long-term validation of a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 from March 2020 to October 2021 in Central Africa, Gabon. PLoS Negl. Trop. Dis. 2022, 16, e0010964. [Google Scholar] [CrossRef]
- Hurtado, L.; Díaz, D.; Escorcia, K.; Flórez, L.; Bello, Y.; Díaz, Y.; Navarro, E.; Pacheco, L.C.; Galán, N.; Maestre, R.; et al. Validación clínica de la prueba RT-LAMP para el diagnóstico rápido del SARS-CoV-2. Biomedica 2022, 42, 59–72. [Google Scholar] [CrossRef]
- Nandi, S.S.; Lambe, U.P.; Sawant, S.A.; Gohil, T.; Deshpande, J. Development of a RT-LAMP assay for detection of SARS-CoV-2. Indian J. Emerg. Med. 2021, 155, 148–155. [Google Scholar] [CrossRef]
- Kitajima, H.; Tamura, Y.; Yoshida, H.; Kinoshita, H.; Katsuta, H.; Matsui, C.; Matsushita, A.; Arai, T.; Hashimoto, S.; Iuchi, A.; et al. Clinical COVID-19 diagnostic methods: Comparison of reverse transcription loop-mediated isothermal amplification (RT-LAMP) and quantitative RT-PCR (qRT-PCR). J. Clin. Virol. J. 2021, 139, 104813. [Google Scholar] [CrossRef]
- Prakash, S.; Priyatma; Aasarey, R.; Pandey, P.K.; Mathur, P.; Arulselvi, S. An inexpensive and rapid diagnostic method for detection of SARS-CoV-2 RNA by loop-mediated isothermal amplification (LAMP). MethodsX 2023, 10, 102011. [Google Scholar] [CrossRef]
- Barboza, S.; Borges, W.; Toniolo, T.; Souza, D. Reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay as a rapid molecular diagnostic tool for COVID-19 in healthcare workers. J. Clin. Virol. Plus 2023, 3, 100134. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.L.; Armson, B.; Gonzales, J.L.; Wise, E.L.; Howson, E.L.A.; Vincent-mistiaen, Z.; Fouch, S.; Maltby, C.J.; Grippon, S.; Munro, S.; et al. A highly effective reverse-transcription loop-me diate d isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J. Infect. 2020, 82, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Du, W.F.; Ge, J.H.; Li, J.J.; Tang, L.J.; Yu, R.Q.; Jiang, J.H. Single-step, high-specificity detection of single nucleotide mutation by primer-activatable loop-mediated isothermal amplification (PA-LAMP). Anal. Chim. Acta 2019, 1050, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021, 67, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Howson, E.L.A.; Kidd, S.P.; Armson, B.; Goring, A.; Sawyer, J.; Cassar, C.; Cross, D.; Lewis, T.; Hockey, J.; Rivers, S.; et al. Preliminary optimisation of a simplified sample preparation method to permit direct detection of SARS-CoV-2 within saliva samples using reverse-transcription loop-mediated isothermal amplification (RT-LAMP). J. Virol. Methods 2021, 289, 114048. [Google Scholar] [CrossRef] [PubMed]
- Kundrod, K.A.; Natoli, M.E.; Chang, M.M.; Smith, C.A.; Paul, S.; Ogoe, D.; Goh, C.; Santhanaraj, A.; Price, A.; Eldin, K.W.; et al. Sample-to-answer, extraction-free, real-time RT-LAMP test for SARS-CoV-2 in nasopharyngeal, nasal, and saliva samples: Implications and use for surveillance testing. PLoS ONE 2022, 17, e0264130. [Google Scholar] [CrossRef]
