Comprehensive Analysis for Anti-Cancer Target-Indication Prioritization of Placental Growth Factor Inhibitor (PGF) by Use of Omics and Patient Survival Data
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
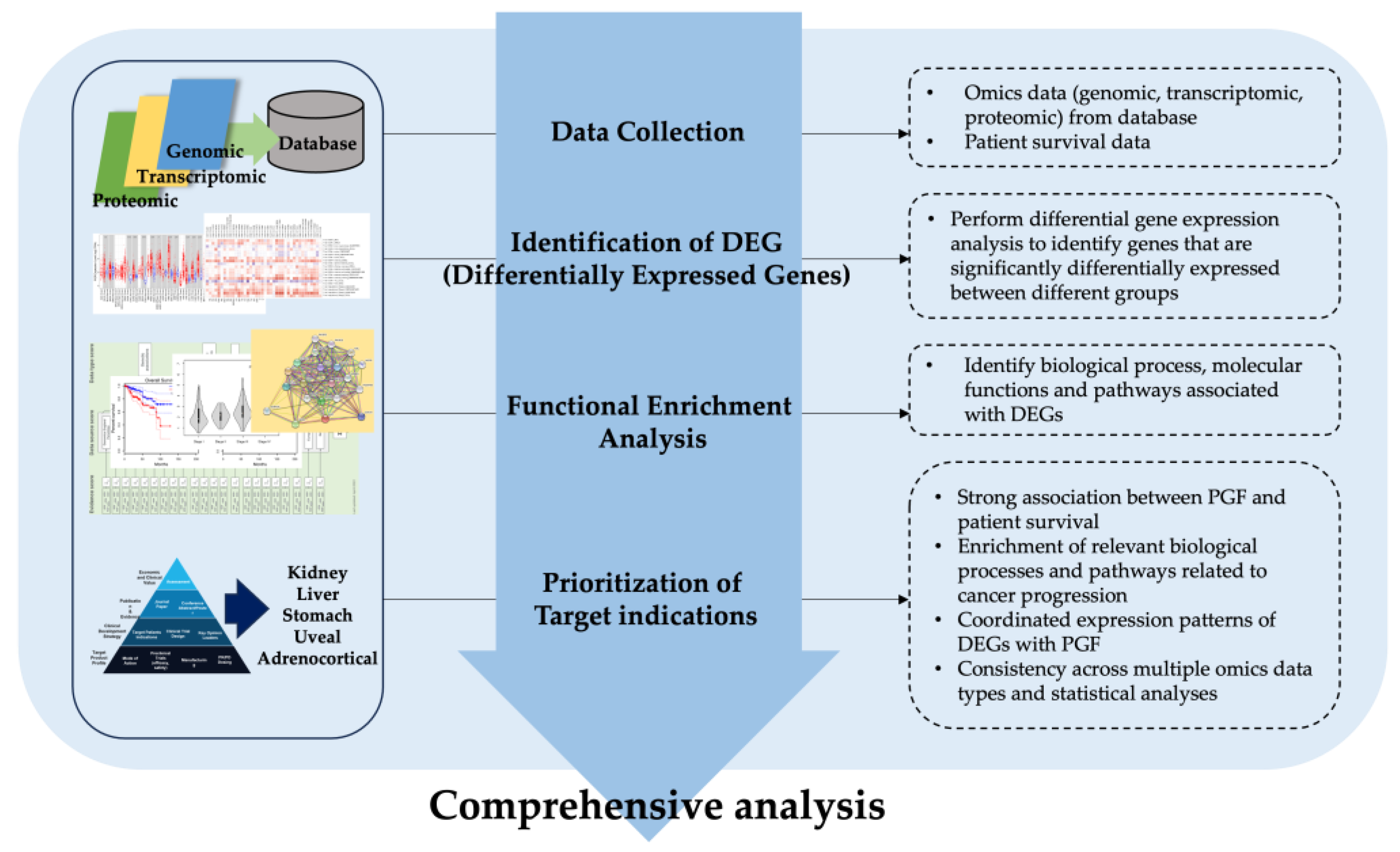
2.1. Comprehensive Analysis Framework
2.2. Data Collection
2.3. Analysis of Differential Gene Expression and Mutation across Cells, Tissues, and Human Cancer Types
2.4. Analysis of PGF Gene Function and Molecular Pathway
2.5. Analysis of Target Disease Association
2.6. Univariate Survival Analysis for Prognostic Effect of PGF Expression
2.7. Multivariate Survival Analysis for Prognostic Effect of PGF Expression and Tumor Infiltrating Lymphocytes
2.8. CAF and MDSCs Level Correlation with PGF Expression
3. Results
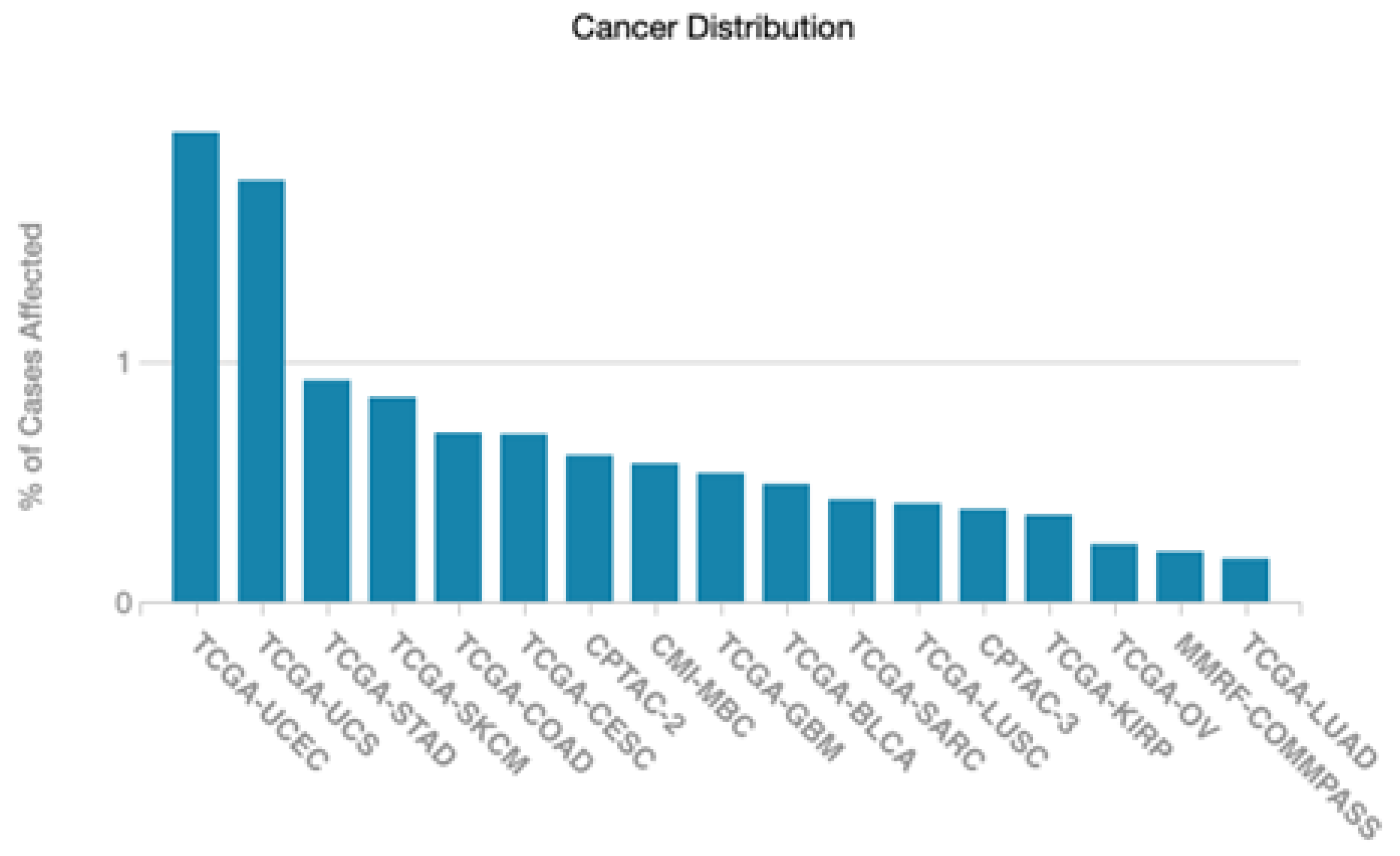
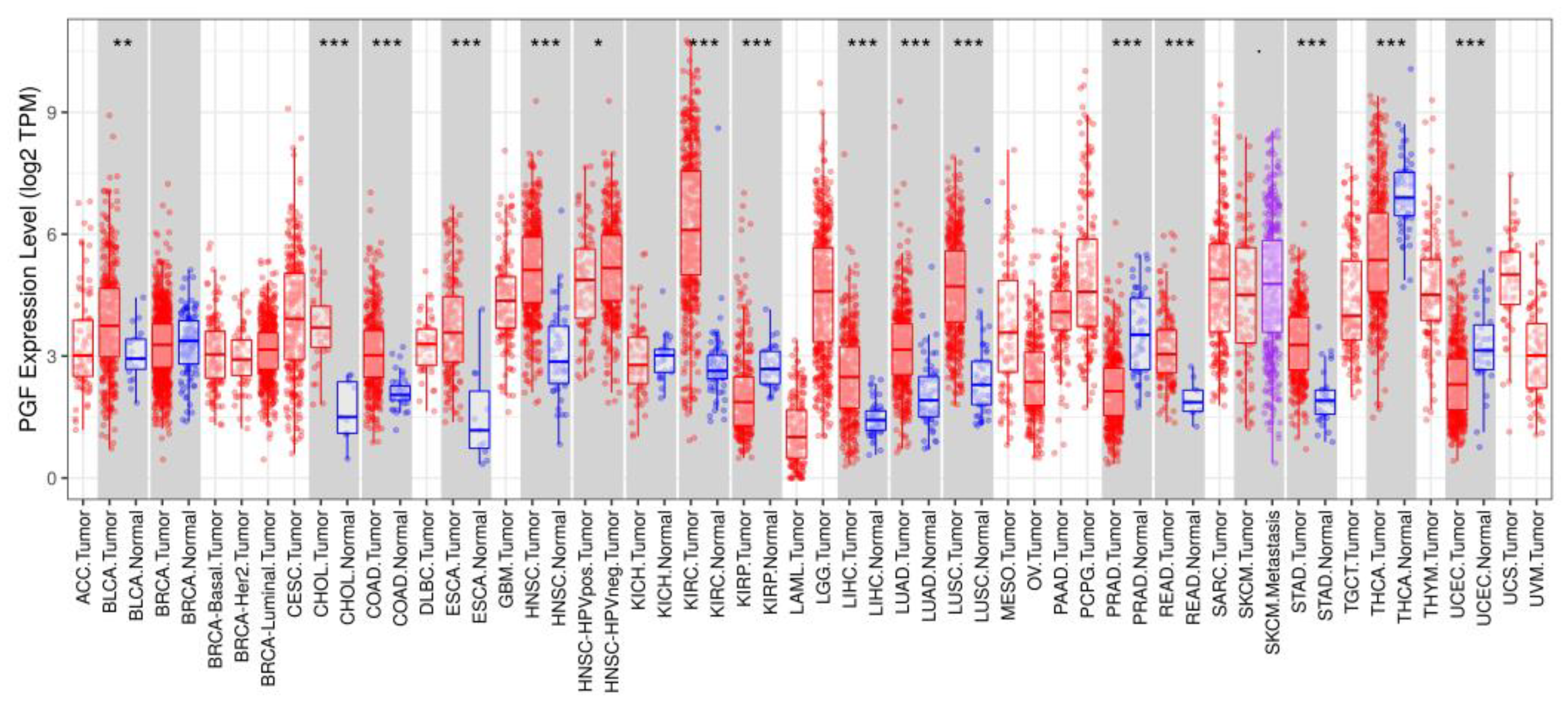
3.1. Differential Gene Expression and Mutation across Cells and Tissues
3.2. PGF Gene Function and Molecular Pathway
3.3. Target Disease Association
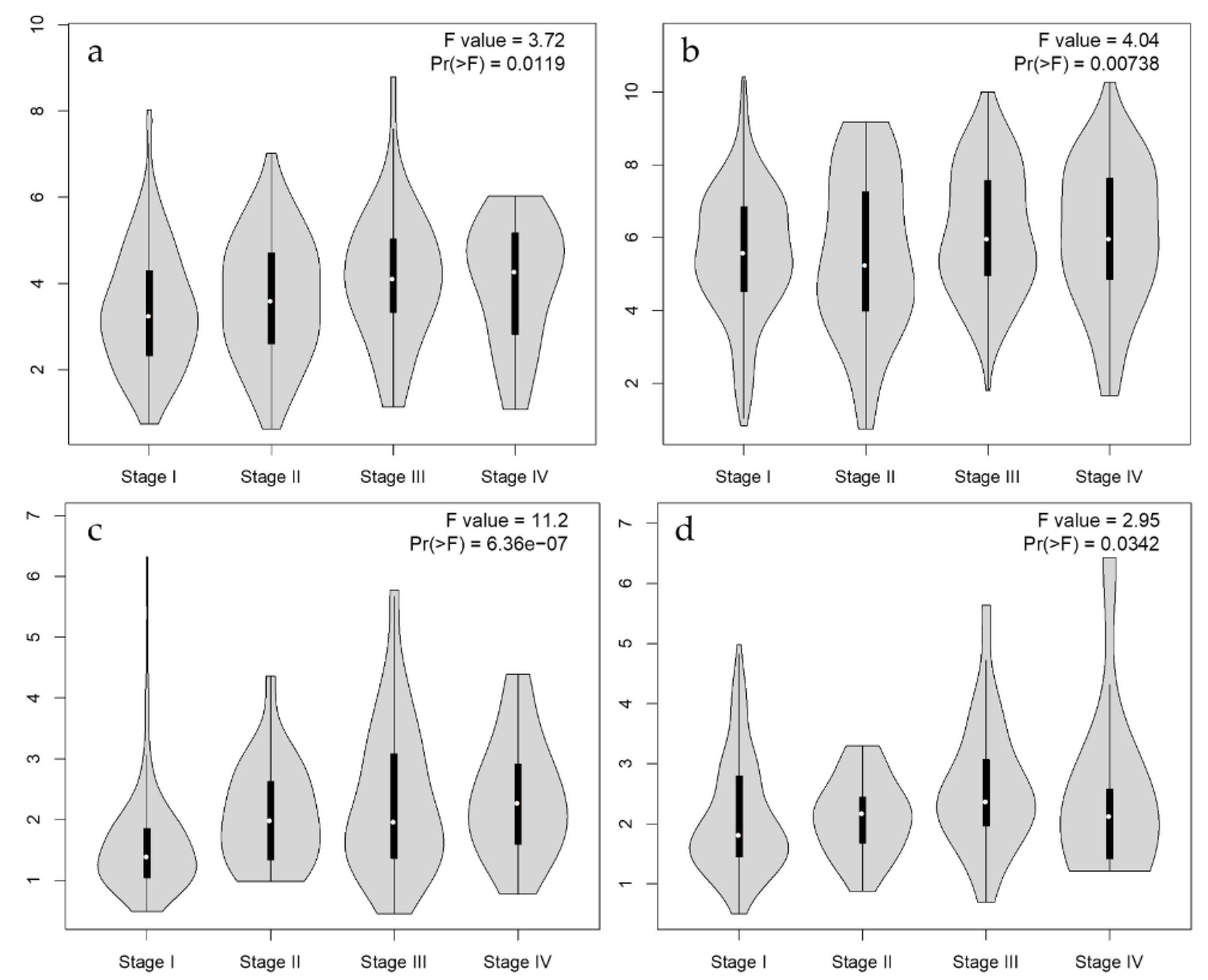
3.4. Univariate Survival Analysis for Prognostic Effect of PGF Expression
3.5. Multivariate Cox Proportional Hazards Model for Predicting Survival Outcome
3.6. CAF and MDSCs Level Correlation with PGF Expression
3.7. Overall Summary Evaluation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luttun, A.; Autiero, M.; Tjwa, M.; Carmeliet, P. Genetic dissection of tumor angiogenesis: Are PlGF and VEGFR-1 novel anti-cancer targets? Biochim. Biophys. Acta 2004, 1654, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimna, A.; Kurpisz, M. Hypoxia-Inducible Factor-1 in Physiological and Pathophysiological Angiogenesis: Applications and Therapies. BioMed Res. Int. 2015, 2015, 549412. [Google Scholar] [CrossRef] [Green Version]
- Korbecki, J.; Siminska, D.; Gassowska-Dobrowolska, M.; Listos, J.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. Chronic and Cycling Hypoxia: Drivers of Cancer Chronic Inflammation through HIF-1 and NF-kappaB Activation: A Review of the Molecular Mechanisms. Int. J. Mol. Sci. 2021, 22, 701. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, H.X.; Wu, C.T.; Wang, W.Q.; Jin, W.; Gao, H.L.; Li, H.; Zhang, S.R.; Xu, J.Z.; Qi, Z.H.; et al. Angiogenesis in pancreatic cancer: Current research status and clinical implications. Angiogenesis 2019, 22, 15–36. [Google Scholar] [CrossRef]
- Dudley, A.C.; Griffioen, A.W. Pathological angiogenesis: Mechanisms and therapeutic strategies. Angiogenesis 2023, 15, 313–347. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Dudley, A.C. Models and molecular mechanisms of blood vessel co-option by cancer cells. Angiogenesis 2020, 23, 17–25. [Google Scholar] [CrossRef]
- Di Luca, M.; Iannella, G.; Montevecchi, F.; Magliulo, G.; De Vito, A.; Cocuzza, S.; Maniaci, A.; Meccariello, G.; Cammaroto, G.; Sgarzani, R.; et al. Use of the transoral robotic surgery to treat patients with recurrent lingual tonsillitis. Int. J. Med. Robot. 2020, 16, e2106. [Google Scholar] [CrossRef] [Green Version]
- Jaradeh, M.; Curran, B.; Poulikidis, K.; Rodrigues, A.; Jeske, W.; Abdelsattar, Z.M.; Lubawski, J.; Walenga, J.; Vigneswaran, W.T. Inflammatory cytokines in robot-assisted thoracic surgery versus video-assisted thoracic surgery. J. Thorac. Dis. 2022, 14, 2000–2010. [Google Scholar] [CrossRef]
- Albonici, L.; Giganti, M.G.; Modesti, A.; Manzari, V.; Bei, R. Multifaceted Role of the Placental Growth Factor (PlGF) in the Antitumor Immune Response and Cancer Progression. Int. J. Mol. Sci. 2019, 20, 2970. [Google Scholar] [CrossRef] [Green Version]
- De Falco, S. The discovery of placenta growth factor and its biological activity. Exp. Mol. Med. 2012, 44, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Saraniti, C.; Speciale, R.; Santangelo, M.; Massaro, N.; Maniaci, A.; Gallina, S.; Serra, A.; Cocuzza, S. Functional outcomes after supracricoid modified partial laryngectomy. J. Biol. Regul. Homeost. Agents 2019, 33, 1903–1907. [Google Scholar] [CrossRef]
- de Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef] [Green Version]
- Lau, D.K.; Mencel, J.; Chau, I. Safety and efficacy review of aflibercept for the treatment of metastatic colorectal cancer. Expert Opin. Drug Saf. 2022, 21, 589–597. [Google Scholar] [CrossRef]
- Croston, G.E. The utility of target-based discovery. Expert Opin. Drug Discov. 2017, 12, 427–429. [Google Scholar] [CrossRef] [Green Version]
- Salehi, Z.; Akrami, H.; Seydi, H. Bioinformatics analysis of regulated MicroRNAs by placental growth factor signaling in cancer stem cells. J. Cancer Res. Ther. 2020, 16, S90–S94. [Google Scholar] [CrossRef]
- Rudolf, T.; Pillich, J.C.; Vladimir, R.; David, W.; Dexter, P. NDEx: A Community Resource for Sharing and Publishing of Biological Networks. In Protein Bioinformatics; Humana Press: New York, NY, USA, 2017; Volume 1558. [Google Scholar]