- Moon, S.H.; Kim, S.-C.; Kim, B.W.; Park, G.-J.; Chai, H.-S.; Kim, Y.M.; Kim, H.S.; Park, H.S. SARS-CoV-2 molecular diagnostic point-of-care testing based on loop-mediated isothermal amplification: A prospective, single-center validation study. Heliyon 2023, 9, e14564. [Google Scholar] [CrossRef]
- Subsoontorn, P.; Lohitnavy, M.; Kongkaew, C. The diagnostic accuracy of isothermal nucleic acid point-of-care tests for human coronaviruses: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 22349. [Google Scholar] [CrossRef]

| Gene | Primer and Probe Sequence (5′ → 3′) | |
|---|---|---|
| N | Forward | TTACAAACATTGGCCGCAAA |
| Reverse | GCGCGACATTCCGAAGAA | |
| Probe | Cy5-ACAATTTGCCCCCAGCGCTTCAG-BHQ-3 | |
| RdRP | Forward | GGCCTCACTTGTTCTTGCTC |
| Reverse | GCCACACATGACCATTTCAC | |
| Probe | HEX-CGTGTTGTAGCTTGTCACACCGTTTC-BHQ-2 | |
| LYSOZYME | Forward | CGCTACTGGTGTAATGATGGC |
| Reverse | ACGGACAACCCTCTTTGC | |
| Probe | Atto 425-GCCTGTCATTTATCCTGCAGTGCTTTGC-BHQ-1 | |
| Gene | Primer and Sequence (5′ → 3′) | |
|---|---|---|
| N | F3 | CGGCAGTCAAGCCTCTTC |
| B3 | TTGCTCTCAAGCTGGTTCAA | |
| FIP | TCCCCTACTGCTGCCTGGAGCGTTCCTCATCACGTAGTCG | |
| BIP | TCTCCTGCTAGAATGGCTGGCATCTGTCAAGCAGCAGCAAAG | |
| LF | TGAATTTCTTGAACTGTTG | |
| LB | TGGCGGTGATGCTGCTCT | |
| GAPDH | F3 | TCCACCCATGGCAAATTCC |
| B3 | AATGAGCCCCAGCCTTCT | |
| FIP | AGGGATCTCGCTCCTGGAAGATTCAAGGCTGAGAACGGGA | |
| BIP | AATCAAGTGGGGCGATGCTGGCCATGGTGGTGAAGACGC | |
| LF | GATGGGATTTCCATTGATGAC | |
| LB | CTGAGTACGTCGTGGAGTCC | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Sorribes, S.; Lara-Hernández, F.; Manzano-Blasco, I.; Abadía-Otero, J.; Albert, E.; Mulet, A.; Briongos-Figuero, L.S.; Gabella-Martín, M.; Torres, I.; Signes-Costa, J.; et al. Sample Treatment with Trypsin for RT-LAMP COVID-19 Diagnosis. Biology 2023, 12, 900. https://doi.org/10.3390/biology12070900
García-Sorribes S, Lara-Hernández F, Manzano-Blasco I, Abadía-Otero J, Albert E, Mulet A, Briongos-Figuero LS, Gabella-Martín M, Torres I, Signes-Costa J, et al. Sample Treatment with Trypsin for RT-LAMP COVID-19 Diagnosis. Biology. 2023; 12(7):900. https://doi.org/10.3390/biology12070900
Chicago/Turabian StyleGarcía-Sorribes, Soraya, Francisco Lara-Hernández, Iris Manzano-Blasco, Jessica Abadía-Otero, Eliseo Albert, Alba Mulet, Laisa Socorro Briongos-Figuero, Miriam Gabella-Martín, Ignacio Torres, Jaime Signes-Costa, and et al. 2023. "Sample Treatment with Trypsin for RT-LAMP COVID-19 Diagnosis" Biology 12, no. 7: 900. https://doi.org/10.3390/biology12070900
APA StyleGarcía-Sorribes, S., Lara-Hernández, F., Manzano-Blasco, I., Abadía-Otero, J., Albert, E., Mulet, A., Briongos-Figuero, L. S., Gabella-Martín, M., Torres, I., Signes-Costa, J., Navarro, D., Martín-Escudero, J.-C., García-García, A.-B., & Chaves, F. J. (2023). Sample Treatment with Trypsin for RT-LAMP COVID-19 Diagnosis. Biology, 12(7), 900. https://doi.org/10.3390/biology12070900