- Carvalho-Silva, D.; Pierleoni, A.; Pignatelli, M.; Ong, C.; Fumis, L.; Karamanis, N.; Carmona, M.; Faulconbridge, A.; Hercules, A.; McAuley, E.; et al. Open Targets Platform: New developments and updates two years on. Nucleic Acids Res. 2019, 47, D1056–D1065. [Google Scholar] [CrossRef] [Green Version]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef] [Green Version]
- GTEx Consortium; Laboratory Data Analysis; Coordinating Center-Analysis Working Group; Statistical Methods Groups-Analysis Working Group; Enhancing, GTEx Groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; et al. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [Green Version]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef]
- Gene Ontology, C. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R., III; Kalocsay, M.; Jane-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Isildak, U.; Somel, M.; Thornton, J.M.; Donertas, H.M. Temporal changes in the gene expression heterogeneity during brain development and aging. Sci. Rep. 2020, 10, 4080. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Liu, F. Cancer-associated fibroblast-derived gene signatures predict radiotherapeutic survival in prostate cancer patients. J. Transl. Med. 2022, 20, 453. [Google Scholar] [CrossRef]
- Dewerchin, M.; Carmeliet, P. Placental growth factor in cancer. Expert Opin. Ther. Targets 2014, 18, 1339–1354. [Google Scholar] [CrossRef]
- Selvaraj, S.K.; Giri, R.K.; Perelman, N.; Johnson, C.; Malik, P.; Kalra, V.K. Mechanism of monocyte activation and expression of proinflammatory cytochemokines by placenta growth factor. Blood 2003, 102, 1515–1524. [Google Scholar] [CrossRef] [Green Version]
- Aoki, S.; Inoue, K.; Klein, S.; Halvorsen, S.; Chen, J.; Matsui, A.; Nikmaneshi, M.R.; Kitahara, S.; Hato, T.; Chen, X.; et al. Placental growth factor promotes tumour desmoplasia and treatment resistance in intrahepatic cholangiocarcinoma. Gut 2022, 71, 185–193. [Google Scholar] [CrossRef]
- Iwasaki, H.; Kawamoto, A.; Tjwa, M.; Horii, M.; Hayashi, S.; Oyamada, A.; Matsumoto, T.; Suehiro, S.; Carmeliet, P.; Asahara, T. PlGF repairs myocardial ischemia through mechanisms of angiogenesis, cardioprotection and recruitment of myo-angiogenic competent marrow progenitors. PLoS ONE 2011, 6, e24872. [Google Scholar] [CrossRef]
- Shin, J.Y.; Yoon, I.H.; Kim, J.S.; Kim, B.; Park, C.G. Vascular endothelial growth factor-induced chemotaxis and IL-10 from T cells. Cell Immunol. 2009, 256, 72–78. [Google Scholar] [CrossRef]
- Oude Munnink, T.H.; Tamas, K.R.; Lub-de Hooge, M.N.; Vedelaar, S.R.; Timmer-Bosscha, H.; Walenkamp, A.M.; Weidner, K.M.; Herting, F.; Tessier, J.; de Vries, E.G. Placental growth factor (PlGF)-specific uptake in tumor microenvironment of 89Zr-labeled PlGF antibody RO5323441. J. Nucl. Med. 2013, 54, 929–935. [Google Scholar] [CrossRef] [Green Version]
- Incio, J.; Tam, J.; Rahbari, N.N.; Suboj, P.; McManus, D.T.; Chin, S.M.; Vardam, T.D.; Batista, A.; Babykutty, S.; Jung, K.; et al. PlGF/VEGFR-1 Signaling Promotes Macrophage Polarization and Accelerated Tumor Progression in Obesity. Clin. Cancer Res. 2016, 22, 2993–3004. [Google Scholar] [CrossRef] [Green Version]
- Krneta, T.; Gillgrass, A.; Poznanski, S.; Chew, M.; Lee, A.J.; Kolb, M.; Ashkar, A.A. M2-polarized and tumor-associated macrophages alter NK cell phenotype and function in a contact-dependent manner. J. Leukoc. Biol. 2017, 101, 285–295. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Liang, Y.-C.; Chiang, B.-L. Placental growth factor down-regulates type 1 T helper immune response by modulating the function of dendritic cells. J. Leukoc. Biol. 2007, 82, 1473–1480. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]

| Database | Resources |
|---|---|
| NDEx Cytoscape Interactome Analysis Database [17] | https://www.ndexbio.org/iquery/ (accessed on 30 March 2022) |
| Open Target Platform [18] | https://platform.opentargets.org (accessed on 30 March 2022) |
| Human Protein Atlas [19] | https://www.proteinatlas.org (accessed on 30 March 2022) |
| GTEx (Genotype Tissue Expression) [20] | https://gtexportal.org/home/ (accessed on 30 March 2022) |
| Drug Gene Interaction Database [21] | https://www.dgidb.org (accessed on 30 March 2022) |
| Gene Ontology Resource Database [22] | http://geneontology.org (accessed on 30 March 2022) |
| TCGA (The Cancer Genome Atlas) Database [23] | https://www.cancer.gov/tcga (accessed on 30 March 2022) |
| STRING Database (Functional Protein Association Network) [24] | https://string-db.org (accessed on 30 March 2022) |
| CCLE (Cancer Cell Line Encyclopedia) Database [25] | https://sites.broadinstitute.org/ccle/ (accessed on 30 March 2022) |
| TIMER (Tumor Immune Estimation Resource) TIMER2.0 [26,27] | https://cistrome.shinyapps.io/timer (accessed on 30 March 2022) http://timer.comp-genomics.org (accessed on 30 March 2022) |
| GEPIA (Gene Expression Profiling Interactive Analysis) [28] | http://gepia.cancer-pku.cn (accessed on 30 March 2022) |
| Abbreviation | Cancer Type |
|---|---|
| LAML | Acute Myeloid Leukemia |
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder Urothelial Carcinoma |
| LGG | Brain Lower Grade Glioma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| LCML | Chronic Myelogenous Leukemia |
| COAD | Colon adenocarcinoma |
| ESCA | Esophageal carcinoma |
| GBM | Glioblastoma multiforme |
| HNSC | Head and Neck squamous cell carcinoma |
| KICH | Kidney Chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma |
| MESO | Mesothelioma |
| MISC | Miscellaneous |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and Paraganglioma |
| READ | Rectum adenocarcinoma |
| PRAD | Prostate adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin Cutaneous Melanoma |
| STAD | Stomach adenocarcinoma |
| TGCT | Testicular Germ Cell Tumors |
| THYM | Thymoma |
| THCA | Thyroid carcinoma |
| UCS | Uterine Carcinosarcoma |
| UCEC | Uterine Corpus Endometrial Carcinoma |
| UVM | Uveal Melanoma |
| Cancer Type | Outcome | HR (High vs. Low) | p(HR) | Logrank Test | No of Patients | |
|---|---|---|---|---|---|---|
| High | Low | |||||
| ACC | OS | 3.6 | 0.0025 | 0.0013 | 38 | 38 |
| DFS | 5.9 | <0.0001 | <0.0001 | 38 | 38 | |
| BLCA | OS | 1.4 | 0.037 | 0.037 | 201 | 201 |
| DFS | 1.1 | 0.58 | 0.58 | 201 | 201 | |
| BRCA | OS | 1.3 | 0.14 | 0.14 | 535 | 535 |
| DFS | 1.6 | 0.011 | 0.01 | 535 | 535 | |
| CESC | OS | 1.5 | 0.1 | 0.099 | 146 | 146 |
| DFS | 0.79 | 0.42 | 0.42 | 146 | 146 | |
| CHOL | OS | 0.5 | 0.16 | 0.15 | 18 | 18 |
| DFS | 0.7 | 0.44 | 0.43 | 18 | 18 | |
| COAD | OS | 1.3 | 0.31 | 0.31 | 135 | 135 |
| DFS | 1.0 | 0.9 | 0.91 | 135 | 135 | |
| DLBC | OS | 2.1 | 0.38 | 0.37 | 23 | 23 |
| DFS | 0.55 | 0.36 | 0.35 | 23 | 23 | |
| ESCA | OS | 1 | 1 | 0.99 | 91 | 91 |
| DFS | 1.3 | 0.24 | 0.25 | 91 | 91 | |
| GBM | OS | 0.75 | 0.12 | 0.11 | 80 | 81 |
| DFS | 0.85 | 0.44 | 0.42 | 80 | 81 | |
| HNSC | OS | 1.2 | 0.27 | 0.27 | 259 | 259 |
| DFS | 1 | 0.95 | 0.96 | 259 | 259 | |
| KICH | OS | 3.9 | 0.092 | 0.069 | 32 | 32 |
| DFS | 10 | 0.026 | 0.0058 | 32 | 32 | |
| KIRC | OS | 1.3 | 0.088 | 0.086 | 258 | 258 |
| DFS | 1.8 | 0.0028 | 0.0025 | 258 | 258 | |
| KIRP | OS | 2.4 | 0.0068 | 0.0052 | 141 | 141 |
| DFS | 2.2 | 0.0083 | 0.0068 | 141 | 141 | |
| LAML | OS | 1.2 | 0.51 | 0.52 | 53 | 53 |
| DFS | 1 | na | 1 | 53 | 53 | |
| LGG | OS | 0.5 | 0.00021 | 0.00015 | 257 | 257 |
| DFS | 0.54 | 0.00011 | <0.0001 | 257 | 257 | |
| LIHC | OS | 1.8 | 0.0012 | 0.00096 | 181 | 181 |
| DFS | 1 | 0.82 | 0.82 | 181 | 181 | |
| LAUD | OS | 1.1 | 0.41 | 0.41 | 239 | 239 |
| DFS | 1.1 | 0.55 | 0.55 | 239 | 239 | |
| LUSC | OS | 0.84 | 0.2 | 0.2 | 241 | 241 |
| DFS | 1.2 | 0.38 | 0.37 | 241 | 241 | |
| MESO | OS | 1.1 | 0.62 | 0.64 | 41 | 41 |
| DFS | 1.1 | 0.75 | 0.74 | 41 | 41 | |
| OV | OS | 1.2 | 0.24 | 0.23 | 212 | 212 |
| DFS | 0.99 | 0.96 | 0.95 | 212 | 212 | |
| PADD | OS | 0.89 | 0.59 | 0.6 | 89 | 89 |
| DFS | 1.2 | 0.42 | 0.43 | 89 | 89 | |
| PCPG | OS | 0.49 | 0.41 | 0.4 | 91 | 91 |
| DFS | 0.71 | 0.48 | 0.48 | 91 | 91 | |
| PRAD | OS | 0.34 | 0.12 | 0.1 | 246 | 246 |
| DFS | 0.87 | 0.5 | 0.5 | 246 | 246 | |
| READ | OS | 1.7 | 0.27 | 0.26 | 46 | 46 |
| DFS | 1.7 | 0.28 | 0.28 | 46 | 46 | |
| SARC | OS | 1.1 | 0.5 | 0.49 | 131 | 131 |
| DFS | 0.98 | 0.92 | 0.91 | 131 | 131 | |
| SKCM | OS | 0.96 | 0.74 | 0.74 | 229 | 229 |
| DFS | 1 | 0.94 | 0.94 | 229 | 229 | |
| STAD | OS | 1.5 | 0.011 | 0.0098 | 191 | 192 |
| DFS | 1.3 | 0.16 | 0.16 | 191 | 192 | |
| TGCT | OS | 2 | 0.58 | 0.58 | 68 | 68 |
| DFS | 1.2 | 0.61 | 0.61 | 68 | 68 | |
| THCA | OS | 1.3 | 0.6 | 0.6 | 255 | 255 |
| DFS | 0.56 | 0.054 | 0.05 | 255 | 255 | |
| THYM | OS | 0.74 | 0.68 | 0.67 | 59 | 59 |
| DFS | 0.33 | 0.034 | 0.026 | 59 | 59 | |
| UCEC | OS | 1.8 | 0.12 | 0.12 | 86 | 86 |
| DFS | 0.87 | 0.68 | 0.68 | 86 | 86 | |
| UCS | OS | 0.67 | 0.25 | 0.25 | 28 | 28 |
| DFS | 0.65 | 0.25 | 0.24 | 28 | 28 | |
| UVM | OS | 3.1 | 0.019 | 0.013 | 39 | 39 |
| DFS | 2.4 | 0.064 | 0.056 | 39 | 39 | |
| Covariate | HR (95% CIs) | p-Value | Covariate | HR (95% CIs) | p-Value |
|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | Cholangiocarcinoma (CHOL) | ||||
| B-cell | 0.00 (0.00–6.63) | 0.177 | B-cell | 2.79 (0.00–1.01) | 0.696 |
| CD8+ T-cell | 0.00 (0.00–1.63) | 0.044 * | CD8+ T-cell | 5.13 (132.61–1.99) | 0.046 * |
| CD4+ T-cell | 0.00 (0.00–1.14) | 0.319 | CD4+ T-cell | 0.00 (0.00–3.01) | 0.374 |
| Macrophage | 4.00 (0.00–5.76) | 0.853 | Macrophage | 0.00 (0.00–7.11) | 0.240 |
| Neutrophil | 3.23 (97,832.71–1.06) | 0.029 * | Neutrophil | 0.00 (0.00–1.67) | 0.538 |
| Dendritic cell | 2.28 (0.00–5.23) | 0.552 | Dendritic cell | 0.00 (0.00–3.51) | 0.722 |
| PGF expression | 1.78 (1.25–2.55) | 0.001 ** | PGF expression | 2.73 (0.10–7.06) | 0.007 ** |
| Glioblastoma Multiforme (GBM) | Low-Grade Glioma (LGG) | ||||
| B-cell | 0.56 (0.32–0.97) | 0.039 | B-cell | 0.45 (0.00–124.67) | 0.784 |
| CD8+ T-cell | 1.38 (0.93–2.02) | 0.101 | CD8+ T-cell | 214.01 (0.24–188,566.24) | 0.121 |
| CD4+ T-cell | 1.08 (0.57–2.04) | 0.794 | CD4+ T-cell | 0.113 (0.00–240.27) | 0.576 |
| Macrophage | 1.16 (0.63–2.16) | 0.621 | Macrophage | 1034.91 (26.29–40734.86) | 0.000 *** |
| Neutrophil | 1.64 (0.74–3.63) | 0.221 | Neutrophil | 0.005 (0.00–7.62) | 0.155 |
| Dendritic cell | 1.51 (1.18–1.93) | 0.001 ** | Dendritic cell | 6.46 (0.15–274.43) | 0.329 |
| PGF expression | 0.87 (0.77–0.97) | 0.015 * | PGF expression | 0.77 (0.67–0.88) | 0.000 *** |
| Liver Hepatocellular Carcinoma (LIHC) | Ovarian serous cystadenocarcinoma (OV) | ||||
| B-cell | 0.02 (0.00–32.78) | 0.311 | B-cell | 0.02 (0.00–13.86) | 0.255 |
| CD8+ T-cell | 0.00 (0.00–0.05) | 0.002 ** | CD8+ T-cell | 0.04 (0.00–2.00) | 0.110 |
| CD4+ T-cell | 0.00 (0.00–2.92) | 0.098 | CD4+ T-cell | 0.00 (0.00–0.00) | 0.000 *** |
| Macrophage | 101.52 (0.35–29,455.42) | 0.110 | Macrophage | 34,041.86 (145.80–7,947,944.83) | 0.000 *** |
| Neutrophil | 3.89 (0.00–235,124.74) | 0.809 | Neutrophil | 2691.62 (0.45–15,895,296.64) | 0.075 |
| Dendritic cell | 57.48 (1.72–1914.82) | 0.024 * | Dendritic cell | 0.45 (0.00–45.54) | 0.735 |
| PGF expression | 0.27 (1.03–1.55) | 0.021 * | PGF expression | 0.73 (0.55–0.97) | 0.034 * |
| Stomach adenocarcinoma (STAD) | Uterine Corpus Endometrial Carcinoma (UCEC) | ||||
| B-cell | 87.85 (1.09–7025.32) | 0.045 * | B-cell | 0.32 (0.00–155.05) | 0.719 |
| CD8+ T-cell | 0.26 (0.01–4.19) | 0.345 | CD8+ T-cell | 0.00 (0.00–0.03) | 0.001 ** |
| CD4+ T-cell | 0.02 (0.00–2.92) | 0.126 | CD4+ T-cell | 0.00 (0.00–0.02) | 0.002 ** |
| Macrophage | 138.70 (5.93–3244.18) | 0.002 ** | Macrophage | 36.13 (0.25–5044.40) | 0.155 |
| Neutrophil | 0.70 (0.00–153.01) | 0.900 | Neutrophil | 5393.34 (3.75–7,742,799.81) | 0.021 * |
| Dendritic cell | 2.28 (0.18–27.87) | 0.51 | Dendritic cell | 4.58 (0.16–126.21) | 0.368 |
| PGF expression | 1.24 (1.00–1.53) | 0.042 * | PGF expression | 1.38 (1.15–1.65) | 0.001 ** |
| PGF Gene Expression | PGF Gene Mutation | DEG | Survival | TME | |||||
|---|---|---|---|---|---|---|---|---|---|
| CNVs | Methyl | RNAseq | Uni | Multi | CAF | MDSC | |||
| ACC | *** | na | *** | *** | ** | ||||
| BLCA | * | ** | ** | ||||||
| BRCA | * | ** | * | ||||||
| CESC | ** | * | * | ||||||
| CHOL | ** | ** | *** | *** | |||||
| COAD | * | *** | ** | ||||||
| DLBC | |||||||||
| ESCA | *** | ** | ** | ||||||
| GBM | ** | * | ** | ||||||
| HNSC | ** | * | ** | ||||||
| KICH | * | ** | * | ||||||
| KIRC | *** | ** | * | ||||||
| KIRP | ** | *** | ** | * | |||||
| LAML | ** | ||||||||
| LGG | *** | * | *** | ||||||
| LIHC | *** | * | ** | *** | ** | ||||
| LUAD | *** | *** | * | ||||||
| LUSC | *** | ** | |||||||
| MESO | |||||||||
| OV | * | ||||||||
| PAAD | ** | ** | |||||||
| PCPG | |||||||||
| PRAD | ** | *** | ** | ||||||
| READ | ** | *** | ** | ||||||
| SARC | ** | * | |||||||
| SKCM | ** | ||||||||
| STAD | ** | *** | * | * | ** | ||||
| TGCT | *** | ||||||||
| THCA | *** | ** | |||||||
| THYM | * | ||||||||
| UCEC | *** | *** | * | *** | |||||
| UCS | *** | * | |||||||
| UVM | *** | *** | na | *** | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Ko, Y.; Shin, Y.; Park, J.; Lee, A.J.; Kim, K.W.; Pyo, J. Comprehensive Analysis for Anti-Cancer Target-Indication Prioritization of Placental Growth Factor Inhibitor (PGF) by Use of Omics and Patient Survival Data. Biology 2023, 12, 970. https://doi.org/10.3390/biology12070970
Kim N, Ko Y, Shin Y, Park J, Lee AJ, Kim KW, Pyo J. Comprehensive Analysis for Anti-Cancer Target-Indication Prioritization of Placental Growth Factor Inhibitor (PGF) by Use of Omics and Patient Survival Data. Biology. 2023; 12(7):970. https://doi.org/10.3390/biology12070970
Chicago/Turabian StyleKim, Nari, Yousun Ko, Youngbin Shin, Jisuk Park, Amy Junghyun Lee, Kyung Won Kim, and Junhee Pyo. 2023. "Comprehensive Analysis for Anti-Cancer Target-Indication Prioritization of Placental Growth Factor Inhibitor (PGF) by Use of Omics and Patient Survival Data" Biology 12, no. 7: 970. https://doi.org/10.3390/biology12070970
APA StyleKim, N., Ko, Y., Shin, Y., Park, J., Lee, A. J., Kim, K. W., & Pyo, J. (2023). Comprehensive Analysis for Anti-Cancer Target-Indication Prioritization of Placental Growth Factor Inhibitor (PGF) by Use of Omics and Patient Survival Data. Biology, 12(7), 970. https://doi.org/10.3390/biology12070970






